Abstract
The Wnt/β-catenin signaling pathway regulates many developmental processes by modulating gene expression. Wnt signaling induces the stabilization of cytosolic β-catenin, which then associates with lymphoid enhancer factor and T-cell factor (LEF-1/TCF) to form a transcription complex that activates Wnt target genes. Previously, we have shown that a specific mitogen-activated protein (MAP) kinase pathway involving the MAP kinase kinase kinase TAK1 and MAP kinase-related Nemo-like kinase (NLK) suppresses Wnt signaling. In this study, we investigated the relationships among NLK, β-catenin, and LEF-1/TCF. We found that NLK interacts directly with LEF-1/TCF and indirectly with β-catenin via LEF-1/TCF to form a complex. NLK phosphorylates LEF-1/TCF on two serine/threonine residues located in its central region. Mutation of both residues to alanine enhanced LEF-1 transcriptional activity and rendered it resistant to inhibition by NLK. Phosphorylation of TCF-4 by NLK inhibited DNA binding by the β-catenin-TCF-4 complex. However, this inhibition was abrogated when a mutant form of TCF-4 was used in which both threonines were replaced with valines. These results suggest that NLK phosphorylation on these sites contributes to the down-regulation of LEF-1/TCF transcriptional activity.
The Wnt family of secretory glycoproteins constitutes a major family of developmentally important signaling molecules that play important roles in embryonic induction, generation of cell polarity, and specification of cell fate (5, 17, 20). Intensive studies of Wnt signaling in Drosophila, Xenopus, and mammalian cells have provided a general understanding of the molecular machinery of this canonical signaling pathway. According to the current view, a cytoplasmic complex containing glycogen synthase kinase 3β (GSK-3β), the adenomatous polyposis coli protein, and axin catalyzes the phosphorylation of the transcriptional coactivator β-catenin in the absence of Wnt signaling. Phosphorylation of β-catenin targets it for ubiquitination and subsequent degradation by a proteasome pathway, which reduces the levels of free cytoplasmic β-catenin. Activation of the Wnt pathway occurs via stimulation of the Frizzled receptors, which then inhibit GSK-3β by an unknown mechanism. Inhibition of GSK-3β prevents β-catenin phosphorylation and its subsequent ubiquitination and results in the accumulation of cytoplasmic β-catenin. β-Catenin then translocates into the nucleus, where it forms a complex with the HMG box class of transcription factors, including lymphoid enhancer factor 1 (LEF-1) and T-cell factor (TCF), and activates the transcription of its target genes (5, 17, 20). In this complex, LEF-1/TCF provides the DNA-binding domain while β-catenin contributes the transactivation domain, allowing the activation of LEF-1/TCF target genes.
The Wnt pathway is strikingly conserved in different species. In Caenorhabditis elegans, Wnt signaling is involved in specifying the differences in cell fate between sister cells generated from the anterior-posterior division in the early embryo (22, 30). Establishment of these anterior-posterior asymmetries correlates with the down-regulation of the activity or level of POP-1, a protein related to vertebrate LEF-1/TCF transcription factors. Several mutations that result in the loss of polarity and POP-1 asymmetry have been shown to map to genes encoding molecules with similarities to known Wnt signaling components (15, 16, 21, 22, 30). For example, establishment of POP-1 asymmetry requires activation of MOM-5, a Frizzled-related Wnt receptor, and MOM-2, a Wnt factor. These factors appear to act through a β-catenin-related protein, WRM-1. However, there is one important difference between the roles of β-catenin and WRM-1: in mammalian cells, β-catenin enters the nucleus in response to signaling, where it binds to and activates LEF-1/TCF proteins (5, 17, 20), whereas in C. elegans, WRM-1 down-regulates, rather than activates, POP-1 (16, 21, 22, 30).
Insights into POP-1 regulation by WRM-1 have come from the analysis of the lit-1 and mom-4 genes. Mutations in lit-1 or mom-4 cause a loss of polarity and POP-1 asymmetry (16, 22, 27). The lit-1 gene encodes a mitogen-activated protein kinase (MAPK)-like protein similar to the Drosophila Nemo protein kinase and the mouse Nemo-like kinase (NLK). The mom-4 gene encodes a MAPK kinase kinase-like protein related to the mammalian protein kinase TAK1. Thus, a MAPK-related pathway acts in concert with Wnt signaling to establish anterior-posterior polarity in C. elegans. LIT-1 interacts directly with WRM-1, and this complex can phosphorylate POP-1 in a LIT-1 kinase domain-dependent manner, indicating that these pathways converge at WRM-1 (22). Furthermore, MOM-4 can activate kinase activity of the LIT-1-WRM-1 complex (27). These results suggest that Wnt signaling and the MAPK-related cascade both act to control polarity by regulating POP-1 and that integration of two signals occurs at the level of complex formation between WRM-1 and LIT-1.
Recent evidence indicates that the Wnt signaling pathway in mammalian cells is also regulated by a MAPK-related pathway composed of MAPK kinase kinase TAK1 and MAPK-related NLK (13). TAK1 functions upstream of NLK and enhances NLK kinase activity. Active NLK phosphorylates LEF-1/TCF and prevents the β-catenin-TCF complex from binding to DNA, thereby inhibiting the ability of β-catenin-TCF to activate transcription. Thus, TAK1 and NLK are negative regulators of LEF-1/TCF activity. Consistent with this, ectopic expression of NLK blocks double-axis formation in Xenopus induced by overexpression of β-catenin but not by the downstream components siamois and twin (13). These results suggest that TAK1 and NLK act in a pathway parallel to the Wnt pathway. Thus, TAK1-NLK and MOM-4-LIT-1 appear to function analogously to regulate Wnt signaling pathways in mammalian cells and C. elegans, respectively.
In C. elegans, WRM-1 and LIT-1 appear to form a stable protein complex in vivo (22). WRM-1 activates the LIT-1 protein kinase, leading to phosphorylation of WRM-1, LIT-1, and POP-1. On the other hand, in mammalian cells, the relationships among NLK, β-catenin, and LEF-1/TCF are not well understood. In the present report, we show that NLK directly interacts with and phosphorylates LEF-1/TCF. β-Catenin forms a complex with NLK in a LEF-1/TCF-dependent manner. We identify two NLK phosphorylation sites in LEF-1/TCF and present data suggesting that phosphorylation of these sites contributes to the down-regulation of LEF-1/TCF transcriptional activity.
