Abstract
The INK4 family of cyclin-dependent kinase (CDK) inhibitors negatively regulates cyclin D-dependent CDK4 and CDK6 and thereby retains the growth-suppressive function of Rb family proteins. Mutations in the CDK4 gene conferring INK4 resistance are associated with familial and sporadic melanoma in humans and result in a wide spectrum of tumors in mice. Whereas loss of function of other INK4 genes in mice leads to little or no tumor development, targeted deletion of p18INK4c causes spontaneous pituitary tumors and lymphoma late in life. Here we show that treatment of p18 null and heterozygous mice with a chemical carcinogen resulted in tumor development at an accelerated rate. The remaining wild-type allele of p18 was neither mutated nor silenced in tumors derived from heterozygotes. Hence, p18 is a haploinsufficient tumor suppressor in mice.
The Rb family proteins, pRB, p107, and p130, play critical roles in controlling mammalian G1 cell cycle progression (33). Emerging from mitosis or present in quiescent cells as hypophosphorylated forms, these proteins negatively regulate the activity of E2F transcription factors to prevent S-phase entry. Extracellular mitogens induce the expression of D-type cyclins and activate cyclin D-dependent kinases CDK4 and CDK6, leading to phosphorylation and functional inactivation of Rb proteins. Conversely, inhibition of CDK4 and CDK6, resulting from either lack of cyclin D synthesis or binding with an INK4 protein, retains Rb proteins in their growth-suppressive states and prevents the G1/S transition. Disruption of this pathway, consisting of INK4-cyclin D/CDK4/6-Rb-E2F, deregulates G1 cell cycle progression and is believed to be a common event for the development of most types of cancer (29).
Mutational analysis of human tumors has provided compelling evidence supporting the critical importance of this pathway in tumor suppression. The Rb gene is frequently mutated in familial retinoblastomas and osteosarcomas, as well as in sporadic lung, prostate, bladder, and breast cancers (reviewed in references 20 and 33). The p16INK4a locus is frequently deleted or mutated in wide range of human tumors (13, 22; compiled in reference 27). Chromosomal translocation and genomic amplification of cyclin D1 and CDK4 genes leading to elevated level of either protein and its associated kinase activities have been observed in cancers of the breast, head and neck, esophagus, lymphoid system, parathyroid, and brain (reviewed in references 9, 10, and 29). Finally, missense mutations in the CDK4 locus rendering resistance to INK4 binding have been observed in familial and sporadic melanomas (35, 41).
Genetic study with targeted mice provides further support for a critical function of this pathway in tumor suppression. Mice heterozygous for Rb mutation spontaneously develop tumors in several neuroendocrine organs, including the characteristic intermediate lobe of the pituitary (12, 19, 21, 26, 34). Engineering of transgenic mice to overexpress cyclin D1 in mammary glands causes hyperplasia and carcinoma (32), and conversely ablation of cyclin D1 renders mice resistant to breast cancer development (36). Mice carrying an INK4-insensitive mutation (R24C) in the CDK4 locus develop a wide spectrum of tumors, including tumors in several neuroendocrine organs (Leydig cells of testes, granulosa cells of ovaries, pancreatic islet cells, the intermediate lobe of the pituitary, and thyroid) as well as lung adenomas or adenocarcinomas and hepatocellular tumors (30).
However, genetic analysis of mutant mice has yet to clarify the role of individual INK4 genes in tumor suppression. Mice lacking p16INK4a perplexingly display a tumor phenotype that is considerably weaker and different from that predicted from human tumor analysis. Only about one-quarter of mice carrying a deletion of exon 1α of p16INK4a developed spontaneous tumors—mostly sarcomas and lymphomas late in life (16, 28). Such phenotypic discrepancy between CDK4R24C and INK4aΔexon1α mice suggests that an additional INK4 gene or genes, although not frequently mutated in human tumors, must be involved in regulating CDK4 (and possibly CDK6) activity and the Rb pathway in tumor suppression in mice. No obvious tumor phenotypes were detected in mice lacking either p15INK4b (18) or p19INK4d (40). Several observations suggest that p18INK4c may play a major role in mediating the tumor suppression function of the Rb pathway. Mice lacking p18INK4c spontaneously develop intermediate lobe pituitary tumors late in life (5, 18), those that lack both p18INK4c and p27KIP1 develop tumors in several neuroendocrine organs by 3 months of age, and p18INK4c and p21CIP1/WAF1 double knockout mice develop gastric neuroendocrine hyperplasia and lung bronchioalveolar tumors late in life (6). To further evaluate the function of p18INK4c in tumor suppression, we examined the rate and spectrum of tumor development in p18INK4c mutant mice exposed to a chemical carcinogen. Our studies indicate that loss or decrease of p18INK4c function predisposes mice to tumorigenesis and that p18INK4c is haploinsufficient in tumor suppression.
MATERIALS AND METHODS
Mice.
p18INK4c mutant mice were initially generated in a mixed C57BL/6/129SV/D2 genetic background (5). p18INK4c mutant mice with an enriched C57BL/6 genetic background were generated after six successive generations of backcrosses with C57BL/6 wild-type mice (15) and were used in this study. Littermates of different genotypes were produced from interbreeding of heterozygotes. All mice were genotyped by PCR as previously described (5). All animals were maintained in accordance with institutional guidelines and were observed daily for up to 15 months.
Carcinogenesis.
