Abstract
Genetic studies in Saccharomyces cerevisiae have indicated the requirement of DNA polymerase (Pol) ζ for mutagenesis induced by UV light and by other DNA damaging agents. However, on its own, Polζ is highly inefficient at replicating through DNA lesions; rather, it promotes their mutagenic bypass by extending from the nucleotide inserted opposite the lesion by another DNA polymerase. So far, such a role for Polζ has been established for cyclobutane pyrimidine dimers, (6-4) dipyrimidine photoproducts, and abasic sites. Here, we examine whether Polζ can replicate through the 7,8-dihydro-8-oxoguanine (8-oxoG) and O6-methylguanine (m6G) lesions. We chose these two lesions for this study because the replicative polymerase, Polδ, can replicate through them, albeit weakly. We found that Polζ is very inefficient at inserting nucleotides opposite both these lesions, but it can efficiently extend from the nucleotides inserted opposite them by Polδ. Also, the most efficient bypass of 8-oxoG and m6G lesions occurs when Polδ is combined with Polζ, indicating a role for Polζ in extending from the nucleotides inserted opposite these lesions by Polδ. Thus, Polζ is a highly specialized polymerase that can proficiently extend from the primer ends opposite DNA lesions, irrespective of their degree of geometric distortion. Polζ, however, is unusually sensitive to geometric distortion of the templating residue, as it is highly inefficient at incorporating nucleotides even opposite the moderately distorting 8-oxoG and m6G lesions.
DNA lesions often block the progression of the replication fork, but replication through such lesions can be achieved by the action of specialized DNA polymerases. Eukaryotic cells contain a multiplicity of such DNA polymerases (Pols), such as Polη, Polζ, and others. Polη is unique among eukaryotic polymerases (9) in its ability to replicate through a cis-syn thymine-thymine (TT) dimer efficiently and accurately (12, 20), and genetic studies in yeast have indicated a role for Polη in the replication of cyclobutane pyrimidine dimers (CPDs) formed at TC and CC sites (22). Because of the requirement of Polη for the error-free bypass of CPDs, its inactivation in humans (8, 14) causes an increase in the incidence of UV mutagenesis (18, 21) and results in the cancer-prone syndrome, the variant form of xeroderma pigmentosum. Polη can also replicate through other DNA lesions such as 7,8-dihydro-8-oxoguanine (8-oxoG) and O6-methylguanine (m6G) (4, 6). The competency of Polη to replicate through these DNA lesions derives from its proficient ability to both insert nucleotides opposite the lesion site and to extend from the inserted nucleotide.
By contrast to the proficient ability of Polη to replicate through some DNA lesions, replication through certain DNA lesions requires the sequential action of two DNA polymerases, in which one polymerase inserts the nucleotide opposite the lesion site and the other extends from the inserted nucleotide (for a review, see reference 17). Genetic studies in the yeast Saccharomyces cerevisiae have provided evidence for the requirement of Polζ for mutagenesis induced by UV light and by other DNA damaging agents (10, 13), thus indicating a role for Polζ in mutagenic translesion synthesis (TLS). But, on its own, Polζ is unable to replicate through DNA lesions because it is highly inefficient at inserting nucleotides opposite the lesion site. Polζ, however, is very proficient at extending mismatched primer termini on undamaged DNAs and also in extending from nucleotides inserted opposite lesion sites (17). Thus, although Polζ is very inefficient at inserting nucleotides opposite the 3′ T of a cis-syn TT dimer or a (6-4) TT photoproduct, it can proficiently extend from a G nucleotide inserted opposite the 3′ T of these lesions by another DNA polymerase (7, 11). Polζ is also very inefficient at inserting nucleotides opposite an abasic site, but it proficiently extends from nucleotides, particularly an A, inserted opposite an abasic site by another DNA polymerase (5).
Oxygen-free radicals attack bases in DNA, and 8-oxoG is one of the adducts formed. Treatment of DNA with an alkylating agent such as N-methyl-N′-nitro-N-nitrosoguanidine results in the formation of m6G in DNA. Yeast Polη replicates through the 8-oxoG lesion by proficiently inserting a C and by proficiently extending from this nucleotide (6). Polη replicates through the m6G lesion by inserting a C or a T nucleotide opposite the lesion and then extending from the inserted nucleotide (4). Although the replicative yeast DNA polymerase, Polδ, is also able to incorporate nucleotides opposite 8-oxoG and m6G lesions and to extend from the inserted nucleotides, it is quite inefficient at both these steps (4, 6). Nevertheless, because Polδ would be the first polymerase to arrive at the lesion site, it could still carry out the insertion step but then stall because of its inefficient ability to extend from the nucleotide inserted opposite the lesion. In that case, the completion of lesion bypass would necessitate the action of an extender polymerase such as Polζ. Here, we examine the ability of yeast Polζ to replicate through the 8-oxoG and m6G lesions. Using steady-state kinetic analyses, we show that Polζ is highly inefficient at inserting nucleotides opposite both these lesions, but it can efficiently extend from the nucleotide inserted opposite these lesions by Polδ. These results underscore the highly specialized role of Polζ at the extension step in lesion bypass.