MATERIALS AND METHODS
Cell culture and transfection.
Human embryonic kidney 293 and HeLa cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum. 293 cells in 100-mm-diameter plates were transfected with the expression plasmids (10 μg) by calcium phosphate precipitation.
Yeast two-hybrid analysis.
For interaction of two proteins in the yeast two-hybrid system, full-length LEF-1 and full-length NLK were fused to the Gal4 DNA-binding domain and the Gal4 transactivation domain, respectively. These constructs were transformed into yeast strain PJ69-4A, and protein-protein interaction was monitored by growth on selective plates.
Reporter gene assays.
HeLa cells (1.6 × 105/well) were seeded into six-well (35-mm-diameter) plates. Cells were transfected by the calcium phosphate precipitate method at 24 h after seeding with the TOPFLASH reporter gene plasmid along with each expression vector as indicated. The total DNA concentration (1.7 μg/ml) was kept constant by supplementation with empty vector DNAs. Luciferase activity was determined with the Dual-Luciferase Reporter Assay System (Promega). Renilla reniformis luciferase vector (0.05 μg) under the control of the EF-1α promoter was used to normalize transfection efficiencies.
Immunoprecipitation.
The immunoprecipitates and aliquots of total lysates were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to polyvinylidene difluoride membranes (Hybond-P; Amersham). The membranes were immunoblotted with antibodies, and bound antibodies were visualized with horseradish peroxidase-conjugated antibodies to mouse immunoglobulin G by using the enhanced chemiluminescence (ECL) Western blotting system (Amersham).
In vitro kinase assays.
Aliquots of immunoprecipitates were incubated in 10 μl of kinase buffer containing 10 mM HEPES (pH 7.4), 1 mM dithiothreitol, 5 mM MgCl2, and 5 μCi of [γ-32P]ATP at 25°C for 2 min. Samples were resolved by SDS-PAGE, and phosphorylated proteins were visualized by autoradiography.
Gel retardation assays.
As the optimal TCF probe, we used a double-stranded 56-nucleotide oligomer containing three potential LEF-1/TCF-binding sites derived from TOPFLASH. Binding reactions were performed at room temperature for 15 min by incubating 7.5 μl of nuclear extract mixtures and 0.0525 pmol of labeled oligonucleotides in 15 μl of binding buffer [10 mM Tris (pH 7.5), 50 mM NaCl, 1 mM MgCl2, 0.5 mM EDTA, 0.5 mM dithiothreitol, 4% glycerol, 3 μg of poly(dI-dC), 150 μg of bovine serum albumin].
RESULTS
NLK directly interacts with LEF-1/TCF but not with β-catenin.
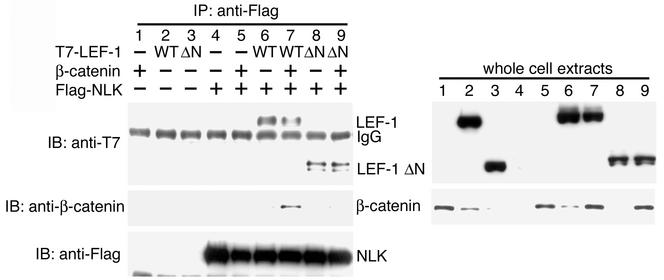
In C. elegans, WRM-1 (β-catenin) directly interacts with and activates LIT-1 (NLK). Active LIT-1 then phosphorylates WRM-1 and POP-1 (LEF-1/TCF) (22). We have previously shown that NLK interacts with the β-catenin-LEF-1/TCF complex (13). To examine whether NLK regulates the β-catenin-LEF-1/TCF complex in mammalian cells in a manner similar to that in C. elegans, we asked if NLK could associate with β-catenin or LEF-1/TCF. Human embryonic kidney 293 cells were cotransfected with various combinations of expression vectors encoding Flag-tagged NLK (Flag-NLK), T7-tagged LEF-1 (T7-LEF-1), and/or β-catenin (Fig. 1). Cell extracts were immunoprecipitated with a monoclonal antibody to Flag, and coprecipitated T7-LEF-1 or β-catenin was detected by immunoblotting with a monoclonal antibody to T7 or anti-β-catenin, respectively. LEF-1 was found to associate with NLK (Fig. 1, top panel, lane 6). In contrast, NLK association with β-catenin was barely detectable (Fig. 1, middle panel, lane 5). We next asked if the presence of LEF-1 could affect the interaction between NLK and β-catenin. Flag-NLK and β-catenin were coexpressed with T7-LEF-1. We observed that coexpression of LEF-1 dramatically increased the interaction between β-catenin and NLK (Fig. 1, middle panel, lane 7). These results suggest that LEF-1 binds directly to NLK, while β-catenin interacts indirectly with NLK via binding to LEF-1. The binding of NLK to LEF-1 was also detected by using a yeast two-hybrid system (data not shown), supporting the idea that the interaction between these two proteins is direct.
FIG. 1.
Association of NLK with β-catenin and LEF-1. 293 cells were transfected with the indicated expression vectors. Cell lysates were immunoprecipitated (IP) with anti-Flag antibody. (Left) Immunoprecipitates were immunoblotted (IB) with anti-T7 antibody (top panel), anti-β-catenin antibody (middle panel), and anti-Flag antibody (bottom panel). (Right) Expression of LEF-1 and β-catenin was monitored with anti-T7 and anti-β-catenin antibodies, respectively. IgG, immunoglobulin G.
β-Catenin binds to a region within the NH2-terminal region of LEF-1/TCF (see Fig. 3A) (2). A small deletion removing the NH2-terminal 62 amino acids of LEF-1 (LEF-1ΔN) abrogated its interaction with β-catenin (data not shown) (2). This deletion mutant allowed us to test the possibility that the interaction of NLK with β-catenin is mediated via LEF-1 by examining the effect of LEF-1ΔN on the interaction between NLK and β-catenin (Fig. 1). We observed that Flag-NLK and T7-LEF-1ΔN could interact when coexpressed (Fig. 1, top panel, lane 8), indicating that the NH2-terminal deletion of LEF-1 does not affect its association with NLK. However, coexpression of LEF-1ΔN did not enhance the association between NLK and β-catenin (Fig. 1, middle panel, lane 9), as had been observed with wild-type LEF-1. These results suggest that β-catenin is recruited to the NLK-LEF-1 complex via interaction with the NH2 terminus of LEF-1.