A cohort of 50 p18+/+, 45 p18+/−, and 45 p18−/− animals were produced from interbreeding of p18 heterozygotes and used for this study. Twenty-seven wild-type, 30 heterozygous, and 20 p18 homozygous mice were treated continuously with 0.0005% dimethylnitrosamine (DMN) in drinking water beginning at the age of 8 weeks as described previously. Mice were monitored daily over the course of the experiment, and all genotypes consumed roughly the same amount of water on a daily basis. Twenty-three p18+/+, 25 p18−/−, and 15 p18+/− mice were monitored in parallel with the treated mice. Cohorts of treated mice were killed and necropsied at 3 and 6 months of age (at least three mice for each genotype and each time point). The remaining mice were necropsied at death or were sacrificed after reaching a moribund state. Tissues of most organs, as well as apparent tumor tissues, were removed, fixed in 10% neutral buffered formalin, and examined histologically by two pathologists after hematoxylin-eosin staining. Lesions were photographed, and additional sections were taken for immunohistochemical analyses.
Immunohistochemistry.
Specimens clearly representative of their respective pathological types were examined immunohistologically with an affinity-purified antibody specific to p18 (2 μg/ml) (7, 8) or to adrenocorticotropic hormone (ACTH; 12 μg/ml) (no. A0579; DAKO). Tissue sections were blocked with normal goat serum in phosphate-buffered saline (PBS), and incubated with primary antibodies for 1 h and biotin-conjugated secondary antibody for 30 min. Immunocomplexes were detected with the Vectastain ABC alkaline phosphatase kit according to the manufacturer's instructions (Vector Laboratories).
Tumor microdissection and genomic DNA amplification.
Tumor microdissection and DNA isolation procedures were described before (1). Ten-micrometer sections adhered on slides were deparaffinized and lightly stained with hematoxylin. Under microscopic observation, the lesions that were clearly separated from normal tissues were scraped from the slides. Particular care was taken to avoid contamination by surrounding tissue. Scraped tissue samples were transferred into a sterile microcentrifuge tube. Forty microliters of digestion buffer (200 μM proteinase K per ml, 0.5% Tween 20, 1 mM EDTA, 20 mM Tris-HCl [pH 7.4]) was added to each tube, and the mixture was then incubated at 42°C for 2 days and then incubated in boiling water for 6 to 7 min. Thirty-five microliters of the supernatant was stored at −20°C until used for PCR assays (1). The murine INK4c gene contains three exons: a noncoding exon, 1, corresponding to the 5′-untranslated region and two coding exons comprising the entire coding sequences (24). Coding exons 2 and 3 were amplified and sequenced with two pairs of primers: p18-E2FW (5′-AGCCTGATTAGGAGCAAAGG-3′) and p18-E2RV (5′-GCTGTCATTTTAGAAACCCAGGC-3′) for exon 2 and p18-E3FW (5′TTGTTGTGGCTCAAGAAGCTG-3′) and p18-E3RV (5′TAGTGAAACGGACAGCCAAC-3′) for exon 3. Reaction products were resolved on a 1.2% agarose gel or subjected to direct DNA sequencing after purification through a QIAquick mini column (QIAGEN). For genotyping, wild-type and targeted exon 3 were amplified with a mixture of three primers: p18E3FW3 (5′-ACTACACAGGTCTTTGTGAAAG-3′), p18E3REV3 (5′-TCTCCGGATTTCCAAGTTTC-3′), and 5′-CCAGCCTCTGAGCCCAGAAAGCGAAGG-3′,a primer corresponding to a neo sequence. The PCR conditions were 95°C for 45 s, 55°C for 45 s, and 72°C for 45 s for each of 30 cycles.
Statistical analyses.
The survival rate was calculated by the CA-Cricket Graph III method, and statistical significance was assessed with Wilcoxon's log-rank test. The range of tumor numbers and sizes (mean ± standard error) were compared with those from an unpaired t test. P < 0.05 was considered to be statistically significant.
RESULTS
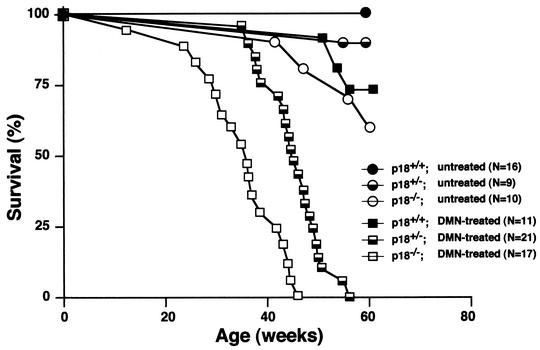
p18INK4c null mice spontaneously developed pituitary intermediate lobe tumors (>75%) (5, 6, 18) or, at a lower penetrance, T-cell lymphomas (∼10%) (15) late in life, with mean latencies of about 12 and 14 months, respectively. Simultaneous loss of three or four alleles of the p18 and p27 genes accelerates tumor growth, resulting in tumor-induced animal death by the ages of 9 and 4 months, respectively (6). We examined the susceptibility of p18 mutant mice to tumorigenesis after continuous low-dose exposure to the chemical carcinogen DMN, which targets mainly the liver and lung (2). As shown in Fig. 1, p18−/− mice and p18+/− mice are more susceptible to tumor-induced mortality after DMN treatment than wild-type mice (P < 0.001). No p18−/− mice in a cohort of 17 survived beyond 45 weeks of exposure to DMN; the mean survival time was 35 weeks. All p18+/− mice (n = 21) died within 58 weeks after exposure to DMN, with a mean survival time of 45 weeks. In contrast, 7 of 11 wild-type mice survived beyond 60 weeks after DMN treatment, and the mean survival time of the 4 mice that died from tumors was 56 weeks. In the untreated control cohorts monitored during the same 60 weeks of the experimental period, four out of nine p18−/− and two out of nine p18+/− mice died. We were unable to establish the exact cause of death of these mice. We detected no malignant tumors in these mice, and only one p18−/− mouse developed a pituitary adenoma. None of 16 wild-type mice died. These results provide further support for the function of the p18 gene in tumor suppression and demonstrate that loss of p18 function sensitizes mice to carcinogen-induced tumorigenesis. Notably, loss of one allele of p18, while not exhibiting an evident tumorigenic effect in untreated mice (5, 6, 18), effectively predisposes hemizygous mice to carcinogenesis.