MATERIALS AND METHODS
Proteins. S. cerevisiae Polζ was expressed in yeast strain Sc334 as a glutathione S-transferase (GST)-Rev3 fusion protein in complex with Rev7 protein as described previously (7). The GST-Rev3/Rev7-containing fractions were concentrated using Microcon 30 (Amicon) and frozen at −70°C. S. cerevisiae Polδ was kindly provided by Peter Burgers.
DNA substrates.
Oligonucleotides were synthesized by Midland Certified Reagent Co. (Midland, Tex.). DNA S1 substrates, shown below in Fig. 1, were generated by annealing a 75-nucleotide (nt) oligomer template, 5′-AGC TAC CAT GCC TGC CTC AAG AAT TCG TAA XAT GCC TAC ACT GGA GTA CCG GAG CAT CGT CGT GAC TGG GAA AAC-3′, containing a G, an 8-oxoG, or an m6G at position 31 (X), to the 40-nt 5′-32P-labeled oligonucleotide primer, N4264 (5′-GTT TTC CCA GTC ACG ACG ATG CTC CGG TAC TCC AGT GTA G-3′). For DNA S2 substrates, shown below in Fig. 2, and also used for steady-state kinetic analysis of extension reactions below in Fig. 3, the 75-nt oligomer templates used for generating the S1 substrates were annealed to the 5′ 32P-labeled oligonucleotide primers, 5′-GTT TTC CCA GTC ACG ACG ATG CTC CGG TAC TCC AGT GTA GGCATN-3′, where N is G, A, T, or C. For steady-state kinetic analysis of the insertion reactions shown below in Fig. 3, the 75-nt templates used for generating the S1 substrates were annealed to the 5′-32P-labeled oligonucleotide primer, N4309 (5′-GTT TTC CCA GTC ACG ACG ATG CTC CGG TAC TCC AGT GTA GGC AT-3′).
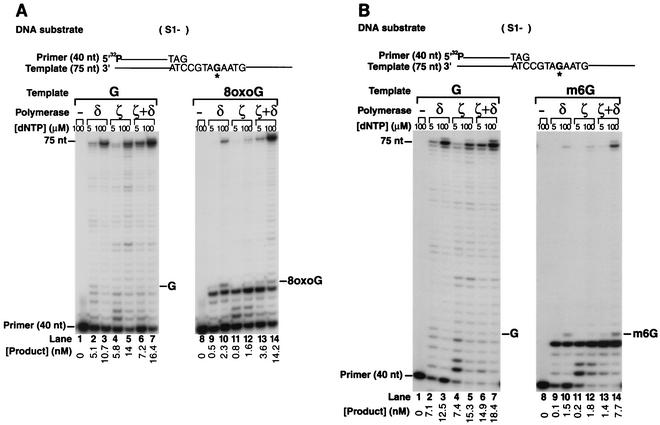
FIG. 1.
Bypass of 8-oxoG and m6G lesions by the combined action of Polδ and Polζ. (A) DNA synthesis on an 8-oxoG-containing DNA template. Lanes 1 to 7, undamaged DNA; lanes 8 to 14, 8-oxoG-containing DNA. Sequences adjacent to the primer-template junction are shown for the 40-nt, 5′-32P-labeled primer and 75-nt template. The position corresponding to the undamaged G or the 8-oxoG site on the template is indicated by ∗G. Yeast Polζ (10 nM), yeast Polδ (10 nM), or a combination of these two enzymes were incubated with the DNA substrate (20 nM) in the presence of each of the four dNTPs (5 or 100 μM) at 30°C for 10 min. The reaction products were resolved on a 15% denaturing polyacrylamide gel and visualized by autoradiography. The gel was analyzed by using a PhosphorImager, and the concentrations of the products of the synthesis past the undamaged G or 8-oxoG are indicated. (B) DNA synthesis on an m6G-containing DNA template. Lanes 1 to 7, undamaged DNA; lanes 8 to 14, m6G-containing DNA. Reactions were carried out as indicated in the legend for panel A, except that the incubation time of the reactions was 15 min.
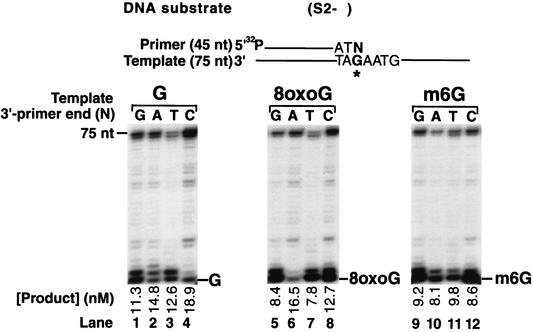
FIG. 2.
