Abstract
Mutations in either the Drosophila melanogaster pelota or pelo gene or the Saccharomyces cerevisiae homologous gene, DOM34, cause defects of spermatogenesis and oogenesis in Drosophila, and delay of growth and failure of sporulation in yeast. These phenotypes suggest that pelota is required for normal progression of the mitotic and meiotic cell cycle. To determine the role of the pelota in mouse development and progression of cell cycle, we have established a targeted disruption of the mouse Pelo. Heterozygous animals are variable and fertile. Genotyping of the progeny of heterozygous intercrosses shows the absence of Pelo−/− pups and suggests an embryo-lethal phenotype. Histological analyses reveal that the homozygous Pelo deficient embryos fail to develop past day 7.5 of embryogenesis (E7.5). The failure of mitotic active inner cell mass of the Pelo−/− blastocysts to expand in growth after 4 days in culture and the survival of mitotic inactive trophoplast indicate that the lethality of Pelo-null embryos is due to defects in cell proliferation. Analysis of the cellular DNA content reveals the significant increase of aneuploid cells in Pelo−/− embryos at E7.5. Therefore, the percent increase of aneuploid cells at E7.5 may be directly responsible for the arrested development and suggests that Pelo is required for the maintenance of genomic stability.
The Pelo gene was originally identified in a mutagenesis screen for spermatogenesis-specific genes of Drosophila melanogaster (7). Spermatogenesis in pelo mutants progresses normally during the four mitotic divisions, and the 16 spermatocytes undergo a premeiotic S-phase and duplicate their DNA content. However, spermatocytes in the mutant arrested prior to full chromosome condensation, spindle pole organization, and nuclear breakdown. Metaphase and anaphase figures of the meiotic divisions, which are clearly recognized in squashed preparations of wild-type testis, were not observed in testis of the pelo mutant. Although meiotic division arrests in pelo spermatocytes, germ cell differentiation continues, resulting in 4N spermatids with head and tail structures. These results indicate that the Pelo is required for the meiotic division during the G2/M transition (10). Beside the effect of the mutation on spermatogenesis, the eyes of the pelo homozygotes are up to 30% smaller than those of wild-type siblings, and the ovaries in mutants are very small. In contrast to the arrest in meiotic divisions in male germ cells, during oogenesis the mitotic divisions are affected. The mitotic zone of ovaries appears disorganized and often contains degenerating cells. Despite the apparent phenotype observed in the gametogenesis of Drosophila mutant, the pelo transcripts are not restricted to germ cells but also detected in early embryonic development.
Analysis of mitotic and meiotic division in the dom34 mutant of Saccharomyces cerevisiae, which has a mutation in the pelota orthologous gene, reveals that the dom34 mutant exhibits a growth delay and fails to undergo sporulation properly (8). Introduction of the Drosophila wild-type pelota transgene into a dom34 mutant was found to result in substantial rescue of the dom34 growth and sporulation defects (10).
The pelota homologous gene has been cloned and sequenced in several species including archaebacteria, Arabidopsis thaliana, yeast, Drosophila, Caenorhabditis elegans, mouse, and human (8, 10, 22, 24, 25). The encoded protein ranges between 347 and 395 amino acids in length. Alignments of the pelota protein sequences reveal that the Pelo is highly conserved during evolution. The amino acid sequence of archaebacteria, yeast, Drosophila, A. thaliana, C. elegans, and human protein are 23, 36, 70, 51, 57, and 95% identical to murine Pelo, respectively. All the Pelo sequences contain a conserved nuclear localization signal and have the eEF1α-like domain at the carboxyl end (8). The eEF1α-like domain is present in several proteins such as the translation elongation factor eEF1α and the translation release factors eRF1 and eRF3 (11). It has been recently shown that the Hbs1 protein in the yeast. S. cerevisiae, which shares structural features with the eEF1α and eRF3 families, specifically interacts with the Dom34 (6). The human and mouse Pelo have been isolated and localized on chromosome 5q11 and chromosome 13, respectively. Expression analyses demonstrated that the Pelo is widely expressed in human and murine tissues as well as during embryonic development (24, 25).
In mammalian species, no mutation or depletion studies on Pelo have been described. In this study, we used the gene targeting method in embryonic stem cells to generate Pelo deficient mice. We describe a lethal phenotype for Pelo null mice and show that Pelo is required for normal progression of mitosis.
MATERIALS AND METHODS
Construction of Pelo targeting vector.