FIG. 1.
Tumor-free survival in untreated wild-type and p18 mutant mice and in mice exposed to DMN. p18+/+, p18+/−, and p18−/− mice were either untreated or fed with DMN (0.0005%) in their drinking water continuously from the age of 8 weeks. Statistically significant differences in survival were detected between cohorts of DMN-treated p18−/− and DMN-treated p18+/− mice (P < 0.001) and between DMN-treated p18+/− and DMN-treated p18+/+ mice (P < 0.05).
Necropsy of DMN-treated p18 mutant animals revealed tumor development predominantly in three tissues: adenoma or carcinoma in the intermediate lobe of the pituitary, adenoma or carcinoma from lung bronchiolar or alveolar cells, and hemangiosarcoma from the sinusoidal endothelial cells of the liver (Table 1 and Fig. 2 to 4). In addition, DMN treatment also caused increased incidence of hyperplasia in these tissues, as well as several other tissues in adrenal, thyroid, and testis in both p18 null and heterozygous mice (Table 1). Liver hemangiosarcoma developed at a higher penetrance (50%) than either lung (35%) or pituitary tumors (25%) (Table 1). The rate of tumor growth was much faster in p18 null mice (averaging 38 weeks for hemangiosarcomas and 41 weeks for lung tumors, respectively) and p18 heterozygous mice (43 and 44 weeks) than in wild-type mice (60 and 58 weeks) (data not shown).
TABLE 1.
Spontanous and DMN-induced tumor and hyperplasia incidence
| Type of tumor in organ | No. of tumors in micea
|
|||||
|---|---|---|---|---|---|---|
| Untreated
|
DMN treated
|
|||||
| WT (n = 18) | p18+/− (n = 12) | p18−/− (n = 14) | WT (n = 13) | p18+/− (n = 20) | p18−/− (n = 20) | |
| Pituitary | ||||||
| Normal | 18 | 10 | 7 | 13 | 16 | 7 |
| Hyperplasia | 0 | 2 | 4 | 0 | 4 | 8 |
| Adenoma | 0 | 0 | 2 | 0 | 0 | 2 |
| Carcinoma | 0 | 0 | 1 | 0 | 0 | 3 |
| Lung | ||||||
| Normal | 18 | 12 | 9 | 10 | 12 | 10 |
| Hyperplasia | 0 | 0 | 2 | 1 | 2 | 3 |
| Adenoma | 0 | 0 | 2 | 2 | 4 | 4 |
| Carcinoma | 0 | 0 | 1 | 0 | 2 | 3 |
| Liver | ||||||
| Normal | 18 | 12 | 11 | 7 | 9 | 10 |
| Hemangiosarcoma | 0 | 0 | 3 | 6 | 11 | 10 |
| Adrenal | ||||||
| Normal | 18 | 12 | 11 | 12 | 16 | 10 |
| Medullary hyperplasia | 0 | 0 | 2 | 0 | 2 | 6 |
| Pheochromocytoma | 0 | 0 | 1 | 0 | 0 | 2 |
| Hemanglosarcoma | 0 | 0 | 0 | 1 | 2 | 2 |
| Thyroid | ||||||
| Normal | 18 | 12 | 12 | 13 | 14 | 15 |
| Hyperplasia | 0 | 0 | 2 | 0 | 6 | 5 |
| Testis | ||||||
| Normal intestitium | 18 | 12 | 8 | 13 | 16 | 10 |
| Hyperplasia | 0 | 0 | 6 | 0 | 4 | 10 |
| Other tumors | ||||||
| Hemangiosarcoma of abdominal | 0 | 0 | 0 | 2 | 2 | 1 |
| Hemangiosarcoma of pancrease | 0 | 0 | 0 | 1 | 0 | 1 |
| Soft tissue sarcoma of abdominal | 0 | 0 | 0 | 1 | 2 | 2 |
| Adenoma of Harderian gland | 0 | 1 | 0 | 0 | 0 | 0 |
WT, wild type.
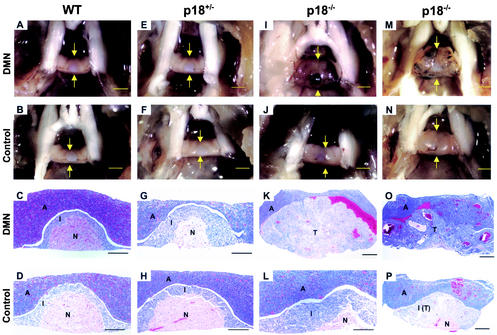
FIG. 2.