Extension of primers with various 3′ ends opposite 8-oxoG and m6G lesions by Polζ. Each of the four different primers, differing only in the 3′-terminal nucleotide, was annealed to the template oligonucleotide containing an undamaged G, an 8-oxoG, or an m6G residue at the same position. In the primer-template pair shown, N designates the position of the variable terminal primer nucleotide and ∗G designates the position of a G, an 8-oxoG, or an m6G residue. Polζ (10 nM) was incubated with the DNA substrate (20 nM) in the presence of each of the four dNTPs (100 μM) at 30°C for 15 min. The gel was analyzed by using a PhosphorImager, and the amount of the product of primer extension opposite from the undamaged G, 8-oxoG, or m6G is indicated.
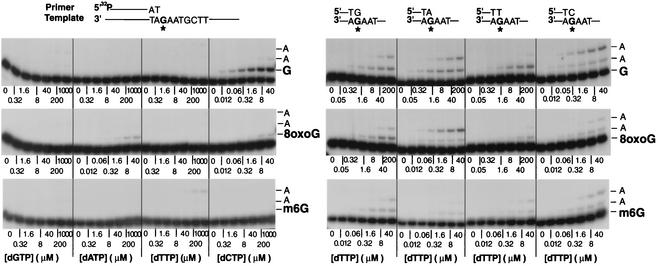
FIG. 3.
Steady-state kinetic analysis of insertion and extension reactions catalyzed by Polζ on 8-oxoG- and m6G-containing DNA templates. A portion of the DNA substrate used for insertion reactions (top left) or for extension reactions (top right) is shown, and the position of an 8-oxoG, m6G, or undamaged G residue in the template oligonucleotide is indicated by an asterisk (top). Polζ (2 nM) was incubated with the primer-template DNA substrate (30 nM) and increasing concentrations of a single deoxynucleotide for 10 min at 30°C. The quenched samples were analyzed by 15% denaturing polyacrylamide gel electrophoresis, and the steady-state kinetic parameters kcat and Km were determined.
DNA polymerase assays.
A standard DNA polymerase reaction mixture (10 μl) contained 25 mM Tris-HCl (pH 7.5), 5 mM MgCl2, 5 mM dithiothreitol, 100 μg of bovine serum albumin/ml, 20 nM 5′-32P-labeled oligonucleotide primer annealed to an oligonucleotide template, and 5 or 100 μM concentrations of each of all four deoxynucleotides (dGTP, dATP, dTTP, and dCTP), as indicated in the figure legends. Reactions were started by the addition of Polζ (10 nM), Polδ (10 nM), or a mixture of Polδ and Polζ and incubated at 30°C for 10 to 15 min followed by quenching by the addition of loading buffer (40 μl) containing EDTA (20 mM), 95% formamide, 0.3% bromphenol blue, and 0.3% cyanol blue. The reaction products were resolved on 15% polyacrylamide gels containing 8 M urea. Quantitation of the results was done using a Molecular Dynamics STORM PhosphorImager and ImageQuant software.
Analysis of steady-state kinetics.
Steady-state kinetic analyses for deoxynucleotide incorporation opposite an 8-oxoG or an m6G, or primer extension opposite from these lesions were performed as described previously (2, 3, 15). Briefly, Polζ (2 nM) was incubated with 30 nM DNA substrate in the presence of increasing concentrations of a single deoxynucleotide for 10 min at 30°C. The reaction products were resolved on 15% polyacrylamide gels containing 8 M urea. Gel band intensities of the substrates and products were quantitated by PhosphorImager, and the observed rate of deoxynucleotide incorporation was plotted as a function of deoxynucleotide triphosphate (dNTP) concentration. The data were fit by nonlinear regression using SigmaPlot 5.0 to the Michaelis-Menten equation describing a hyperbola, v = (Vmax × [dNTP]/(Km+ [dNTP]). The kcat and Km steady-state parameters were obtained from the fit and were used to calculate the frequency of deoxynucleotide incorporation (finc) and the intrinsic efficiency of mispair extension (f0ext) for each primer-template pair by using the following equation: finc or f0ext = (kcat/Km)mispaired/(kcat/Km)paired.
RESULTS
Bypass of the 8-oxoG and m6G lesions by the combined action of Polδ and Polζ.