A genomic fragment containing the Pelo locus was previously cloned from a 129/Sv mouse genomic library and characterized (25). The 16-kb BamHI fragment was subcloned into pGEM 3Zf(+) for restriction mapping and construction of the targeting vector. The 3-kb EcoRI fragment containing the 3′-flanking region of the Pelo gene was isolated and ligated with the EcoRI-digested pPNT vector (29) to yield the clone Pelo1. The 4.8-kb XbaI-SpeI fragment containing the 5′-flanking region and exon 1 of the gene was isolated and inserted in the XhoI-digested clone Pelo1 by blunt-end ligation. The resulting 15-kb targeting vector was linearized with NotI and introduced into the RI embryonic stem (ES) cells by electroporation (32). Cells were plated into nonselective medium in the presence of G418-resistant embryonic mouse fibroblasts. Selection was applied 56 h later using medium containing G418 at 350 μg/ml and ganciclovir at 2 μM. Genomic DNA from individual drug-resistant clones was screened for homologous recombination by Southern blot analysis. DNA was digested with EcoRI, separated on a 0.8% agarose gel, and transferred to a nylon membrane. The 1.3-kb EcoRI-XbaI fragment laying externally 5′of the targeting vector (Fig. 1A) was radioactively labeled and used to probe the Southern blots. Hybridization was carried out at 65°C overnight in a solution containing the following: 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 5× Denhardt's solution, 0.1% sodium dodecyl sulfate, and denatured salmon sperm DNA (100 μg/ml). Filters were washed twice at 65°C to a final stringency of 0.2× SSC-0.1% sodium dodecyl sulfate. Cells from two recombinant ES clones were injected into day 3.5 C57BL/6J blastocysts, and these were transferred into DBA/BL6 pseudopregnant females (20). Germ line-transmitting chimeric males obtained from both lines were backcrossed to C57BL/6J and 129/Sv females. Progeny from all crosses were PCR genotyped using three primers. A single sense primer, F11 (5′-TGAGCCCAGACTGTACGTGAC-3′), was designed from exon 1 to amplify both wild-type and targeted loci. The first antisense primer, R12 (5′-TCTGCACCTTAGCGTGAAGCC-3′), was designed from intron 1 to amplify the wild-type locus. The second antisense primer, Pgk1 (5′-CCATTTGTCACGTCCTGCACG-3′), was designed from the Pgk promoter to amplify the targeted locus. Thermal cycling was carried out for 35 cycles, denaturation at 94°C for 30 s, annealing at 58°C for 1 min, and extension at 72°C for 1 min.
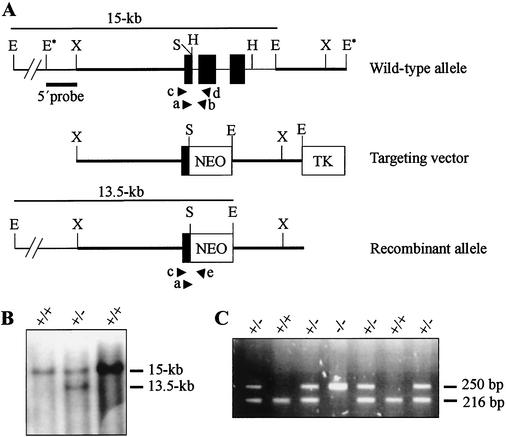
FIG. 1.
Targeted disruption of the Pelo gene. (A) The structures of the wild-type, targeting vector, and recombinant allele are shown together with the relevant restriction sites. The 3.2-kb SpeI-EcoRI fragment containing exons 2 and 3 of the gene was replaced by a pgk-neo selection cassette (NEO). The probe used and the predicted length of EcoRI restriction fragments in Southern blot analysis are shown. Primers F11 (a), R12 (b), F12 (c), R13 (d), and pgk1 (e), used to amplify the wild-type and mutant allele by PCR, are indicated. Abbreviations: TK, Thymidine kinase cassette; E, EcoRI; E*, EcoRI site from polylinker of phage clone; H, HindIII; S, SpeI; X, XbaI. (B) Southern blot analysis of the recombinant ES cell clones. Genomic DNA extracted from ES cell clones was digested with EcoRI and probed with the 5′probe shown in panel A. The wild-type Pelo allele generated a 15-kb EcoRI fragment, whereas the targeted allele yielded a 13.5-kb EcoRI fragment, as indicated in panel A. (C) Second-round PCR products from E3.5 embryos, which were derived from Pelo heterozygous intercrosses and electrophoresed on 2% agarose gel. PCR primers F11 and R12 detect the wild-type allele and generate a 216-bp band; primers F11 and pgk amplify a 250-bp fragment of the targeting allele.
Embryo dissection and histological analysis.
Timed matings were performed with Pelo+/− mice on a mixed genetic background (C57BL/J × 129/Sv). Females with copulation plugs were considered to be at day 0.5 of gestation. Pregnant females were sacrificed at different times of gestation, and the embryos were dissected free of maternal tissues, examined, photographed, and genotyped by PCR. For histological preparations, embryos in deciduae were fixed in 4% paraformaldehyde in phosphate-buffered saline, embedded in paraffin, sectioned at 6 μm, and stained with hematoxylin and eosin.
In vitro culture of E3.5 embryos.