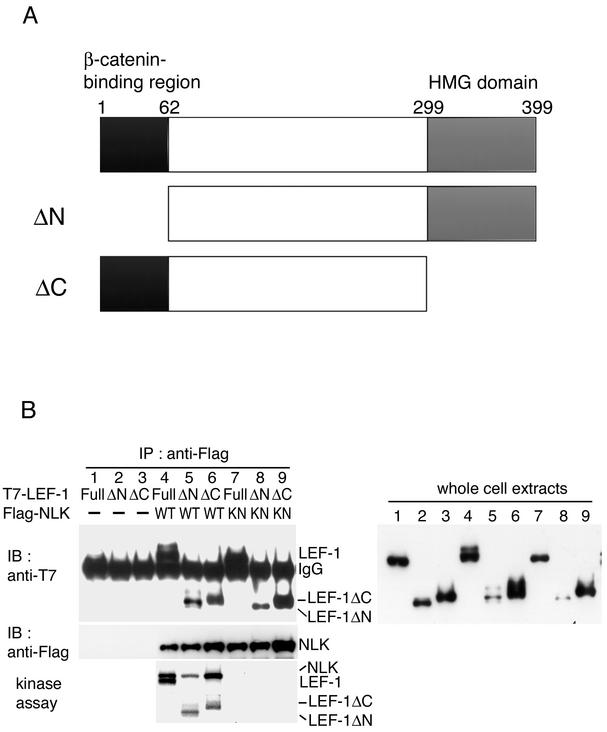
FIG. 3.
Association of NLK with truncated mutant forms of LEF-1. (A) Schematic diagram of the structure of truncated mutant forms of LEF-1. (B) Association of NLK with truncated mutant forms of LEF-1. 293 cells were transfected with the indicated expression vectors. Cell lysates were immunoprecipitated (IP) with anti-Flag antibody. (Left) Immunoprecipitates were immunoblotted (IB) with anti-T7 antibody (top panel) and anti-Flag antibody (middle panel). Immunoprecipitated complexes were also incubated with [γ-32P]ATP and analyzed by autoradiography (bottom panel). (Right) Expression of LEF-1 was monitored with anti-T7 antibody. IgG, immunoglobulin G.
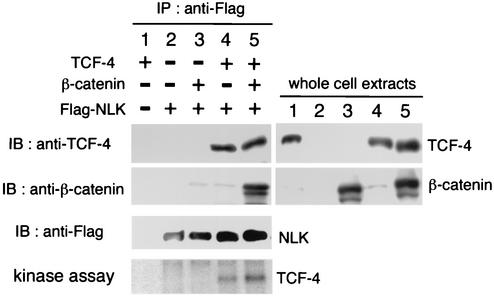
We have previously shown that NLK associates with TCF-4 (13). We confirmed this interaction by coexpressing Flag-NLK and TCF-4 in 293 cells (Fig. 2, top panel, lane 4). We further examined whether the association of β-catenin with NLK is affected by expression of TCF-4. 293 cells were cotransfected with Flag-NLK and β-catenin either with or without TCF-4. We observed that, in the absence of exogenous TCF-4, little β-catenin was detected in the NLK immunocomplexes precipitated with the anti-Flag antibody (Fig. 2, second panel, lane 3). However, when TCF-4 was coexpressed, the amount of β-catenin detected in the immunocomplexes increased (Fig. 2, second panel, lane 5). This supports the possibility that LEF-1/TCF forms a bridge between β-catenin and NLK.
FIG. 2.
Interaction of NLK with TCF-4. 293 cells were transfected with the indicated expression vectors. Cell lysates were immunoprecipitated (IP) with anti-Flag antibody. (Left) Immunoprecipitates were immunoblotted (IB) with anti-TCF-4 antibody (top panel), anti-β-catenin antibody (second panel), and anti-Flag antibody (third panel). Immunoprecipitated complexes were also incubated with [γ-32P]ATP and analyzed by autoradiography (bottom panel). (Right) Expression of TCF-4 and β-catenin was monitored with anti-TCF-4 and anti-β-catenin antibodies, respectively.
To determine if β-catenin binding has any effect on NLK activity, we analyzed the ability of NLK to phosphorylate TCF-4 by in vitro kinase assay (Fig. 2). 293 cells were cotransfected with Flag-NLK and TCF-4 in the absence or presence of β-catenin. Cells were then subjected to immunoprecipitation with anti-Flag antibody, and the precipitates were incubated with [γ-32P]ATP. As observed previously (13), NLK phosphorylated TCF-4 in vitro even in the absence of β-catenin transfection (Fig. 2, bottom panel, lane 4). However, cotransfection of β-catenin enhanced NLK phosphorylation of TCF-4 (Fig. 2, bottom panel, lane 5). This suggests that NLK may phosphorylate the β-catenin-TCF-4 complex more efficiently than TCF-4. In a parallel experiment, LEF-1 was not observed to cause such enhancement (data not shown).
Identification of NLK phosphorylation sites in LEF-1/TCF.
The LEF-1 protein contains a COOH-terminal HMG DNA-binding domain and an NH2-terminal β-catenin-binding domain (Fig. 3A). Although LEF-1/TCF family members share overall sequence similarity, these two regions are particularly conserved (18). In order to define the domain in the LEF-1 protein that mediates its interaction with NLK, we constructed truncation mutant forms of LEF-1 lacking either the NH2-terminal β-catenin-binding domain (LEF-1ΔN) or the COOH-terminal HMG DNA-binding domain (LEF-1ΔC), both tagged with the T7 epitope (Fig. 3A). We tested their ability to interact with Flag-NLK by immunoprecipitation, followed by immunoblotting (Fig. 3B). In this assay, NLK was found to associate with both LEF-1ΔN and LEF-1ΔC (Fig. 3, top panel, lanes 5 and 6). This result indicates that the middle region of LEF-1 contributes to NLK interaction. A kinase-negative form of NLK, NLK(K155M), also interacted with LEF-1, LEF-1ΔN, and LEF-1ΔC (Fig. 3, top panel, lanes 7 to 9).