The bypass of an 8-oxoG and an m6G lesion was examined in running start reactions by using a 75-nt template containing an 8-oxoG or an m6G residue 45 nt from the 3′ end, which was primed with a 5′-32P-labeled 40-nt oligomer. Polymerase reactions were carried out at low (5 μM) as well as at high (100 μM) nucleotide concentrations to monitor lesion bypass under both restrictive and unrestrictive conditions. First, we examined the ability of yeast Polζ to replicate past the 8-oxoG lesion in the DNA template (Fig. 1A). 8-oxoG is a strong block to DNA synthesis by Polζ, as even at 100 μM dNTP, compared to synthesis through the corresponding undamaged G residue (Fig. 1A, lane 5), Polζ replicated through only ∼12% of the 8-oxoG lesions (Fig. 1A, lane 12). Although Polζ was severely inhibited from inserting a nucleotide opposite 8-oxoG, which is indicated by a strong stall site right before the 8-oxoG residue, Polζ was able to extend opposite from the 8-oxoG residue, which is shown by an absence of a stall site opposite this lesion (Fig. 1A, lane 12). This observation raised the possibility that Polζ is able to stimulate the bypass of 8-oxoG by extending from the nucleotide previously inserted opposite 8-oxoG by another DNA polymerase. Yeast Polδ, the replicative DNA polymerase, is able to replicate though the 8-oxoG lesion; however, compared to replication through an undamaged G (Fig. 1A, lanes 2 and 3), it replicates through the 8-oxoG lesion inefficiently (Fig. 1A, lanes 9 and 10). Remarkably, the most efficient bypass of the 8-oxoG lesion occurred when Polζ was combined with Polδ, and these polymerases together replicated through the 8-oxoG lesion almost as efficiently as through the undamaged G residue (Fig. 1A, compare lanes 13 and 14 with lanes 6 and 7).
Polζ replicated through an m6G residue also, but again, the bypass was highly inefficient (Fig. 1B, compare lanes 11 and 12 with lanes 4 and 5). However, the absence of a stall site opposite m6G in contrast to a strong stall site right before this lesion indicated that Polζ was able to extend the 3′ primer end opposite from m6G, and only the nucleotide incorporation opposite m6G was severely inhibited (Fig. 1B, lanes 11 and 12). Polδ shows some ability to incorporate nucleotides opposite the m6G residue; however, it is quite inefficient in bypassing this lesion (Fig. 1B, compare lanes 9 and 10 with lanes 2 and 3). Importantly, the combination of Polζ with Polδ promoted m6G bypass, and together they replicated through the m6G lesion ∼40% as efficiently as through the undamaged G residue (Fig. 1B, compare lane 14 with lane 7). The key result here is that the two DNA polymerases, Polδ and Polζ, together are much more effective in 8-oxoG and m6G bypass than either polymerase alone. Moreover, these observations suggested that since Polδ would be the first DNA polymerase to encounter the DNA lesion during its role in DNA replication, it would contribute to the insertion of nucleotides opposite the 8-oxoG and m6G lesions, while Polζ could carry out the subsequent extension step from the ensuing primer end.
Extension of primer ends situated opposite the 8-oxoG and m6G lesions by Polζ.
Next, we examined the ability of Polζ to extend from the various 3′ primer termini situated across from an 8-oxoG or an m6G site in the 75-nt template in the presence of a 100 μM concentration of each of the four dNTPs (Fig. 2). Polζ is a promiscuous extender of primer-terminal mispairs opposite undamaged DNA templates (Fig. 2, lanes 1 to 4) and, importantly, it is also very efficient at extending from primer ends opposite from the 8-oxoG and m6G residues (Fig. 2, lanes 5 to 8 and lanes 9 to 12, respectively). Polζ extended from an A opposite the 8-oxoG site ∼90% as efficiently as from a C opposite undamaged G (Fig. 2, compare lane 6 with lane 4). The extension from a C, a G, and a T opposite from 8-oxoG was also efficient, occurring with 67, 44, and 41% of the efficiency of extension from the C opposite undamaged G. Polζ extended from a T, a G, a C, and an A opposite from the m6G residue with 40 to 50% of the efficiency of extension from the C opposite undamaged G (Fig. 2, compare lanes 9 to 12 with lane 4).
We have previously shown that yeast Polδ predominantly inserts an A residue opposite 8-oxoG (6) and a T or a C opposite m6G (4). Thus, efficient bypass of 8-oxoG and m6G lesions could be carried out by incorporating an A residue opposite 8-oxoG and a T or a C residue opposite m6G, respectively, by Polδ, and by the subsequent extension of the resulting 3′ primer end by Polζ.
Steady-state kinetic analysis of nucleotide insertion and extension reactions across from the 8-oxoG and m6G lesions by Polζ.
To characterize further the spectrum and the efficiencies of nucleotide insertion and extension reactions across from 8-oxoG and m6G lesions by Polζ, we measured the steady-state kinetic parameters of these reactions. The kinetics of insertion of a single deoxynucleotide opposite an 8-oxoG and an m6G and the kinetics of addition of the next correct nucleotide to various 3′ primer termini situated across from 8-oxoG and m6G were determined as a function of deoxynucleotide concentration under steady-state conditions. We incubated Polζ with the 8-oxoG- and m6G-containing DNA substrates and increasing concentrations of a single nucleotide (Fig. 3). The rate of nucleotide incorporation was plotted as a function of nucleotide concentration. The steady-state apparent kcat and Km values for each nucleotide incorporation as well as for the extension of each primer terminus were obtained from the curve fitted to the Michaelis-Menten equation by using nonlinear regression. The frequency of nucleotide incorporation, finc, and the relative efficiency of extension, f0ext, were calculated as the ratio of the efficiency (kcat/Km) of incorrect nucleotide incorporated or extended from, to the efficiency (kcat/Km) of correct nucleotide incorporated or extended from, respectively. As shown in Table 1, Polζ incorporated an A or a C opposite 8-oxoG about 100-fold less efficiently than the incorporation of a C opposite undamaged G in the template. However, Polζ extended from the primer end situated opposite the 8-oxoG very efficiently (Table 2). Compared to the extension from a C opposite undamaged G in the template, Polζ extended from an A, a C, a G, and a T 3′ primer ends situated opposite the 8-oxoG residue with frequencies of 7.3 × 10−1, 1.7 × 10−1, 4.1 × 10−2, and 4 × 10−3, respectively (Table 2). Thus, Polζ extends from an A opposite 8-oxoG about as efficiently as from a C opposite undamaged G in the template.