DMN treatment accelerated pituitary tumorigenesis in p18-deficient mice. Pituitary glands (pointed by arrows) from p18+/+, p18+/−, and p18−/− mice, either untreated or exposed to DMN, were microscopically examined at 6 months (A to L) and 9 months (M to P) of age either directly (top two rows; bars, 1 mm) or after hematoxylin and eosin (H/E) staining (lower two rows; bars, 200 μm). WT, wild type. Anterior lobe (A), intermediate lobe (I), neurohypophysis (N), and tumor (T) are indicated.
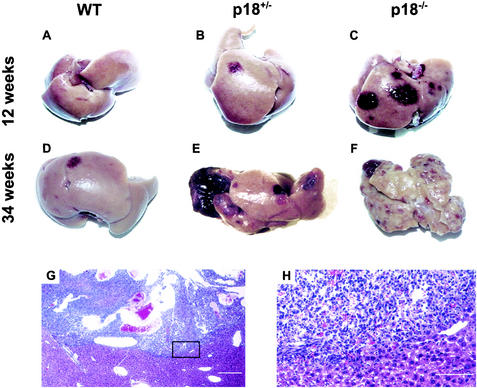
FIG. 4.
Mutation in the p18 gene sensitized mice to DMN-induced liver tumors. (A to F) Livers from p18−/−, p18+/−, and p18+/+ mice exposed to DMN were examined at the ages of 12 weeks (A to C) and 34 weeks (D to F). Normal gross liver architecture was retained in DMN-treated mutant mice at a young age despite the development of multiple hemangiosarcomas (red spots pointed by the arrows), but was not observed in p18 null mice after 34 weeks of age. WT, wild type. (G) Microscopic examination of hemangiosarcoma that developed in the liver of a 40-week-old p18−/− mouse exposed to DMN. Note that irregular vascular spaces have destroyed the normal hepatic parenchyma. (H) Higher magnification of the boxed area in (G) showing proliferating malignant endothelial cells replacing normal hepatic parenchyma.
p18 null mice spontaneously develop hyperplasia and adenomas in the intermediate lobe of the pituitary (5, 6, 18). DMN treatment accelerated pituitary adenoma growth, from a mean latency of 50 weeks in untreated p18 null mice to 32 weeks in treated mice (Table 1) (data not shown). The cells from the pituitary adenomas stained strongly for ACTH, confirming their intermediate lobe origin (data not shown). A carcinogenic effect of DMN has not been previously reported in the pituitary. This result indicates a broader tumorigenic effect of DMN in nontarget tissues sensitized by other events, such as tumor suppressor gene mutation. Pituitary adenomas that developed in DMN-treated p18 null mice were also more aggressive, often invading the adjacent anterior lobe and obliterating the neurohypophysis (Fig. 2K and O). Although no pituitary tumors were found in the DMN-treated p18+/− mice when they succumbed to liver tumors before the age of 56 weeks, their intermediate lobes were often enlarged compared with those of untreated p18+/− mice (Fig. 2G).
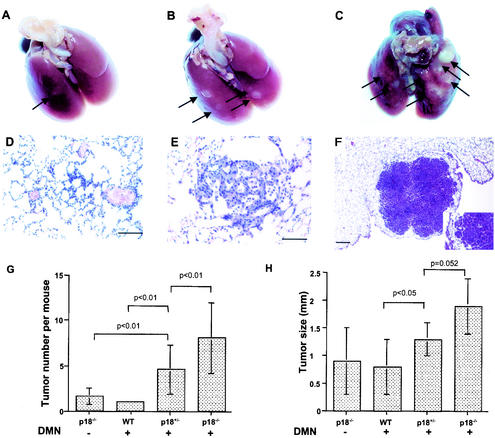
Lung bronchiolar or alveolar adenomas or adenocarcinomas were observed in 7 of 20 (35%) p18−/− mice, 6 of 20 (30%) p18+/− mice, and 2 of 13 (15%) wild-type mice exposed to DMN between 35 and 58 weeks (Table 1 and Fig. 3). In untreated control cohorts, lung tumors were found in 3 of 14 p18−/− mice (21%), but no wild-type (n = 18) or p18+/− (n = 12) mice. DMN treatment also caused an increased incidence of lung bronchiolar or alveolar hyperplasia (Fig. 3D). The lung adenomas were composed of atypical cells with nuclear pleomorphism and were characterized by the compression of adjacent alveolar tissue (Fig. 3E). Bronchiolar and alveolar adenocarcinomas invaded adjacent alveolar tissue (Fig. 3F). Loss of one or both alleles of p18 not only sensitized the mutant mice to DMN-induced lung tumor development, but also increased both the number and the size of lung tumors in each mouse. On average, 4.6 ± 2.7 and 8.1 ± 3.9 lung tumors developed in each DMN-treated p18+/− and p18−/− mouse, comparing with 1 tumor in each wild-type mouse (Fig. 3G). The average tumor sizes were 1.3 ± 0.3 and 1.9 ± 0.5 mm in DMN-treated p18+/− and p18−/− mice, compared with 0.8 ± 0.5 mm in wild-type mice (Fig. 3H).
FIG. 3.