To determine whether NLK can phosphorylate LEF-1ΔN and/or LEF-1ΔC, we analyzed the NLK immunoprecipitates for LEF-1 phosphorylation. As shown in Fig. 3B, NLK phosphorylated both LEF-1ΔN and LEF-1ΔC in vitro (Fig. 3, bottom panel, lanes 5 and 6) and this phosphorylation was dependent on the kinase activity of NLK (Fig. 3, bottom panel, lanes 8 and 9). These results suggest that the NLK phosphorylation sites are also located in the middle region between the β-catenin-binding domain and the DNA-binding domain of LEF-1.
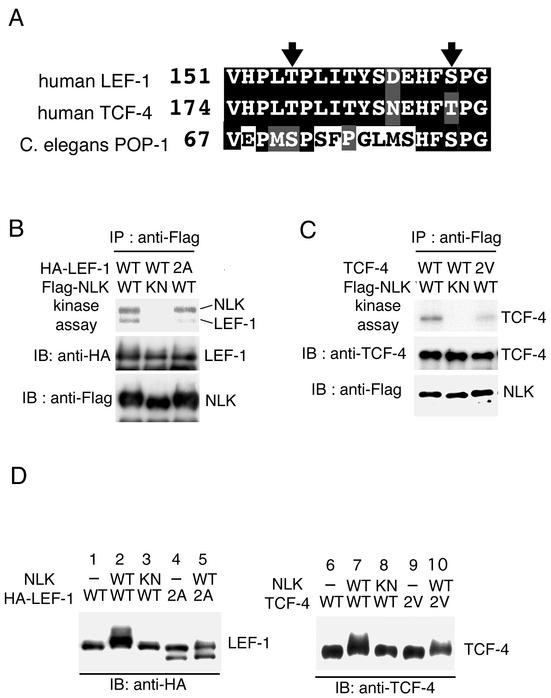
S/TP motifs are consensus sites for MAPK phosphorylation (31). Examination of the primary sequences of the LEF-1/TCF middle regions revealed two potential NLK phosphorylation sites that are conserved among human LEF-1, human TCF-4, and C. elegans POP-1 (Fig. 4A). To test whether these residues are indeed the sites of NLK phosphorylation, we altered them by site-directed mutagenesis. We constructed a mutant form of LEF-1 [LEF-1(2A)] in which Thr-155 and Ser-166 were replaced with alanines and examined whether this mutated LEF-1(2A) protein is phosphorylated by NLK in vitro (Fig. 4B). Extracts prepared from 293 cells expressing wild-type HA-LEF-1 or HA-LEF-1(2A) were mixed with extracts prepared from 293 cells expressing Flag-NLK or kinase-negative Flag-NLK(K155M). Mixed extracts were immunoprecipitated with anti-Flag antibody and immunoblotted with anti-HA antibody. Precipitation of LEF-1(2A) with NLK was normal (Fig. 4B, middle panel, lane 3), suggesting that the LEF-1(2A) mutation does not affect its interaction with NLK in vitro. We next performed kinase assays by using immunoprecipitated Flag-NLK or Flag-NLK(K155M) from mixed lysates. Wild-type LEF-1 was efficiently phosphorylated by active but not inactive NLK (Fig. 4B, top panel, lanes 1 and 2). On the other hand, the mutated LEF-1(2A) protein was only weakly phosphorylated by NLK (Fig. 4B, top panel, lane 3). These results suggest that NLK catalyzes the phosphorylation of Thr-155 and Ser-166 in LEF-1 in vitro.
FIG. 4.
NLK phosphorylation sites in LEF-1 and TCF-4. (A) Comparison of amino acid sequences. The amino acid residues in the region of the putative NLK phosphorylation sites are aligned. Arrows indicate the putative NLK phosphorylation sites. (B and C) NLK phosphorylation of LEF-1 and TCF-4 in vitro. 293 cells were separately transfected with wild-type (WT) HA-LEF-1, HA-LEF-1(2A), wild-type TCF-4, TCF-4(2V), wild-type Flag-NLK, or Flag-NLK(K155M) (KN). Cell lysates expressing Flag-NLK were mixed with those expressing LEF-1 or TCF-4 as indicated. Mixtures of cell lysates were immunoprecipitated (IP) with anti-Flag antibody. Immunoprecipitated complexes were incubated with [γ-32P]ATP and analyzed by autoradiography (top panels). Immunoprecipitates were immunoblotted (IB) with anti-HA antibody (B) or anti-TCF-4 antibody (C) (middle panels) and anti-Flag antibody (bottom panels). (D) NLK phosphorylation of LEF-1 and TCF-4 in vivo. 293 cells were transfected with the indicated expression vectors. Cell lysates were immunoblotted with anti-HA antibody (left panel) and anti-TCF-4 antibody (right panel).
We next investigated whether these NLK phosphorylation sites are functionally conserved in TCF-4. The corresponding sites in TCF-4 (Thr-178 and Thr-189) were mutated to valines. The mutant form of TCF-4, designated TCF-4(2V), was analyzed for NLK-mediated phosphorylation by immune complex kinase assay as described above. In vitro kinase reactions showed that phosphorylation of TCF-4(2V) by NLK was greatly decreased in comparison with that of wild-type TCF-4 (Fig. 4C, top panel, lanes 1 and 3). This suggests that NLK phosphorylates at least the Thr-178 and Thr-189 sites of TCF-4 in vitro. The fact that some weak phosphorylation was still observed with the LEF-1(2A) and TCF-4(2V) mutant forms suggests that LEF-1 and TCF-4 may contain additional NLK phosphorylation sites.