TABLE 1.
Steady-state kinetic parameters of nucleotide insertion reactions opposite 8-oxoG and m6G template residues by yeast Polζ
| DNA substrate | Incoming nucleotide | kcat (min−1) | Km (μM) | kcat/Km (min−1 μM−1) | Relative incorporation efficiency |
|---|---|---|---|---|---|
| Insertion opposite G | |||||
| 5′---CAT | dGTP | 0.06 ± 0.008 | 420 ± 70 | 0.00014 | 1.1 × 10−4 |
| ---GTAGAAT--- | dATP | 0.02 ± 0.01 | 370 ± 100 | 0.000054 | 4.3 × 10−5 |
| dTTP | 0.03 ± 0.006 | 450 ± 80 | 0.000067 | 5.4 × 10−5 | |
| dCTP | 0.44 ± 0.02 | 0.36 ± 0.03 | 1.23 | 1 | |
| Insertion opposite 8oxoG | |||||
| 5′---CAT | dGTP | NDa | ND | ||
| ---GTAGAAT | dATP | 0.11 ± 0.02 | 6.3 ± 0.4 | 0.017 | 1.4 × 10−2 |
| 8oxo | dTTP | ND | ND | ||
| dCTP | 0.09 ± 0.03 | 7.5 ± 1 | 0.012 | 9.7 × 10−3 | |
| Insertion opposite m6G | |||||
| 5′---CAT | dGTP | 0.02 ± 0.007 | 430 ± 80 | 0.000047 | 3.8 × 10−5 |
| ---GTAGAAT | dATP | ND | ND | ||
| m6 | dTTP | 0.08 ± 0.008 | 46 ± 11 | 0.0017 | 1.4 × 10−3 |
| dCTP | 0.02 ± 0.004 | 370 ± 90 | 0.000054 | 4.4 × 10−5 |
ND, not detected.
TABLE 2.
Steady-state kinetic parameters of extension reactions opposite from 8-oxoG and m6G template residues by yeast Polζ
| DNA substrate | Incoming nucleotide | kcat (min−1) | Km (μM) | kcat/Km (min−1 μM−1) | Relative extension efficiency |
|---|---|---|---|---|---|
| Extension from G, A, T and C opposite G | |||||
| 5′---CATG | |||||
| ---GTAGAAT--- | dTTP | 0.22 ± 0.01 | 7.2 ± 0.8 | 0.03 | 5 × 10−2 |
| 5′---CATA | |||||
| ---GTAGAAT--- | dTTP | 0.2 ± 0.02 | 2.4 ± 0.3 | 0.083 | 1.4 × 10−1 |
| 5′---CATT | |||||
| ---GTAGAAT--- | dTTP | 0.16 ± 0.02 | 7.8 ± 0.9 | 0.02 | 3.3 × 10−2 |
| 5′---CATC | |||||
| ---GTAGAAT--- | dTTP | 0.23 ± 0.02 | 0.38 ± 0.05 | 0.6 | 1 |
| Extension from G, A, T and C opposite 8oxoG | |||||
| 5′---CATG | |||||
| ---GTAGAAT---- | |||||
| 8oxo | dTTP | 0.19 ± 0.02 | 7.6 ± 0.9 | 0.025 | 4.1 × 10−2 |
| 5′---CATA | |||||
| ---GTAGAAT---- | |||||
| 8oxo | dTTP | 0.21 ± 0.01 | 0.48 ± 0.05 | 0.44 | 7.3 × 10−1 |
| 5′---CATT | |||||
| ---GTAGAAT---- | |||||
| 8oxo | dTTP | 0.08 ± 0.03 | 34 ± 7 | 0.0024 | 4 × 10−3 |
| 5′---CATC | |||||
| ---GTAGAAT---- | |||||
| 8oxo | dTTP | 0.15 ± 0.02 | 1.5 ± 0.2 | 0.1 | 1.7 × 10−1 |
| Extension from G, A, T and C opposite m6G | |||||
| 5′---CATG | |||||
| ---GTAGAAT---- | |||||
| m6 | dTTP | 0.18 ± 0.03 | 2.0 ± 0.1 | 0.09 | 1.5 × 10−1 |
| 5′---CATA | |||||
| ---GTAGAAT---- | |||||
| m6 | dTTP | 0.07 ± 0.01 | 6.9 ± 0.5 | 0.01 | 1.7 × 10−2 |
| 5′---CATT | |||||
| ---GTAGAAT---- | |||||
| m6 | dTTP | 0.08 ± 0.009 | 4.3 ± 0.6 | 0.019 | 3.1 × 10−2 |
| 5′---CATC | |||||
| ---GTAGAAT---- | |||||
| m6 | dTTP | 0.16 ± 0.03 | 1.4 ± 0.1 | 0.11 | 1.9 × 10−1 |
Polζ is also very inefficient at incorporating nucleotides opposite the m6G lesion. Polζ incorporated nucleotides opposite m6G with frequencies ranging from 1.4 × 10−3 to 4.4 × 10−5, and even the incorporation of a T, the most efficiently incorporated nucleotide opposite m6G by Polζ, was ∼700-fold less efficient than the incorporation of C opposite undamaged G (Table 1). In contrast, Polζ extended the primer ends opposite from the m6G lesion quite efficiently. Polζ extended from a C opposite m6G ∼5-fold less efficiently than from a C opposite an undamaged G in the template and, intriguingly, Polζ could also extend from a G placed opposite m6G quite efficiently. Overall, the steady-state kinetic analyses indicate that Polζ is quite inefficient in inserting nucleotides opposite the 8-oxoG and m6G lesions, but it is quite efficient at extending primer ends opposite from these lesions.