Pelo+/− mice were mated and checked for copulation plugs. Embryos were flushed from the uteri of plugged females at day 3.5 and placed in gelatinized 96-well dishes. Embryos were cultured in ES cell medium without the addition of leukemia inhibitory factor and photographed every day. After culturing, DNA was prepared by incubation of individual embryos with 10 μl of lysis buffer (proteinase K [300 μg/ml], 100 mM KCl, 20 mM Tris at pH 8.0, 4 mM MgCl2, 0.9% NP-40, 0.9% Triton X-100) for 4 h at 60°C, and this was followed by incubation at 90°C for 30 min. Three microliters of embryonic DNA was used in the first round of a nested-PCR strategy. The first-round primers F12 (5′-TGCTCTCTCTGGGCTCTGTCC-3′), R13 (5′-TTCCCGGAACATCCTTGTGTG-3′), and Pgk1 were used in a duplex reaction, using the following conditions for the first round: 95°C for 2 min, 55°C for 60 s, 72°C for 90 s, in a final reaction volume of 25 μl. For the second round, 1 μl of the first-round amplified product was used in a reaction with primers F11, R12, and Pgk1. The second-round amplification was carried out as described for PCR genotyping of Pelo mutant mice.
Determination of DNA content.
For determination of DNA content in cells of embryos at E7.5, paraffin sections (5 μm thick) were placed in 5 N hydrochloric acid for 60 min, stained with Feulgen stain for 60 min using manufacturer's protocols (CAS DNA staining kit [cell analysis system]; Pharmingen-Becton Dickinson, Hamburg, Germany), rinsed in acid alcohol, cleaned in xylene, and marked under a phase-contrast microscope before CAS image analysis. The quantification of the DNA was based on assigning on optical density to each pixel of an image and summing the optical density values for each nucleus. Calibration was performed using rat hepatocytes and mouse lymphocytes. Cells with a DNA index of 0.83 to 1.22, 1.22 to 1.83, 1.83 to 2.22, 2.22 to 3.84, and 3.84 to 4.23 were defined according to the system of Auer et al. (1) as diploid (2N), aneuploid (2N to 4N), tetraploid (4N), aneuploid (4N to 8N), and octaploid (8N), respectively.
RESULTS
Targeted disruption of Pelo gene.
To disrupt the Pelo gene, a targeting vector was designed in which the 3.2-kb SpeI/EcoRI fragment (Fig. 1A) was deleted and replaced with the neomycin phosphotransferase (neo) gene under the control of the phosphoglycerate kinase promoter (Pgk). Homologous recombination at the Pelo locus resulted in the deletion of exons 2 and 3 (Fig. 1A). RI ES cells were transfected with the targeting vector and selected for homologous recombination events. Drug-resistant clones were selected, and DNA was isolated and screened by Southern blot analysis using the 5′external probe (Fig. 1A). Hybridization with the 5′external probe visualized a 15-kb EcoRI fragment for the wild-type allele and a 13.5-kb fragment for the targeted allele (Fig. 1B). Cells from three ES clones were injected into host blastocysts of the C57BL/J6 strain, and male chimeras were obtained for two of them. Two independent lines of heterozygous mice were generated in both a 129/Sv inbred and a C57BL/6J × 129/Sv mixed genetic background. The phenotype described below was consistently seen in both backgrounds.
Pelo mutation results in early embryonic lethality.
Heterozygous males and females displayed no overt phenotype and were fertile. Heterozygous animals were intercrossed, and the offspring were genotyped by PCR analysis. No viable Pelo−/− mice were identified among 240 mice. These results indicated that Pelo deficiency results in embryonic lethality.
To assess the consequences of the Pelo mutation for embryonic development, embryos were collected from heterozygous intercrosses at different days of gestation. None of 31 studied E8.5, E9.5 and E10.5 was Pelo−/− (Table 1). However, empty deciduae, which could account for the resorption of Pelo−/− embryos, were detected at E8.5. Resorbed embryos were also noticed at E9.5 and E10.5. To investigate whether the Pelo deficient embryos die during preimplantation stages, E2.5 and E3.5 were isolated and their genotypes were determined by PCR (Fig. 1C). At E2.5 and E3.5, Pelo−/− embryos were detected in the expected ratio as defined by Mendelian distribution (Table 1). These results suggest that Pelo−/− embryos die between E3.5 and E8.5.
TABLE 1.