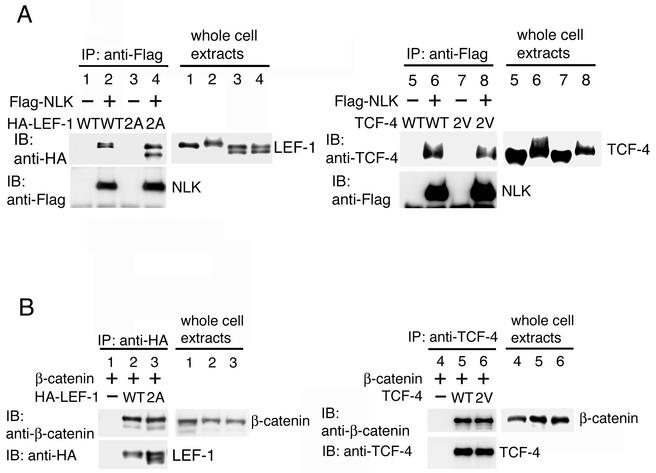
NLK-catalyzed in vivo phosphorylation of LEF-1 at Thr-155 and Ser-166 and TCF-4 at Thr-178 and Thr-189 was also investigated by examining the electrophoretic mobility of LEF-1 and TCF-4 on SDS-PAGE (Fig. 4D). Western blot analysis revealed that the LEF-1 and TCF-4 proteins migrated more slowly on SDS-PAGE when wild-type NLK was coexpressed (Fig. 4D, lanes 2 and 7). These mobility shifts were shown to be due to phosphorylation (13). Expression of the kinase-negative mutant NLK(K155M) failed to cause mobility shifts of LEF-1 (lane 3) and TCF-4 (lane 8). Furthermore, when LEF-1(2A) or TCF-4(2V) was coexpressed along with NLK, a fractional shift was observed (lanes 5 and 10), indicating that the full mobility shift requires phosphorylation of both sites. These results are consistent with the idea that NLK phosphorylates LEF-1 and TCF-4 at these serine/threonine residues in vivo. However, these results do not completely rule out the possibility that these residues play a role in the association of LEF-1 and TCF-4 with NLK. To address this possibility, we compared the abilities of the wild-type and mutated forms of LEF-1 and TCF-4 to associate with NLK in vivo (Fig. 5A). 293 cells were cotransfected with Flag-NLK and wild-type HA-LEF-1, HA-LEF-1(2A), wild-type TCF-4, or TCF-4(2V). Cell extracts were immunoprecipitated with anti-Flag antibody and immunoblotted with anti-HA or anti-TCF-4 antibodies to detect LEF-1 or TCF-4, respectively, in the NLK immunoprecipitates. This analysis revealed that NLK association with LEF-1(2A) (Fig. 5, top panel, lane 4) and TCF-4(2V) (Fig. 5, top panel, lane 8) was normal, suggesting that the LEF-1(2A) and TCF-4(2V) mutations abrogate phosphorylation per se and do not otherwise affect the interaction with NLK in vivo.
FIG. 5.
Effects of NLK phosphorylation site mutation in LEF-1 and TCF-4 on interactions with NLK and β-catenin. (A) Interaction of NLK with LEF-1 and TCF-4. 293 cells were transfected with the indicated expression vectors. Cell lysates were immunoprecipitated (IP) with anti-Flag antibody. Immunoprecipitates were immunoblotted (IB) with anti-HA antibody (left) or anti-TCF-4 antibody (right) (upper panels) and anti-Flag antibody (lower panels). Expression of HA-LEF-1 and TCF-4 was monitored with anti-HA antibody (left) and anti-TCF-4 antibody (right), respectively (right panels). (B) Interaction of β-catenin with LEF-1 and TCF-4. 293 cells were transfected with the indicated expression vectors. Cell lysates were immunoprecipitated with anti-HA antibody (left) or anti-TCF-4 antibody (right). Immunoprecipitates were immunoblotted with anti-β-catenin antibody (upper panels) and anti-HA antibody (left) or anti-TCF-4 antibody (right) (lower panels). Expression of β-catenin was monitored with anti-β-catenin antibody (right panels). WT, wild type.
NLK phosphorylation sites are important for LEF-1/TCF activity.
We next examined whether the two NLK phosphorylation sites have any effect on LEF-1/TCF activity in vivo. The abilities of the LEF-1(2A) and TCF-4(2V) mutants to bind β-catenin were assayed and compared with those of wild-type LEF-1 and TCF-4, respectively (Fig. 5B). β-Catenin was coexpressed with wild-type HA-LEF-1, HA-LEF-1(2A), wild-type TCF-4, or TCF-4(2V) in 293 cells. Cell lysates were immunoprecipitated with anti-HA or anti-TCF-4 antibodies, and interacting β-catenin proteins were detected by Western blot analysis with anti-β-catenin antibody. We found that the mutated forms of LEF-1 and TCF-4 associated with β-catenin to the same extent as each of the respective wild-type forms, indicating that mutations at the sites of NLK phosphorylation probably do not alter the global three-dimensional structure of LEF-1 or TCF-4.
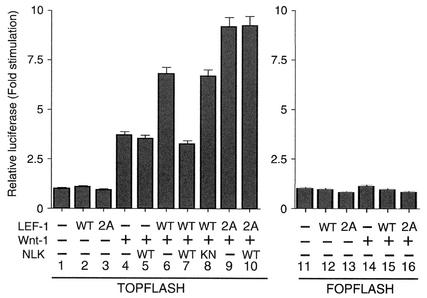
Next, we examined whether the LEF-1(2A) mutation has altered transcriptional activity (Fig. 6). LEF-1/TCF has been shown to form a complex with β-catenin in response to Wnt stimulation (2, 4, 18, 34). To examine LEF-1/TCF transcriptional activity, a luciferase reporter driven by a LEF-1/TCF-responsive promoter, TOPFLASH (32, 33), was cotransfected together with a Wnt-1 expression vector into HeLa cells, which contain low levels of endogenous LEF-1/TCF (10). Stimulation with Wnt-1 induced expression of this reporter gene about fourfold over that of the vector-transfected cells (Fig. 6, lanes 1 and 4). In contrast, no activity was observed when a FOPFLASH reporter, which lacks LEF-1/TCF-binding sites, was cotransfected (Fig. 6, lane 14). Expression of LEF-1 alone did not augment the activation of the reporter gene (lane 2). However, coexpression of LEF-1 and Wnt-1 enhanced Wnt-1-induced activation of the reporter (Fig. 6, lane 6). Expression of the mutant form LEF-1(2A) alone did not activate the reporter (Fig. 6, lane 3), but coexpression of LEF-1(2A) with Wnt-1 stimulated Wnt-1-induced reporter activity more strongly than did expression of wild-type LEF-1 (Fig. 6, lane 9). When NLK was coexpressed with Wnt-1 and LEF-1, it inhibited reporter activity (Fig. 6, lane 7), whereas coexpression of the kinase-negative mutant form NLK(K155M) did not (Fig. 6, lane 8). This indicates that NLK inhibits LEF-1-dependent activation of the LEF-1/TCF reporter and that this inhibition requires NLK kinase activity. However, NLK did not inhibit reporter activity induced by coexpression of LEF-1(2A) with Wnt-1 (Fig. 6, lane 10). These results suggest that the repression of LEF-1 activity by NLK requires the two phosphorylation sites in LEF-1. NLK overexpression failed to suppress the endogenous LEF-1/TCF-dependent transcription (lanes 4 and 5). This suggests that HeLa cells may contain an LEF-1/TCF complex that is resistant to NLK-mediated repression.