Mutation in p18 gene sensitized mice to DMN-induced lung tumors. (A to C) The rate of lung tumor development varied between age-matched (11 months) p18+/+, p18+/−, and p18−/− mice after DMN exposure. Gross alveolar and bronchiolar adenomas (white spots) are identified by arrows. Note that p18 mutant lungs developed more tumors. (D to F) Alveolar or bronchiolar origin of lung tumor development. Lungs of DMN-treated mice at different ages were microscopically examined after hematoxylin-eaosin staining and exhibited alveolar or bronchiolar hyperplasia (D), papillary alveolar or bronchiolar adenoma with mucinous cell differentiation (E), and alveolar or bronchiolar carcinoma (F). The inset in panel F is a higher magnification showing tumor infiltration of adjacent pulmonary parenchyma. (G) The number of lung tumors that developed in each mouse exposed to DMN increases with p18 gene mutation. A statistically significant difference in tumor multiplicity was seen in mice of a different p18 genotype following exposure to DMN. WT, wild type. (H) The average size of DMN-induced lung tumors was larger in p18 mutant mice than in wild-type mice.
Neither pituitary nor pulmonary tumors were considered to be the cause of death in mice bearing them. The increased mortality of p18 mutant mice was mainly attributable to hemorrhage from hepatic hemangiosarcomas. When clinically normal DMN-treated mice of three genotypes were sacrificed and examined at 12 weeks of age, multiple liver hemangiosarcomas were present in p18 null and p18 hemizygous mice, although they were small and displayed no evidence of internal hemorrhaging (Fig. 4B and C). At 34 weeks of age, most p18 null mice were debilitated, while p18+/− mice remained relatively healthy despite the development of multiple hemangiosarcomas and retained normal liver histology (Fig. 4E). Only one DMN-treated wild-type mouse developed hemangiosarcomas by 34 weeks of age (Fig. 4D). The livers in 34-week-old DMN-treated p18 null mice were pale, necrotic, and contained multiple hemangiosarcomas, and normal liver tissue was difficult to observe (Fig. 4F). These hemangiosarcomas formed poorly delineated vascular channels lined by pleomorphic endothelial cells with frequent mitotic figures. Many liver hemangiosarcomas had progressed to become large blood-filled cavernous tumors that distended the liver capsule (Fig. 4). Tumors frequently ruptured, causing massive hemorrhage into the body cavity. In addition to the liver, hemangiosarcomas also developed in the adrenal glands, abdominal cavity, and pancreas (Table 1).
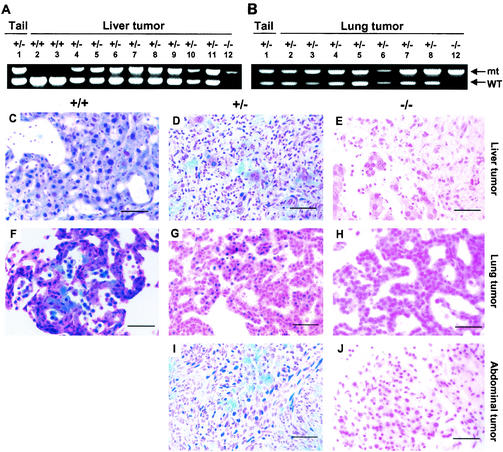
To determine the status of the p18 gene in DMN-induced tumors, we microscopically dissected tumor cells from paraffin-embedded samples and analyzed the p18 gene by PCR. Of 15 tumors from p18 hemizygous mice (8 liver hemangiosarcomas and 7 lung adenomas) and 2 liver hemangiosarcomas from wild-type mice, the wild-type allele was retained in all 17 tumors (Fig. 5A and B). To determine whether p18 expression was silenced in tumors, we determined p18 protein expression by immunohistochemistry with an affinity-purified antibody to p18INK4c. Of the same set of 15 tumors from p18 hemizygous mice and 2 liver hemangiosarcomas from wild-type mice, p18 staining was evident in all of the tumors (a representative tumor of each organ is shown in Fig. 5). In addition, we have also examined p18INK4c protein expression in two abdominal sarcomas (Fig. 5I) and one adrenal hemangiosarcoma (data not shown) arising from p18+/− mice and observed positive p18 expression by immunohistochemical staining. To determine whether expressed p18 alleles suffered mutations during tumor development, we amplified p18 from genomic DNA prepared from microscopically dissected tumor cells from all 17 liver and lung tumors induced in either wild-type or p18 heterozygous mice and sequenced the entire coding region. No mutation was detected in the 10 tumor samples we examined, including 6 liver hemangiosarcomas and 3 lung adenomas from p18 hemizygous mice and 1 liver hemangiosarcoma from a wild-type mouse (data not shown).
FIG. 5.
p18 is a haploinsufficient tumor suppressor. (A and B) Retention of the remaining wild-type p18 allele in liver (A) and lung (B) tumors from wild-type and p18 heterozygous mice. Genomic DNA was extracted from microdissected tumor cells or tails of p18+/− or p18−/− mice and subjected to PCR amplification with two pairs of primers flanking either the wild-type (WT) or mutated (mt) allele of the p18 gene. PCR products were resolved by agarose gel (1.2%) electrophoresis. The entire coding regions of p18 from liver tumor samples 2 to 11 and from lung tumor samples 2 to 8 were sequenced, and no mutation was found. (C to J) The expression of p18INK4c protein in three different types of tumors (liver hemangiosarcoma, lung adenoma, and abdominal sarcoma) from mice of three different p18 genotypes was determined by immunohistological staining with an affinity-purified polyclonal antibody specific to p18INK4c protein (8). Bars in all panels are 50 μm.