Genotypes of offspring and embryos from heterozygous matings
| Stage | No. of progeny with genotype:
|
Total no. of progeny | ||
|---|---|---|---|---|
| +/+ | +/− | −/− | ||
| 3 wk old | 83 | 157 | 0 | 240 |
| Embryo, E8.5-E10.5 | 12 | 19 | 0 | 31 |
| Blastocyst, E3.5 | 10 | 27 | 11 | 48 |
| Morula, E2.5 | 8 | 13 | 6 | 27 |
| Blastocysts grown in culture | ||||
| Normal at E3.5+7 | 9 | 20 | 0 | 29 |
| ICM atrophied by E3.5+7 | 0 | 0 | 5 | 5 |
To determine the causes and time of embryonic lethality, deciduae at E6.5, E7.5, and E8.5 were dissected from uteri of heterozygous females mated to heterozygous males and characterized by light microscopy. The mean number of dissected 6.5-, 7.5-, and 8.5-day-old deciduae was not significantly different from that of the wild-type females mated with wild-type males. Furthermore, the dissected deciduae from heterozygous intercrosses showed no obvious morphological differences (data not shown). These results indicate that the implantation of Pelo deficient embryos is not affected. The embryos at E7.5 and E8.5 were isolated from deciduae, examined by light microscopy (Fig. 2A and B), and then genotyped by PCR. Approximately 28% of E7.5 (8 of 28) were scored as morphologically abnormal and genotyped as Pelo−/−. Embryos with normal morphology were genotyped as wild type or heterozygous. Mutant embryos were found to be smaller and developmentally retarded compared to their normal littermates (Fig. 2A and B). The boundary between the embryonic and the extraembryonic region could be distinguished, but both tissues were undersized. These observations indicate that Pelo−/− embryos have the capacity to generate some of the features of a gastrulated embryo despite impaired growth, and manifest their phenotype between days 6.5 and 7.5 of embryonic development.
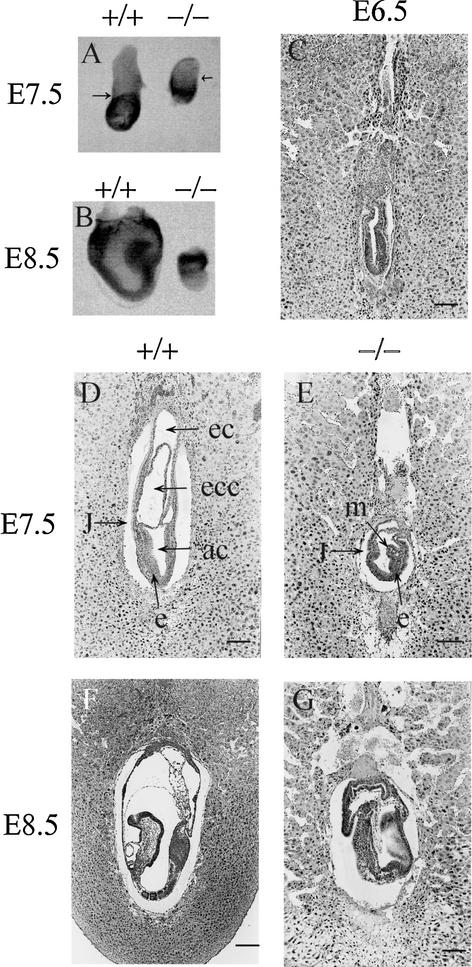
FIG. 2.
Morphological and histological analysis of embryos derived from heterozygous intercrosses. (A) Pelo−/− embryos at E7.5 are smaller than wild-type littermates. The arrows point to the junction between the embryonic and extraembryonic regions. (B) E8.5 Pelo−/− embryos have failed to progress and are starting to become resorbed. (C) Histological analysis of E6.5 derived from heterozygous intercrosses does not recognize developmental delay in Pelo−/− embryos. (D) E7.5 wild-type embryos clearly display the distinct germ layers as well as three cavities. (E) E7.5 Pelo−/− embryos are developmentally delayed and have a layer of columnar ectoderm lining the amniotic cavity and a layer of mesodermal cells. The junction between the embryonic and extraembryonic region is clearly seen in the E7.5 mutant embryos. (F) E8.5 wild-type embryo. (G) The E8.5 Pelo−/− embryo is less organized than its wild-type littermates. Abbreviations: ec, ectodermal cavity; ecc, exocoelomic cavity; ac, amniotic cavity; m, mesoderm; e, ectoderm; j, junction between embryonic and extraembryonic regions. (Bar: 120 μm [C to G] or 200 μm [F].)
To determine the lethality profile of Pelo−/− mutants, embryos from timed heterozygous intercross matings were histologically analyzed at different days of gestation. At E6.5, all 26 embryos examined from three pregnancies exhibited normal egg cylinder morphology and differentiation of the ectodermal and the endodermal cell layers (Fig. 2C). Twenty-one embryos from two pregnancies were analyzed at E7.5, and seven of them were much smaller than their littermates. The embryonic germ layers (ectoderm, mesoderm, and endoderm) could be histologically identified. However, the extraembryonic region was particularly reduced and lacked distinct exocoelomic and chorionic cavities (Fig. 2D and E). At E8.5, Pelo mutants were abnormally small (3 of 17) or were entirely resorbed (2 of 17). Unlike their normal littermates, which had developed head folds, a foregut, initial somites, and an extended allantois (Fig. 2F), Pelo−/− embryos did not progress further in development than that shown in E7.5 mutant embryos (Fig. 2G).
Pelota mutation affects proliferation of ICM of blastocysts in vitro.