FIG. 6.
Effects of NLK phosphorylation site mutation in LEF-1 on transcriptional activation. HeLa cells were transfected with a luciferase reporter plasmid (0.1 μg) and expression vectors encoding wild-type (WT) LEF-1 (0.2 μg), Wnt-1 (1 μg), LEF-1(2A) (0.2 μg), wild-type NLK (0.05 μg), and NLK(K155M) (KN; 0.05 μg). After 24 h of incubation, cells were harvested and luciferase activity was measured. The values shown are the averages of one representative experiment in which each transfection was performed in duplicate.
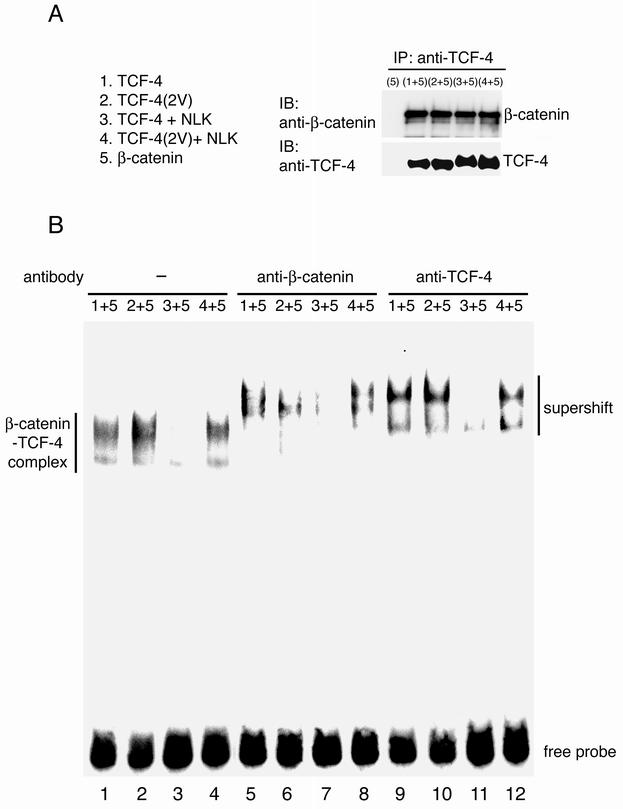
We have previously shown that NLK phosphorylates TCF-4 and prevents the β-catenin-TCF-4 complex from binding DNA (13). We therefore used gel mobility shift assays to determine if the phosphorylation sites in TCF-4 affect the DNA-binding properties of the β-catenin-TCF-4 complex. TCF-4 and β-catenin were separately transfected into 293 cells, and nuclear extracts were prepared from cells expressing TCF-4 or β-catenin. These nuclear extracts were mixed and immunoprecipitated with anti-TCF-4 antibody. Western blot analysis confirmed that neither mutation of TCF-4 to TCF-4(2V) nor phosphorylation of TCF-4 by NLK affected its interaction with β-catenin in vitro (Fig. 7A). These mixtures of nuclear extracts were subjected to a gel mobility shift assay using a probe containing a TCF-binding motif (Fig. 7B). A specific band was detected when mixtures of nuclear extracts containing wild-type TCF-4 and β-catenin were incubated with the probe (Fig. 7B, lane 1). As expected, this band was supershifted by anti-β-catenin (lane 5) and anti-TCF-4 (lane 9) antibodies, confirming that it contained the β-catenin-TCF-4-DNA complex. In contrast, when nuclear extracts containing β-catenin were mixed with those prepared from cells coexpressing wild-type TCF-4 and NLK, the formation of the β-catenin-TCF-4-DNA complex was greatly reduced (Fig. 7B, lane 3). This indicates that NLK expression interferes with the assembly of the complex. Thus, as observed previously (13), NLK phosphorylation of TCF-4 prevents the β-catenin-TCF-4 complex from binding DNA. Next, we tested whether the TCF-4(2V) mutation would affect the DNA-binding properties of the β-catenin-TCF-4 complex. As shown in lane 2, we found that the TCF-4(2V) mutation actually enhanced DNA-binding activity. Moreover, the mutant TCF-4(2V)-β-catenin complex remained bound to DNA even when NLK was expressed (Fig. 7B, lane 4), suggesting that NLK phosphorylation of the two sites on wild-type TCF-4 causes decreased DNA binding. These results support a model whereby phosphorylation of Thr-178 and Thr-189 in TCF-4 mediates the inhibitory effect of NLK.
FIG. 7.
Effect of NLK phosphorylation site mutation in TCF-4 on β-catenin-TCF-4 complex DNA-binding activity. (A) Association of TCF-4 with β-catenin. 293 cells were transfected with expression vectors 1 to 5. Nuclear extracts expressing β-catenin (expression vector 5) were mixed with those expressing TCF-4 (expression vector 1), TCF-4(2V) (expression vector 2), TCF-4 and NLK (expression vector 3), or TCF-4(2V) and NLK (expression vector 4). Mixtures of these nuclear extracts were immunoprecipitated (IP) with anti-TCF-4 antibody. Immunoprecipitates were immunoblotted (IB) with anti-β-catenin antibody (upper panel) and anti-TCF-4 antibody (lower panel). (B) DNA-binding activity of β-catenin-TCF-4 complex. Gel retardation-supershift assays were performed on mixtures of nuclear extracts prepared from 293 cells transfected with the indicated expression vectors. Samples were incubated with the optimal TCF retardation probe. Antibodies were added as indicated.
DISCUSSION
Comparison of Wnt signaling between C. elegans and vertebrates.
The current models describing Wnt/β-catenin signaling in vertebrates and Drosophila suggest that signaling stabilizes the β-catenin protein, making it available for binding to LEF-1/TCF-related transcription factors. The β-catenin-LEF-1/TCF complex, in turn, activates Wnt target genes in the nucleus (2, 4, 18, 34). In the early C. elegans embryo, Wnt signaling controls anterior-posterior cell polarity in the EMS blastomere. The β-catenin-related factor WRM-1 and the NLK-related kinase LIT-1 are effectors of this signaling pathway. POP-1, a LEF-1/TCF-related protein, functions to repress gene expression, and Wnt signaling down-regulates the activity of POP-1 (15, 16, 21, 22, 30). Rocheleau et al. (22) have demonstrated that the WRM-1 protein binds to and activates LIT-1. This activation leads to the phosphorylation of POP-1 and apparent changes in its subcellular localization (22). Thus, WRM-1 and POP-1, rather than working as cofactors, have opposing functions. POP-1 plays a negative role in Wnt signaling and is inhibited by WRM-1 through LIT-1-mediated phosphorylation of POP-1 (22).