DISCUSSION
We have shown previously that Polζ is highly inefficient at inserting nucleotides opposite the 3′ T of a cis-syn TT dimer or a (6-4) TT photoproduct and also that it is unable to insert nucleotides opposite an abasic site (5, 7, 11). The inefficient ability of Polζ to incorporate nucleotides opposite a CPD or (6-4) photoproduct could be explained by its inability to accommodate these distorting lesions in its active site, and the inability to incorporate nucleotides opposite an abasic site may suggest the need of Polζ for a templating base. Here, we show that Polζ is also very inefficient at inserting nucleotides opposite the 8-oxoG and m6G lesions. This is rather surprising, given the fact that Polδ can insert an A opposite 8-oxoG and a T or a C opposite m6G (4, 6). Compared to the insertion of a C opposite undamaged template G, Polζ shows an ∼100-fold-reduced efficiency for the insertion of an A opposite 8-oxoG, and it inserts a T or a C opposite the m6G lesion with an efficiency that is reduced by ∼700-fold or more. The highly inefficient incorporation of nucleotides opposite the 8-oxoG or m6G lesions by Polζ, where Polδ is able to insert the nucleotides which cause the least amount of geometric distortion, suggests that Polζ is very sensitive to any modification of the templating base.
By contrast to its high degree of sensitivity to geometric distortions of the templating base, Polζ can tolerate a wide range of distortions at the primer terminus. Thus, it can efficiently extend from mispaired primer termini on undamaged as well as on damaged DNAs. Here we show that Polζ proficiently extends from an A opposite 8-oxoG, which is the nucleotide inserted opposite this lesion by Polδ. Accordingly, the most proficient replication through the 8-oxoG lesion occurs when Polδ is combined with Polζ. Thus, Polζ could contribute to the mutagenic bypass of this DNA lesion. Polζ extends from a C opposite m6G only ∼5-fold less well than it extends from a C opposite undamaged G. Polζ is, however, less efficient (∼30-fold) at extending from a T opposite m6G, but Polζ extends quite well from a G opposite this lesion. Thus, Polζ could contribute to the error-free bypass of m6G lesions by extending from the C nucleotide inserted by Polδ. For the m6G lesion also, the most proficient bypass occurs in the presence of both Polδ and Polζ.
Yeast Polδ exhibits a strong stall site just before the 8-oxoG and m6G lesions, suggesting that this polymerase is inhibited at the nucleotide incorporation step. It is possible that some of this block derives from the subsequent removal of the incorporated nucleotide by Polδ's 3′ → 5′ exonuclease activity. In that case, in the presence of Polζ, because of the proficient extension of the primer terminus opposite the lesion, the forward synthesis reaction could counteract the backward exonucleolytic reaction of Polδ, and that may account for the synergistic enhancement of lesion bypass that occurs when Polδ is combined with Polζ.