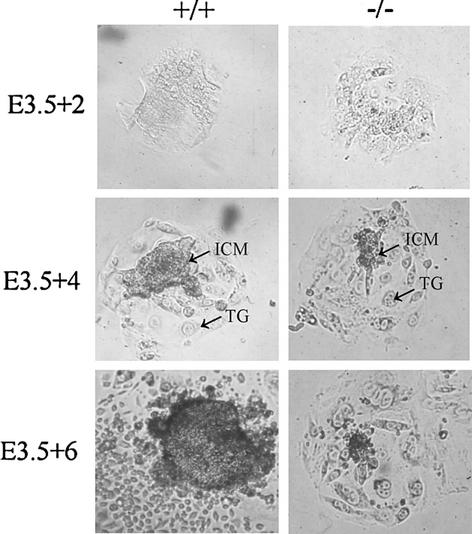
The fact that differentiation of the three germ layers is initiated in the Pelo−/− embryos suggests that the developmental arrest is not due to the lack of differentiation but rather results from a proliferation defect. To assay the growth and survival potential of the Pelo−/− embryos, E3.5 obtained from timed heterozygous intercrosses were cultured individually in 96 well plates for 7 days. The blastocyst outgrowth was monitored daily, and DNA of embryos was extracted on the last day of culture and genotyped by PCR assay. Twenty Pelo heterozygous, five Pelo homozygous, and nine wild-type embryos were assayed (Table 1). Pelo−/− blastocysts displayed no overt anomalies. After 2 days in culture, Pelo+/+, Pelo+/−, and Pelo−/− blastocysts successfully attached to culture dish, hatched from the zona pellucida and initiated growth (Fig. 3). Proliferating inner cell masses (ICMs) surrounded by trophoblast were observed in the three genotyped outgrowths during the first 3 days in culture; the ICM of Pelo−/− embryos was indistinguishable from that of Pelo+/− and Pelo+/+. However, while the ICM cells of Pelo+/+ and Pelo+/− embryos continued to expand throughout the 7-day culture period, Pelo−/− ICM cells failed to expand subsequent to day 4 and invariably died by day 6 in culture (Fig. 3). In contrast, Pelo−/− trophoblast cells remained attached to the culture dish and continued to grow in size through 7 days of culture. Trophoblast cells are derived from cells that become mitotically inactive at about E4.5, undergo repeated rounds of S-phase and, generate polyploid nuclei and a large cytoplasm (23). The impaired growth and the death of mitotic active ICM of Pelo−/− blastocysts and the survival of mitotic inactive trophoblast cells of Pelo−/− blastocysts indicate that Pelo is essential for the normal mitotic division and for early embryonic development in the mouse.
FIG. 3.
Growth of Pelo+/+ and Pelo−/− embryos in culture. Blastocysts (E3.5) derived from heterozygous intercrosses were cultured in vitro in 96-well plates for 7 days and photographed at the 2nd (3.5 + 2), 4th (E3.5 + 4), and 6th day (E3.3 + 6) of culture. TG, trophoplast giant cells.
Pelo is required for progression of the mitotic cell cycle.
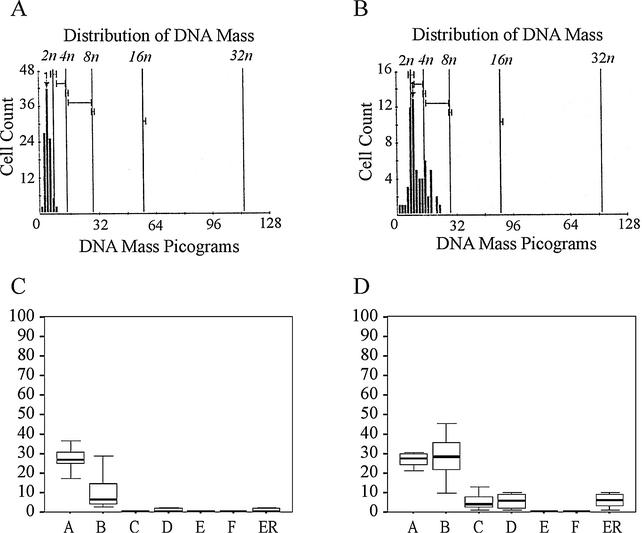
In Drosophila, the mitotic divisions appear to progress normally in pelo mutants. In contrast, the meiotic cell cycle of spermatogenic cells is arrested during the G2/M transition of the first meiotic division. The duplication of DNA during S phase of the cell cycle and the failure of chromosomes to segregate in the arrested cells during M phase were found to result in 4N spermatids (10). To look for evidence of DNA polyploidy in murine Pelo-deficient cells, DNA content in cells of histological sections from 8 developmentally impaired and 10 normal E7.5 embryos was measured with the CAS imaging system. To ensure a representative selection of the cells, DNA ploidy was evaluated in 50 to 150 nuclei from at least 10 different areas of the embryos. As shown in Fig. 4A, diploid DNA histogram patterns were observed in all wild-type embryos. In contrast, all the developmentally delayed embryos display an aneuploid and tetraploid DNA histogram (Fig. 4B). The percentage of the aneuploid (2N to 4N) (P < 0.001) and polyploid cells (>4N) (P < 0.05) in developmentally impaired embryos was significantly higher than that in the normal littermates. In contrast, there were no significant differences in percentage of the diploid cells between Pelo−/− and wild-type embryos (Fig. 4C and D). The significant increase of cells with aneuploid DNA content in Pelo mutant embryos indicates that Pelo deficiency lead to genomic instability.