DISCUSSION
In this study, we provide further evidence supporting the role of p18INK4c gene in tumor suppression in mice; mutation in one or both alleles of p18 gene predisposed mice to carcinogen-induced tumorigenesis. Our results also establish that the p18 gene is haploinsufficient in tumor suppression. In both liver and lung tumors where we could isolate relatively pure tumor samples (17 samples), we found no loss of the wild-type p18 allele or mutations in the p18 coding region. Clear p18 protein expression was seen in tumor cells.
p18 joins a small number of tumor suppressor genes that have been definitively demonstrated to be haploinsufficient through the analysis of genetically targeted mice. These include p53 (31), p27 (4), Dmp1 (11), Nf1 (39), and Pten (17). How widespread haploinsufficiency is among tumor suppressor genes is a topic of current interest (3, 25). The determination of this issue not only may influence our view on the notion that inactivation mutations in tumor suppressor genes are recessive, but also has a clinical implication. While both alleles of a haplosufficient tumor suppressor gene are inactivated in tumors, a haploinsufficient tumor suppressor gene likely retains at least one allele in tumors and thus represents a potential target of therapeutic intervention. A single wild-type allele, according to the classical two-hit model of tumor suppressor genes, is sufficient to suppress tumor development and must be inactivated for tumorigenesis (14). This model is supported by the observations that germ line inactivation mutations have occurred in cancer-prone families, and tumors that developed in these family members are frequently associated with inactivation mutations of the remaining wild-type allele. Although experimentally suitable for identifying tumor suppressor genes with hereditary mutations in human tumors, the criterion of biallelic or complete functional loss excludes genes that are haploinsufficient, such as p18. For these haploinsufficient tumor suppressor genes, genomic mutations in human tumors may be rare, and hemizygous loss and decrease of expression level are difficult to determine. Our results add to the call for further investigations into genes that are not frequently mutated in human tumors, but exhibit clear tumor suppression function in mice.
Various degrees of haploinsufficiency have been seen with demonstrated tumor suppressor genes, with p27 and Dmp1 exhibiting either no or rare loss of the remaining wild-type allele in tumors from heterozygous mice, and thus a strong haploinsufficiency, and p53 and Pten displaying a noticeable frequency of loss of the wild-type allele and a weak effect. Moreover, the mutation rate in human tumors correlates with the various degrees of haploinsufficiency observed in mice. While both p53 and Pten are frequently mutated in human tumors, mutations in p18, p27, and Dmp1 appear to be rare. Is p18 a completely haploinsufficient tumor suppressor gene? It is important to note that loss of two alleles of p18 is clearly more tumorigenic than loss of one. The mean survival times of DMN-treated p18−/− and p18+/− mice are 35 and 45 weeks, respectively. The average numbers and size of tumors developed in DMN-treated p18−/− mice are 8.1 tumors per lung and 1.9 mm per tumor, compared with 4.6 tumors per lung and 1.3 mm per tumor in DMN-treated p18+/− mice. These results indicate that the haploinsufficiency of p18 in tumor suppression is incomplete and there must be a gradient of p18 tumor suppression activity, with expression of one allele exhibiting an intermediate effect between full function and complete functional loss. The biochemical basis for such gradient haploinsufficiency of p18 is not clear, but likely relates to a change in the level of CDK4 and CDK6 kinase activity, because these two enzymes are the only identified targets of p18 function.
Biochemically, all four INK4 proteins inhibit CDK4 and CDK6 almost indistinguishably, implying that members of the INK4 gene family, through activating or retaining the growth suppression activity of pRb proteins, could have a function in tumor suppression and are candidate tumor suppressors. Yet, mutations in human tumors have only been found in the p16INK4a gene and are rare in the other three INK4 genes (23, 37, 38). The finding that p18 is a haploinsufficient tumor suppressor may explain the lack of widespread mutations in the p18 gene in human tumors. It suggests a need for quantitative examination of p18 protein levels in order to accurately assess the role of p18 in the suppression of human tumors. Loss of function of p18 alone exhibited a relative weak tumor phenotype; about half of p18 null mice developed pituitary tumors, and 10% developed T-cell lymphomas after 1 year of age, but no obvious tumor phenotype developed in p18 heterozygous mice (5, 15). A clear tumor suppression function for p18 was revealed by the analysis of p18 p27 double mutant mice (6) and after carcinogen exposure (this study). These findings suggest the possibility that the role of other INK4 genes in tumor suppression could have been underestimated, and further investigations are needed to more completely explore this issue.
Acknowledgments
We thank Chad McCall for reading the manuscript and Joe He for helping with figure preparation.
Y.X. is supported in part by a U.S. Department of Defense Career Development Award (DAMD17-99-1-9574). This study was supported by NIH grants CA65572 and CA68377 to Y.X.