These components are conserved in vertebrates. In the present study, we have characterized the relationships among β-catenin, LEF-1/TCF, and NLK. The C. elegans LIT-1 protein interacts directly with WRM-1, and this complex can phosphorylate POP-1 in a manner that requires the LIT-1 kinase domain (22). This indicates that WRM-1 is required for POP-1 phosphorylation. In contrast, in vertebrates, LEF-1/TCF binds directly to NLK and β-catenin interacts indirectly with NLK via LEF-1/TCF to form a larger complex. The presence of β-catenin enhances the interaction between NLK and TCF-4 and thereby promotes phosphorylation of TCF-4 by NLK. These findings indicate that the components of the Wnt pathway in C. elegans and vertebrates are similar but have different activities. In particular, C. elegans WRM-1 appears to possess important functional differences from other family members. Indeed, WRM-1, which is only 19% identical to vertebrate β-catenin, does not bind to the POP-1 β-catenin-binding domain per se but weakly binds full-length POP-1 (14, 19, 22). Such a physical interaction might be stabilized by LIT-1 (22). On the other hand, β-catenins can stably bind to LEF-1/TCF (2).
Understanding how upstream signals control LIT-1 will require much more genetic and biochemical investigation. C. elegans Wnt signaling also controls anterior-posterior cell polarity in postembryonic T blast cells. T-cell polarity is controlled by LIN-44 (Wnt), LIN-17 (Frizzled receptor), and POP-1 (7, 8, 9, 26). Although LIT-1 controls both EMS and T-cell polarities, neither WRM-1 nor any other β-catenin homolog is involved in T-cell polarity (9). Our observation that NLK interacts directly with LEF-1/TCF makes it likely that LIT-1 would interact directly with POP-1 in the control of T-cell polarity. Herman has proposed that the LIN-44 signal, acting through LIT-1 kinase, functions to modify POP-1 (9). It is therefore possible that a Wnt-like protein may activate LIT-1 and NLK in C. elegans and vertebrates, respectively.
Conserved LEF-1/TCF phosphorylation sites.
In this study, we found that the primary sites of NLK phosphorylation are located in the core region between the β-catenin-binding domain and the DNA-binding domain in the LEF-1 and TCF-4 proteins. Specifically, these sites mapped, in vitro and in vivo, to Thr-155 and Ser-166 of LEF-1 and Thr-178 and Thr-189 of TCF-4. Site-directed mutagenesis of these conserved serine and threonine residues abrogated NLK-mediated phosphorylation in vitro and in vivo. Similar to other LEF-1/TCF proteins, C. elegans POP-1 contains a conserved NH2-terminal β-catenin-binding domain and a more central HMG box that binds DNA. Sequence comparisons indicate that Ser-71 and Ser-82 of C. elegans POP-1 are located in analogous positions. Although phosphorylation of these residues in POP-1 has not been demonstrated, it is likely that these serine residues are also targets of phosphorylation by LIT-1. Whether LIT-1 mediates the phosphorylation of Ser-71 and Ser-82 of POP-1 remains to be investigated. It is likely that functionally conserved serine and threonine residues are also present between the β-catenin-binding domain and the DNA-binding domain of LEF-1/TCF that regulate its activity.
Role of NLK phosphorylation in LEF-1/TCF.
We have previously demonstrated that NLK can regulate the transcriptional activity of a β-catenin-LEF-1/TCF complex (13). Mechanistically, NLK directly interacts with and phosphorylates TCF-4. This phosphorylation does not disrupt the formation of a complex between LEF-1/TCF and β-catenin, yet it does prevent DNA binding of the β-catenin-TCF-4 complex and thereby inhibits β-catenin-mediated transcription through LEF-1/TCF-binding sites in Wnt pathway-responsive promoters. In vivo, coinjection of NLK RNA into early Xenopus embryos results in the inhibition of double-axis formation by β-catenin. Since Xenopus NLK is expressed ubiquitously as both a maternal and a zygotic transcript in Xenopus embryos (11), it has the potential both to regulate the axis-determining activity of LEF-1/TCF and to modulate other potential functions of LEF-1/TCF at later stages of development, such as neural patterning or mesoderm development (11).
In this report, we identify two NLK phosphorylation sites in LEF-1 and TCF-4 and present data suggesting that phosphorylation of these sites contributes to the down-regulation of the ability of LEF-1/TCF transcriptional activity. Mutational replacement of these serine and threonine residues with alanine or valine prevents NLK-mediated negative regulation of LEF-1 and TCF-4. Such regulation of DNA binding by phosphorylation of sequence-specific transcription factors has been well documented in several cases and can act both positively and negatively (3). Several mechanisms could explain how serine/threonine phosphorylation of TCF-4 by NLK affects the DNA-binding activity of the β-catenin-TCF-4 complex. In many cases in which phosphorylation inhibits DNA binding, the phosphorylation sites are located either within or near the DNA-binding domain. In such cases, it is likely that phosphorylation interferes with DNA binding by electrostatic repulsion between phosphate groups on the protein and the DNA. Although we have not determined the mechanism for inhibition of the β-catenin-TCF-4 complex DNA binding by NLK-mediated threonine phosphorylation, it is presumably not caused by direct phosphorylation of the DNA-binding domain, since the sites of NLK phosphorylation are located distal to the DNA-binding domain. In cases in which the phosphorylation sites are separate from the DNA-binding domain, it is most likely that phosphorylation alters the conformation of the protein in such a way that its DNA-binding activity is altered.
Negative regulation of Wnt/β-catenin signaling.