The results with the DNA lesions studied so far indicate that the role of Polζ is to extend from the nucleotide inserted opposite the lesion site by another DNA polymerase from which the inserter polymerase is unable to extend (Table 3). Thus, for a TT dimer, Polη mediates the error-free bypass of this lesion by inserting an A opposite the 3′ T and then extending the primer end by inserting an A opposite the 5′ T of the lesion. However, if a wrong nucleotide, such as a G, were to be inserted opposite the 3′ T by Polη, then the extension step would become dependent upon Polζ because of its more proficient ability to extend from this primer terminus (11) than that of Polη (19). Opposite the 3′ T of a (6-4) TT photoproduct, Polη inserts a G about eightfold more efficiently than an A, but it cannot extend from either of these nucleotides (7). Polζ is ∼3-fold more efficient at extending from a G than from an A opposite this lesion (7, 11). Thus, the sequential action of Polη and Polζ could effect the mutagenic bypass of a (6-4) TT photoproduct (Table 3). In fact, genetic studies in yeast have corroborated the involvement of Polη in the mutagenic bypass of the (6-4) TT lesion, as the incidence of 3′ T→C mutations in a plasmid carrying a site-specific (6-4) TT lesion is greatly reduced in the absence of Polη (1). Because of the inability of any other polymerase to extend from the nucleotide inserted opposite the 3′ T of a (6-4) photoproduct, TLS through this lesion, whether mutagenic or error free, would be entirely dependent upon Polζ (Table 3). Similarly, TLS through an abasic site would also have the absolute requirement of Polζ for the extension step, as no other polymerase is able to perform this task (Table 3).
TABLE 3.
Role of yeast Polζ in extending primer ends opposite from DNA lesions
| DNA lesion | Nucleotide inserted | Inserter polymerase | Extender polymerase | Mode of lesion bypass |
|---|---|---|---|---|
| TT dimer | A | Polη | Polη | Error free |
| G | Polη | Polζ | Mutagenic | |
| (6-4) TT photoproduct | G | Polη | Polζ | Mutagenic |
| A | ? | Polζ | Error free | |
| Abasic site | A | Polδ | Polζ | |
| G | Polη | Polζ | Mostly mutagenic | |
| C | Rev1 | Polζ | ||
| 8-oxoG | C | Polη | Polη | Error free |
| A | Polδ | Polζ or Polη | Mutagenic | |
| m6G | C or T | Polη | Polη | Error free and mutagenic |
| T or C | Polδ | Polζ or Polη | Mutagenic and error free |
Although yeast Polδ can replicate through the 8-oxoG lesion, it stalls at both the insertion and extension steps (6) (Fig. 1A); therefore, we expect it to make only a marginal contribution to the bypass of this DNA lesion. In yeast, Polη plays a predominant role in the error-free bypass of 8-oxoG (Table 3), as the frequency of G:C to T:A transversions is greatly increased in a strain lacking both Polη and the Ogg1 DNA glycosylase which functions in the removal of 8-oxoG paired with C (6). The increased frequency of G:C to T:A transversions that ensue from the replicative bypass of 8-oxoG presumably result from the insertion of an A opposite this lesion by Polδ (6). Although either Polη or Polζ could extend from the A nucleotide inserted opposite 8-oxoG by Polδ (Table 3), we expect Polζ to be more effective at this step since it is almost as efficient at extending from the A opposite 8-oxoG as it is in extending from a C opposite G, whereas Polη extends from an A opposite 8-oxoG with an ∼6-fold-reduced efficiency (6).
Genetic studies in yeast have indicated the requirement of Polη and Polδ for the mutagenic bypass of the m6G lesion (4). While Polη replicates through the m6G lesion fairly efficiently by inserting a C or T residue and by extending from the inserted nucleotide, Polδ is quite inefficient at bypassing this lesion, as it is inhibited at both the insertion and extension steps (4) (Fig. 1B). However, because of the proximity of Polδ to the lesion site, it could still be able to carry out the nucleotide incorporation step but stall at the extension step. In that case, the subsequent extension could depend upon Polζ or Polη (Table 3).
In summary, Polζ is very adept at extending primer ends opposite from a diverse array of DNA lesions. It is the only yeast polymerase that is able to proficiently extend from nucleotides inserted opposite a (6-4) dipyrimidine photoproduct or an abasic site, which explains its in vivo requirement for TLS through these DNA lesions (10, 16). For the extension of primer ends opposite from the 8-oxoG and m6G lesions, however, Polζ could compete with Polη. Polζ and Polη differ remarkably in their roles in lesion bypass, as Polζ is a highly specialized polymerase that acts primarily to extend the primer ends opposite from DNA lesions, regardless of the degree of geometric distortion they cause, whereas Polη bypasses moderately distorting lesions by carrying out both the insertion and extension steps.
Acknowledgments
This work was supported by National Institutes of Health grant GM19261.