REFERENCES
- 1.Bai, F., Y. Nakanishi, K. Takayama, X. H. Pei, H. Tokiwa, and N. Hara. 1998. Ki-ras mutation and cell proliferation of lung lesions induced by 1-nitropyrene in A/J mice. Mol. Carcinog. 22:258-264. [DOI] [PubMed] [Google Scholar]
- 2.Clapp, N. K., A. W. Craig, and R. E. Toya, Sr. 1968. Pulmonary and hepatic oncogenesis during treatment of male RF mice with dimethyl-nitrosamine. J. Natl. Cancer Inst. 41:1213-1227. [PubMed] [Google Scholar]
- 3.Cook, W. D., and B. J. McCaw. 2000. Accommodating haploinsufficient tumor suppressor genes in Knudson's model. Oncogene 19:3434-3438. [DOI] [PubMed] [Google Scholar]
- 4.Fero, M., E. Randel, K. E. Gurley, J. M. Roberts, and C. J. Kemp. 1998. The murine gene p27Kip1 is haplo-insufficient for tumor suppression. Nature 396:177-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Franklin, D. S., V. L. Godfrey, H. Lee, G. I. Kovalev, R. Schoonhoven, S. Chen-Kiang, L. Su, and Y. Xiong. 1998. CDK inhibitors p18INK4c and p27KIP1 mediate two separate pathways to collaboratively suppress pituitary tumorigenesis. Genes Dev. 12:2899-2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Franklin, D. S., V. L. Godfrey, D. A. O'Brien, C. Deng, and Y. Xiong. 2000. Functional collaboration between different cyclin-dependent kinase inhibitors suppresses tumor growth with distinct tissue specificity. Mol. Cell. Biol. 20:6147-6158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Franklin, D. S., and Y. Xiong. 1996. Induction of p18INK4c and its predominant association with CDK4 and CDK6 during myogenic differentiation. Mol. Biol. Cell 7:1587-1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guan, K.-L., C. W. Jenkins, Y. Li, M. A. Nichols, X. Wu, C. L. O'Keefe, A. G. Matera, and Y. Xiong. 1994. Growth suppression by p18, a p16INK4/MTS1- and p14INK4B/MTS2-related CDK6 inhibitor, correlates with wild-type pRb function. Genes Dev. 8:2939-2952. [DOI] [PubMed] [Google Scholar]
- 9.Hall, M., and G. Peters. 1996. Genetic alterations of cyclins, cyclin-dependent kinases, and Cdk inhibitors in human cancer. Adv. Cancer Res. 68:67-108. [DOI] [PubMed] [Google Scholar]
- 10.Hunter, T., and J. Pines. 1994. Cyclins and cancer II: cyclin D and CDK inhibitors come of age. Cell 79:573-582. [DOI] [PubMed] [Google Scholar]
- 11.Inoue, K., F. Zindy, D. H. Randle, J. E. Rehg, and C. J. Sherr. 2001. Dmp1 is haplo-insufficient for tumor suppression and modifies the frequencies of Arf and p53 mutations in Myc-induced lymphomas. Genes Dev. 15:2934-2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jacks, T., A. Fazeli, E. M. Schmitt, R. T. Bronson, M. A. Goodell, and R. A. Weinberg. 1992. Effects of an Rb mutation in the mouse. Nature 359:295-300. [DOI] [PubMed] [Google Scholar]
- 13.Kamb, A., N. A. Gruis, J. Weaver-Feldhaus, Q. Liu, K. Harshman, S. V. Tavitgian, E. Stockert, R. S. Day, B. E. Johnson, and M. H. Skolnick. 1994. A cell cycle regulator potentially involved in genesis of many tumor types. Science 264:436-440. [DOI] [PubMed] [Google Scholar]
- 14.Knudson, A. G., Jr. 1985. Hereditary cancer, oncogenes, and antioncogenes. Cancer Res. 45:1437-1443. [PubMed] [Google Scholar]
- 15.Kovalev, G. I., D. S. Franklin, V. M. Coffield, Y. Xiong, and L. Su. 2001. An important role of CDK inhibitor p18(INK4c) in modulating antigen receptor-mediated T cell proliferation. J. Immunol. 167:3285-3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krimpenfort, P., K. C. Quon, W. J. Mooi, A. Loonstra, and A. Berns. 2001. Loss of p16Ink4a confers susceptibility to metastatic melanoma in mice. Nature 413:83-86. [DOI] [PubMed] [Google Scholar]
- 17.Kwabi-Addo, B., D. Giri, K. Schmidt, K. Podsypanina, R. Parsons, N. Greenberg, and M. Ittmann. 2001. Haploinsufficiency of the Pten tumor suppressor gene promotes prostate cancer progression. Proc. Natl. Acad. Sci. USA 98:11563-11568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Latres, E., M. Malumbres, R. Sotillo, J. Martin, S. Ortega, J. Martin-Caballero, J. M. Flores, C. Cordon-Cardo, and M. Barbacid. 2000. Limited overlapping roles of p15INK4b and p18INK4c cell cycle inhibitors in proliferation and tumorigenesis. EMBO J. 19:3496-3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee, Y.-H. P., C.-Y. Chang, N. Hu, Y.-C. J. Wang, C.-C. Lai, K. Herrup, W.-H. Lee, and A. Bradley. 1992. Mice deficient for Rb are nonviable and show defects in neurogenesis and haematopoiesis. Nature 359:288-294. [DOI] [PubMed] [Google Scholar]
- 20.Lohmann, D. R. 1999. RB1 gene mutations in retinoblastoma. Hum. Mutat. 14:283-288. [DOI] [PubMed] [Google Scholar]