FIG. 4.
Increase of polyploid cells in E7.5 mutant embryos. (A and B) DNA histogram profile of the cells in a wild-type (A) and a developmentally impaired (B) embryo. (C and D) Distribution of the cells with 2N (bar A), 2N to 4N (bar B), 4N (bar C), 4N to 8N (bar D), 8N (bar F), and >5N (bar ER) DNA content in normal (C) and abnormal embryos (D).
DISCUSSION
We describe here the generation of a null mutation of the Pelo gene by homologous recombination in the mouse. Heterozygous Pelo+/− mice show no apparent abnormalities in development or fertility, indicating that one functional copy of the gene is sufficient for normal development. Genotyping of the progeny of heterozygous intercrosses indicates the absence of Pelo−/− pups and suggests an embryo-lethal phenotype. Early embryonic failure can be detected past E7.5. The development of the three germ layers in the Pelo-deficient E7.5 embryos indicates that the lethality of the Pelo−/− embryos is not due to disruption of differentiation. The failure of mitotic ICM cells to extend and the survival of postmitotic trophoblast giant cells in blastocyst outgrowth experiments reveal that the murine Pelo is essential for some aspects of the mitotic cell cycle. Gastrulation in mouse embryo takes place at about day 6.5 of gestation and is characterized by a dramatic increase of cell number, which correlates with reduction of the cell cycle to as few as 4 to 6 h (27). The ability of the Pelo−/− embryos to undergo cell division in vitro and in vivo till the start of gastrulation at E6.5 suggests that Pelo deficient cells are intrinsically impaired by Pelo mutation. Therefore, the phenotype of Pelo−/− embryos become more apparent when the embryonic growth rate increases and the length of the cell cycle decreases. The rapid mitotic division at gastrulation requires regulatory mechanisms to ensure the correct ordering of cell cycle events or the repair of DNA damage. In the absence of the regulators, instability in the genome accumulate and progression through the cell cycle slows down or arrests and leads to embryonic lethality (12, 16). The other interpretation for survival of the Pelo−/− embryos until gastrulation might be that prior to day 6.5, Pelo is not essential or its function can be compensated by other proteins during early embryonic cell division.
The essential role of mouse Pelo in the mitotic division of embryonic development was unexpected, because mitotically dividing cells of dom34 mutant yeast and pelo mutant Drosophila were found to be variable albeit slower in growth of the mutant yeast relative to the wild type (8, 10). There is a possible explanation for this discrepancy. The mammalian Pelo might have maintained the same function in the progression of the cell cycle as the yeast and Drosophila homologous gene, but this function is essential in mitotic division of mammalian cells but not in that of yeast and Drosophila. Differences in the viability between mammalian and yeast cells, which have mutations in genes controlling the mitotic cell cycle, have been reported. The Chk1-, Mad2-, and Bub3-deficient mouse embryos die at an early developmental stage, while the corresponding mutants of yeast are viable under normal growth conditions (9, 13, 14, 15, 28, 31).
Studies of the Pelo Drosophila mutant demonstrated that the failure of spermatogenesis is due to the arrest of the first meiotic division during the G2/M transition and subsequently results in 4N spermatids. The increase in the percentage of cells exhibiting aneuploidy in the Pelo deficient embryos at E7.5 may be responsible for the arrested development of the Pelo−/− embryos. Such polyploid genomes may be the result of damage in the spindle assembly, disruption of centrosome structure or chromosomal missegregation, which is often correlated with a failure of complete cytokinesis (5, 30). It is interesting that the eEF1α is associated with the mitotic apparatus, particularly with centrosomes and other microtubule organizing centers and may play an important role in the microtubule nucleation (17, 21, 26). Mutation of the DSUP35 gene encoding the Drosophila homolog of the yeast eRF3 affects the meiotic spindle assembly and results in abnormal chromosome segregation during the meiotic divisions (2). With regard to the eEF1α-like domain in the Pelo protein and accumulation of aneuploid cells in the Pelo null embryos, it is likely that the Pelo is involved in the mitotic spindle structure and function. Early developmental arrest of the Pelo deficient embryos and subsequently the failure to establish a Pelo−/− cell line prevented us to define the cause of aneuploidy observed in cells of the mutant embryos. Therefore, it would be feasible to use conditional gene targeting for identification of the role of the Pelo in the mammalian mitotic and meiotic division.
Postimplantation lethality and failure of ICM to proliferate in vitro were reported for knockout mice, which lack genes controlling the mitotic cell cycle. However, the cause of proliferation arrest varied among these different mutant mice. Lacking of the checkpoint protein Mad2 and Bub3 results in chromosome missegregation (7, 11, 12). Extensive chromosomal fragmentation was observed in Atr-deficient cells (6). Aneuploid cells have also been associated with mutation in the Brca1 (33). Determination of the causes of the aneuploidy in the Pelo−/− cells would be assist to link the Pelo to one of the regulatory mechanisms that control the normal progression of the cell cycle.