REFERENCES
- 1.Bresson, A., and R. P. P. Fuchs. 2002. Lesion bypass in yeast cells: Pol η participates in a multi-DNA polymerase process. EMBO J. 21:3881-3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Creighton, S., L. B. Bloom, and M. F. Goodman. 1995. Gel fidelity assay measuring nucleotide misinsertion, exonucleolytic proofreading, and lesion bypass efficiencies. Methods Enzymol. 262:232-256. [DOI] [PubMed] [Google Scholar]
- 3.Goodman, M. F., S. Creighton, L. B. Bloom, and J. Petruska. 1993. Biochemical basis of DNA replication fidelity. Crit. Rev. Biochem. Mol. Biol. 28:83-126. [DOI] [PubMed] [Google Scholar]
- 4.Haracska, L., S. Prakash, and L. Prakash. 2000. Replication past O6-methylguanine by yeast and human DNA polymerase η. Mol. Cell. Biol. 20:8001-8007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haracska, L., I. Unk, R. E. Johnson, E. Johansson, P. M. J. Burgers, S. Prakash, and L. Prakash. 2001. Roles of yeast DNA polymerases δ and ζ and of Rev1 in the bypass of abasic sites. Genes Dev. 15:945-954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haracska, L., S.-L. Yu, R. E. Johnson, L. Prakash, and S. Prakash. 2000. Efficient and accurate replication in the presence of 7,8-dihydro-8-oxoguanine by DNA polymerase η. Nat. Genet. 25:458-461. [DOI] [PubMed] [Google Scholar]
- 7.Johnson, R. E., L. Haracska, S. Prakash, and L. Prakash. 2001. Role of DNA polymerase η in the bypass of a (6-4) TT photoproduct. Mol. Cell. Biol. 21:3558-3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnson, R. E., C. M. Kondratick, S. Prakash, and L. Prakash. 1999. hRAD30 mutations in the variant form of xeroderma pigmentosum. Science 285:263-265. [DOI] [PubMed] [Google Scholar]
- 9.Johnson, R. E., S. Prakash, and L. Prakash. 1999. Efficient bypass of a thymine-thymine dimer by yeast DNA polymerase, Polη. Science 283:1001-1004. [DOI] [PubMed] [Google Scholar]
- 10.Johnson, R. E., C. A. Torres-Ramos, T. Izumi, S. Mitra, S. Prakash, and L. Prakash. 1998. Identification of APN2, the Saccharomyces cerevisiae homolog of the major human AP endonuclease HAP1, and its role in the repair of abasic sites. Genes Dev. 12:3137-3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson, R. E., M. T. Washington, L. Haracska, S. Prakash, and L. Prakash. 2000. Eukaryotic polymerases ι and ζ act sequentially to bypass DNA lesions. Nature 406:1015-1019. [DOI] [PubMed] [Google Scholar]
- 12.Johnson, R. E., M. T. Washington, S. Prakash, and L. Prakash. 2000. Fidelity of human DNA polymerase η. J. Biol. Chem. 275:7447-7450. [DOI] [PubMed] [Google Scholar]
- 13.Lawrence, C. W., and D. C. Hinkle. 1996. DNA polymerase ζ and the control of DNA damage induced mutagenesis in eukaryotes. Cancer Surveys 28:21-31. [PubMed] [Google Scholar]
- 14.Masutani, C., R. Kusumoto, A. Yamada, N. Dohmae, M. Yokoi, M. Yuasa, M. Araki, S. Iwai, K. Takio, and F. Hanaoka. 1999. The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase η. Nature 399:700-704. [DOI] [PubMed] [Google Scholar]
- 15.Mendelman, L. V., J. Petruska, and M. F. Goodman. 1990. Base mispair extension kinetics. Comparison of DNA polymerase α and reverse transcriptase. J. Biol. Chem. 265:2338-2346. [PubMed] [Google Scholar]
- 16.Nelson, J. R., P. E. M. Gibbs, A. M. Nowicka, D. C. Hinkle, and C. W. Lawrence. 2000. Evidence for a second function for Saccharomyces cerevisiae Rev1p. Mol. Microbiol. 37:549-554. [DOI] [PubMed] [Google Scholar]
- 17.Prakash, S., and L. Prakash. 2002. Translesion DNA synthesis in eukaryotes: a one- or two-polymerase affair. Genes Dev. 16:1872-1883. [DOI] [PubMed] [Google Scholar]
- 18.Wang, Y.-C., V. M. Maher, D. L. Mitchell, and J. J. McCormick. 1993. Evidence from mutation spectra that the UV hypermutability of xeroderma pigmentosum variant cells reflects abnormal, error-prone replication on a template containing photoproducts. Mol. Cell. Biol. 13:4276-4283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Washington, M. T., R. E. Johnson, L. Prakash, and S. Prakash. 2001. Accuracy of lesion bypass by yeast and human DNA polymerase η. Proc. Natl. Acad. Sci. USA 98:8355-8360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Washington, M. T., R. E. Johnson, S. Prakash, and L. Prakash. 2000. Accuracy of thymine-thymine dimer bypass by Saccharomyces cerevisiae DNA polymerase η. Proc. Natl. Acad. Sci. USA 97:3094-3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Waters, H. L., S. Seetharam, M. M. Seidman, and K. H. Kraemer. 1993. Ultraviolet hypermutability of a shuttle vector propagated in xeroderma pigmentosum variant cells. J. Investig. Dermatol. 101:744-748. [DOI] [PubMed] [Google Scholar]
- 22.Yu, S.-L., R. E. Johnson, S. Prakash, and L. Prakash. 2001. Requirement of DNA polymerase η for error-free bypass of UV-induced CC and TC photoproducts. Mol. Cell. Biol. 21:185-188. [DOI] [PMC free article] [PubMed] [Google Scholar]