- 21.Nikitin, A. Y., M. I. Juarez-Perez, S. Li, L. Huang, and W. H. Lee. 1999. RB-mediated suppression of spontaneous multiple neuroendocrine neoplasia and lung metastases in Rb+/− mice. Proc. Natl. Acad. Sci. USA 96:3916-3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nobori, T., K. Mlura, D. J. Wu, A. Lois, K. Takabayashi, and D. A. Carson. 1994. Deletion of the cyclin-dependent kinase-4 inhibitor gene in multiple human cancers. Nature 368:753-756. [DOI] [PubMed] [Google Scholar]
- 23.Okamoto, A., S. P. Hussain, K. Hagiwara, E. A. Spillare, M. R. Rusin, D. J. Demetrick, M. Serrano, G. J. Hannon, M. Shiseki, M. Zariwala, Y. Xiong, D. H. Beach, J. Yokota, and C. C. Harris. 1995. Mutations in the p16INK4/MTS1/CDKN2, p15INK4B/MTS2, and p18 genes in primary and metastatic lung cancer. Cancer Res. 55:1448-1451. [PubMed] [Google Scholar]
- 24.Phelps, D. E., K.-M. Hsiao, Y. Li, N. Hu, D. S. Franklin, E. Westphal, E. Y.-H. P. Lee, and Y. Xiong. 1998. Coupled transcriptional and translational control of the CDK inhibitor p18INK4c expression during myogenesis. Mol. Cell. Biol. 18:2334-2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quon, K. C., and A. Berns. 2001. Haplo-insufficiency? Let me count the ways. Genes Dev. 15:2917-2921. [DOI] [PubMed] [Google Scholar]
- 26.Robanus-Maandag, E., M. Dekker, M. van der Valk, M. L. Carrozza, J. C. Jeanny, J. H. Dannenberg, A. Berns, and H. te Riele. 1998. p107 is a suppressor of retinoblastoma development in pRb-deficient mice. Genes Dev. 12:1599-1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ruas, M., and G. Peters. 1998. The p16INK4a/CDK2A tumor suppresor and its relatives. Biochim. Biophys. Acta 1378:F115-F177. [DOI] [PubMed] [Google Scholar]
- 28.Sharpless, N. E., N. Bardeesy, K. H. Lee, D. Carrasco, D. H. Castrillon, A. J. Aguirre, E. A. Wu, J. W. Horner, and R. A. DePinho. 2001. Loss of p16Ink4a with retention of p19Arf predisposes mice to tumorigenesis. Nature 413:86-91. [DOI] [PubMed] [Google Scholar]
- 29.Sherr, C. J. 1996. Cancer cell cycle. Science 274:1672-1677. [DOI] [PubMed] [Google Scholar]
- 30.Sotillo, R., P. Dubus, J. Martin, E. de La Cueva, S. Ortega, M. Malumbres, and M. Barbacid. 2001. Wide spectrum of tumors in knock-in mice carrying a Cdk4 protein insensitive to INK4 inhibitors. EMBO J. 20:6637-6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Venkatachalam, S., Y. P. Shi, S. N. Jones, H. Vogel, A. Bradley, D. Pinkel, and L. A. Donehower. 1998. Retention of wild-type p53 in tumors from p53 heterozygous mice: reduction of p53 dosage can promote cancer formation. EMBO J. 17:4657-4667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang, T. C., R. D. Cardiff, L. Zukerberg, E. Lees, A. Arnold, and E. V. Schmidt. 1994. Mammary hyperplasia and carcinoma in MMTV-cyclin D1 transgenic mice. Nature 369:669-671. [DOI] [PubMed] [Google Scholar]
- 33.Weinberg, R. A. 1995. The retinoblastoma protein and cell cycle control. Cell 81:323-330. [DOI] [PubMed] [Google Scholar]
- 34.Williams, B. O., E. M. Schmitt, L. Remington, R. T. Bronson, D. M. Albert, R. A. Weinberg, and T. Jacks. 1994. Extensive contribution of Rb-deficient cells to adult chimeric mice with limited histopathological consequences. EMBO J. 13:4251-4259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wolfel, T., M. Hauer, J. Schneider, M. Serrano, C. Wolfel, E. Klehmann-Hieb, E. De Plaen, T. Hankeln, M. Buschenfelde, and D. Beach. 1995. A p16INK4a-insensitive CDK4 mutant targeted by cytolytic T lymphocytes in a human melanoma. Science 269:1281-1284. [DOI] [PubMed] [Google Scholar]
- 36.Yu, Q., Y. Geng, and P. Sicinski. 2001. Specific protection against breast cancers by cyclin D1 ablation. Nature 411:1017-1021. [DOI] [PubMed] [Google Scholar]
- 37.Zariwala, M., E. Liu, and Y. Xiong. 1996. Mutational analysis of the p16 family cyclin-dependent kinase inhibitors p15INK4b and p18INK4c in tumor-derived cell lines and primary tumors. Oncogene 12:451-455. [PubMed] [Google Scholar]
- 38.Zariwala, M., and Y. Xiong. 1996. Lack of mutation in the cyclin-dependent kinase inhibitor, p19INK4d, in tumor-derived cell lines and primary tumors. Oncogene 13:2033-2038. [PubMed] [Google Scholar]
- 39.Zhu, Y., P. Ghosh, P. Charnay, D. K. Burns, and L. F. Parada. 2002. Neurofibromas in NF1: Schwann cell origin and role of tumor environment. Science 296:920-922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zindy, F., J. van Deursen, G. Grosveld, C. J. Sherr, and M. F. Rousel. 2000. INK4d-deficient mice are fertile despite testicular atrophy. Mol. Cell. Biol. 20:372-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zuo, L., J. Weger, Q. Yang, A. M. Goldstein, M. A. Tucker, G. J. Walker, N. Hayward, and N. C. Dracopoli. 1996. Germline mutations in the p16INK4a binding domain of CDK4 in familial melanoma. Nat. Genet. 12:97-99. [DOI] [PubMed] [Google Scholar]