Aneuploidy is the most common genetic aberration in different types of cancer (18). It is interesting that disruption of the Pelo gene led to a small increase in the incidence of large benign tumor in our heterozygous animals. The particular increase of benign tumor suggests that the deficiency of Pelo has an effect on the rate of tumor initiation but not on the rate of progression to malignancy. Chromosomal localization of the human PELO is to 5q11 (24). Loss of heterozygosity around this region has been associated with different tumor types (3, 19). Further mutational analysis of PELO in these cancer types is under investigation.
Acknowledgments
We acknowledge the assistance of M. Schindler, H. Riedesel, and S. Wolf with the generation and breeding of knockout mice and H.-G. Sydow, I. Paprotta, and A. Winkler for excellent technical assistance and secretarial help.
This work was supported by Deutsche Forschungsgemeinschaft to W.E. (SFB 271).
REFERENCES
- 1.Auer, G. U., T. O. Caspersson, and A. S. Wallgren. 1980. DNA content and survival in mammary carcinoma. Anal. Quant. Cytol. 2:161-165. [PubMed] [Google Scholar]
- 2.Basu, J., B. C. Williams, Z. Li, E. V. Williams, and M. L. Goldberg. 1998. Depletion of a Drosophila homolog of yeast Sup35p disrupts spindle assembly, chromosome segregation, and cytokinesis during male meiosis. Cell Motil. Cytoskeleton 39:286-302. [DOI] [PubMed] [Google Scholar]
- 3.Benachenhou, N., S. Guiral, I. Gorska-Flipot, D. Labuda, and D. Sinnett. 1999. Frequent loss of heterozygosity at the DNA mismatch-repair loci hMLH1 and hMSH3 in sporadic breast cancer. Br. J. Cancer 79:1012-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown, E. J., and D. Baltimore. 2000. ATR disruption leads to chromosomal fragmentation and early embryonic lethality. Genes Dev. 14:397-402. [PMC free article] [PubMed] [Google Scholar]
- 5.Bunz, F., A. Dutriaux, C. Lengauer, T. Waldman, S. Zhou, J. P. Brown, J. M. Sedivy, K. W. Kinzler, and B. Vogelstein. 1998. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science 282:1497-1501. [DOI] [PubMed] [Google Scholar]
- 6.Carr-Schmid, A., C. Pfund, E. A. Craig, and T. G. Kinzy. 2002. Novel G-protein complex whose requirement is linked to the translational status of the cell. Mol. Cell. Biol. 22:2564-2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Castrillon, D. H., P. Gönczy, S. Alexander, R. Rawson, C. G. Eberhart, S. Viswanathan, S. DiNardo, and S. A. Wasserman. 1993. Toward a molecular genetic analysis of spermatogenesis in Drosophila melanogaster: characterization of male-sterile mutant generated by single E-element mutagenesis. Genetics 135:489-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davis, L., and J. Engebrecht. 1998. Yeast dom34 mutants are defective in multiple developmental pathways and exhibit decreased levels of polyribosomes. Genetics 149:45-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dobles, M., V. Liberal, M. L. Scott, R. Benezra, and P. K. Sorger. 2000. Chromosome missegregation and apoptosis in mice lacking the mitotic checkpoint protein Mad2. Cell 101:635-645. [DOI] [PubMed] [Google Scholar]
- 10.Eberhart, C. G., and S. A. Wasserman. 1995. The pelota locus encodes a protein required for meiotic cell division: an analysis of G2/M arrest in Drosophila spermatogenesis. Development 121:3477-3486. [DOI] [PubMed] [Google Scholar]
- 11.Frolova, L., X. Le Goff, H. H. Rasmussen, S. Cheperegin, G. Drugeon, M. Kress, I. Arman, A. L. Haenni, J. E. Celis, M. Philipp, J. Justesen, L. Kisselev. 1994. A highly conserved eukaryotic protein family possessing properties of polypeptide chain release factor. Nature 372:701-703. [DOI] [PubMed] [Google Scholar]
- 12.Hartwell, L. H., and T. A. Weinert. 1989. Checkpoints: controls that ensure the order of cell cycle events. Science 246:629-635. [DOI] [PubMed] [Google Scholar]
- 13.He, X., T. E. Patterson, and S. Sazer. 1997. The Schizosaccharomyces pombe spindle checkpoint protein mad2p blocks anaphase and genetically interacts with the anaphase-promoting complex. Proc. Natl. Acad. Sci. USA 94:7965-7970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoyt, M. A., I. Totis, and B. T. Roberts. 1991. S. Cerevisiae gene required for cell cycle arrest in response to loss of microtubule function. Cell 66:507-517. [DOI] [PubMed] [Google Scholar]
- 15.Kalitsis, P., E. Earle, K. J. Fowler, and K. H. Choo. 2000. Bub3 gene disruption in mice reveals essential mitotic spindle checkpoint function during early embryogenesis. Genes Dev. 14:2277-2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kinzler, K. W., and B. Vogelstein. 1997. Cancer-susceptibility genes. Gatekeepers and caretakers. Nature 386:761-763. [DOI] [PubMed] [Google Scholar]
- 17.Kuriyama, R., P. Savereide, P. Lefebvre, and S. Dasgupta. 1990. The predicted amino acid sequence of a centrosphere protein in dividing sea urchin eggs is similar to elongation factor (EF-1α). J. Cell Sci. 95:231-236. [DOI] [PubMed] [Google Scholar]
- 18.Lengauer, C., K. Kinzler, and B. Vogelstein. 1997. Genetic instability in colorectal cancers. Nature 386:623-627. [DOI] [PubMed] [Google Scholar]
- 19.Murty, V. V., V. E. Reuter, G. J. Bosl, and R. S. Chaganti. 1996. Deletion mapping identifies loss of heterozygosity at 5q15.1-15.2,5q11 and 5q34-35 in human male germ cell tumors. Oncogene 12:2719-2723. [PubMed] [Google Scholar]
- 20.Nagy, A., and J. Rossant. 1993. Production of completely ES cell-derived fetuses, p. 147-179. In A. L. Joyner (ed.), Gene targeting: a practical approach. IRL Press, Oxford, England.
- 21.Ohta, K., M. Toriyama, M. Miyazaki, M. Murofushi, S. Hosoda, S. Endo, and H. Sakai. 1990. The mitotic apparatus-associated 51-kDa protein from sea urchin eggs is a GTP-binding protein and is immunologically related to yeast polypeptide elongation factor 1α. J. Biol. Chem. 265:3240-3247. [PubMed] [Google Scholar]
- 22.Ragan, M. A., J. M. Logsdon, C. W. Sensen, R. L. Charlebois, and W. F. Doolittle. 1996. An archaebacterial homolog of pelota, a meiotic cell division protein in eukaryotes. FEMS Microbiol. Lett. 144:151-155. [DOI] [PubMed] [Google Scholar]
- 23.Rugh, R. 1990. The mouse, p. 44-101. Oxford University Press, London, England.
- 24.Shamsadin, R., I. M. Adham, G. von Beust, and W. Engel. 2000. Molecular cloning, expression and chromosomal localization of the human pelota gene PELO. Cytogenet. Cell Genet. 90:75-78. [DOI] [PubMed] [Google Scholar]
- 25.Shamsadin, R., I. M. Adham, and W. Engel. 2002. Mouse pelota gene (Pelo): cDNA cloning, genomic structure, and chromosomal localization. Cytogenet. Genome Res. 97:95-99. [DOI] [PubMed] [Google Scholar]
- 26.Shiina, N., Y. Gotoh, N. Kubomura, A. Iwamatsu, and E. Nishida. 1994. Microtubule severing by elongation factor 1α. Science 266:282-285. [DOI] [PubMed] [Google Scholar]
- 27.Snow, M. 1977. Gastrulation in the mouse: growth and regionalization of the epiblast. J. Embryol. Exp. Morphol. 42:293-303. [Google Scholar]
- 28.Takai, H., K. Tominaga, N. Motoyama, Y. A. Minamishima, H. Nagahama, T. Tsukiyama, K. Ikeda, K. Nakayama, M. Nakanishi, and K. Nakayama. 2000. Aberrant cell cycle checkpoint function and early embryonic death in Chk1−/− mice. Genes Dev. 14:1439-1447. [PMC free article] [PubMed] [Google Scholar]
- 29.Tybulewicz, V. L., C. E. Crawford, P. T. Bronson, and R. C. Mulligan. 1991. Neonatal lethality and lymphopenia in mice with a homozygous disruption of the c-abl protooncogene. Cell 65:1153-1163. [DOI] [PubMed] [Google Scholar]
- 30.Waldman, T., C. Lenghauer, K. W. Kinzler, and B. Vogelstein. 1996. Uncoupling of the S phase and mitosis induced by anticancer agents in cell lacking p21. Nature 381:713-716. [DOI] [PubMed] [Google Scholar]
- 31.Walworth, N., S. Davey, and D. Beach. 1993. Fission yeast chk1 protein kinase links the rad checkpoint pathway to cdc2. Nature 363:368-371. [DOI] [PubMed] [Google Scholar]
- 32.Wurst, W., and A. L. Joyner. 1993. Production of targeted embryonic stem cell clones, p. 33-61. In A. L. Joyner (ed.), Gene targeting: a practical approach. IRL Press, Oxford, England.
- 33.Xu, X., Z. Weaver, S. P. Linke, C. Li, J. Gotay, X.-W. Wang, C. C. Harris, T. Ried, and C.-X. Deng. 1999. Centrosome amplification and a defective G2-M cell cycle checkpoint induce genetics instability in BRCA1 exon 11 isoform-deficient cells. Mol. Cell 3:389-395. [DOI] [PubMed] [Google Scholar]