The canonical Wnt/β-catenin signal transduction cascade is subject to multiple levels of negative control. In the cytoplasm, the amount of β-catenin is negatively regulated by degradation of β-catenin in the axin complex (1, 12). In the nucleus, several proteins negatively regulate β-catenin-dependent gene expression by interfering with complex formation among β-catenin, LEF-1/TCF, and DNA. Members of the Sox protein family have been shown to interact with the same Armadillo repeat region of β-catenin as LEF-1/TCF, thereby repressing its signaling activity (36). ICAT also binds to β-catenin and represses β-catenin-mediated transactivation (29). Specific binding partners of TCF are homologues of Groucho, a transcriptional corepressor (6, 23). TCF binding to target DNA sequences is suppressed by its association with HMG box repressor protein 1 (25) and I-mfa domain proteins (28).
In addition to the above cofactors, LEF-1/TCF also interacts with several other classes of molecules that modify LEF-1/TCF. For example, LEF-1 interacts with PIASy, which catalyzes the covalent modification of LEF-1 through conjugation of multiple small ubiquitin-like modifier peptides, suggesting that PIASy functions as an E3 small ubiquitin-like modifier ligase. Sumoylation of LEF-1 inhibits its function as a transcription factor and leads to sequestration of LEF-1 in PML nuclear bodies (24). In Drosophila, the CREB-binding protein interacts with the HMG domain of TCF (also known as pangolin) and acetylates a conserved lysine in the Armadillo-binding domain of TCF. This acetylation lowers the affinity of TCF for β-catenin (35). Furthermore, in both C. elegans and mammalian cells, the Nemo-like kinases LIT-1 and NLK, respectively, phosphorylate LEF-1/TCF. Phosphorylation inhibits LEF-1/TCF signaling by disruption of DNA binding (13) and by inducing redistribution of POP-1 from the nucleus to the cytoplasm (22). Thus, it is likely that β-catenin signaling through LEF-1/TCF is inhibited by several mechanisms at the level of LEF-1/TCF and β-catenin in the nucleus. Since β-catenin functions as an oncogene, there may be several mechanisms that protect against abnormal cellular proliferation by inhibiting β-catenin signaling. It will be interesting to know how these different mechanisms are coordinated to regulate Wnt/β-catenin signaling in a tissue- and stage-specific manner.
Acknowledgments
We thank T. Akiyama, H. Clevers, A. Kikuchi, and M. Waterman for materials; E. Nishida for helpful discussions; and M. Lamphier for critical reading of the manuscript.
This research was supported by special grants for CREST and Advanced Research on Cancer from the Ministry of Education, Culture and Science of Japan (K.M.).
REFERENCES
- 1.Aberle, H., A. Bauer, J. Stappert, A. Kispert, and R. Kemler. 1997. β-Catenin is a target for the ubiquitin-proteasome pathway. EMBO J. 16:3797-3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Behrens, J., J. P. von Kries, M. Kuhl, L. Bruhn, D. Wedlich, R. Grosschedl, and W. Birchmeier. 1996. Functional interaction of β-catenin with the transcription factor LEF-1. Nature 382:638-642. [DOI] [PubMed] [Google Scholar]
- 3.Brivanlou, A. H., and J. E. Darnell, Jr. 2002. Signal transduction and the control of gene expression. Science 295:813-818. [DOI] [PubMed] [Google Scholar]
- 4.Brunner, E., O. Peter, L. Schweizer, and K. Basler. 1997. Pangolin encodes a Lef-1 homologue that acts downstream of Armadillo to transduce the wingless signal in Drosophila. Nature 385:829-833. [DOI] [PubMed] [Google Scholar]
- 5.Cardigan, K. M., and R. Nusse. 1997. Wnt signaling: a common theme in animal development. Genes Dev. 11:3286-3305. [DOI] [PubMed] [Google Scholar]
- 6.Cavallo, R. A., R. T. Cox, M. M. Moline, J. Roose, G. A. Polevoy, H. Clevers, M. Peifer, and A. Bejsovec. 1998. Drosophila Tcf and Groucho interact to repress wingless signalling activity. Nature 395:604-608. [DOI] [PubMed] [Google Scholar]
- 7.Herman, M. A., and H. R. Horvitz. 1994. The Caenorhabditis elegans gene lin-44 controls the polarity of asymmetric cell divisions. Development 120:1035-1047. [DOI] [PubMed] [Google Scholar]
- 8.Herman, M. A., L. L. Vassilieva, H. R. Horvitz, J. E. Shaw, and R. K. Herman. 1995. The C. elegans gene lin-44, which controls the polarity of certain asymmetric cell divisions, encodes a Wnt protein and acts cell nonautonomously. Cell 83:101-110. [DOI] [PubMed] [Google Scholar]
- 9.Herman, M. C. 2001. C. elegans POP-1/TCF functions in a canonical Wnt pathway that controls cell migration and in a noncanonical Wnt pathway that controls cell polarity. Development 128:581-590. [DOI] [PubMed] [Google Scholar]
- 10.Hovanes, K., T. W. Li, and M. L. Waterman. 2000. The human LEF-1 gene contains a promoter preferentially active in lymphocytes and encodes multiple isoforms derived from alternative splicing. Nucleic Acids Res. 28:1994-2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hyodo-Miura, J., S. Urushiyama, S. Nagai, M. Nishita, N. Ueno, and H. Shibuya. 2002. Involvement of NLK and Sox11 in neural induction in Xenopus development. Genes Cells 7:487-496. [DOI] [PubMed] [Google Scholar]
- 12.Ikeda, S., S. Kishida, H. Yamamoto, H. Murai, S. Koyama, and A. Kikuchi. 1998. Axin, a negative regulator of the Wnt signaling pathway, forms a complex with GSK-3β and β-catenin and promotes GSK-3β-dependent phosphorylation of β-catenin. EMBO J. 17:1371-1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ishitani, T., J. Ninomiya-Tsuji, S. Nagai, M. Nishita, M. Meneghini, N. Barker, M. Waterman, B. Bowerman, H. Clevers, H. Shibuya, and K. Matsumoto. 1999. The TAK1-NLK-MAPK-related pathway antagonizes signalling between β-catenin and transcription factor TCF. Nature 399:798-802. [DOI] [PubMed] [Google Scholar]
- 14.Korswagen, H. C., M. A. Herman, and H. C. Clevers. 2000. Distinct β-catenins mediate adhesion and signalling functions in C. elegans. Nature 406:527-532. [DOI] [PubMed] [Google Scholar]
- 15.Lin, R., R. J. Hill, and J. R. Priess. 1998. POP-1 and anterior-posterior fate decisions in C. elegans embryos. Cell 92:229-239. [DOI] [PubMed] [Google Scholar]
- 16.Meneghini, M. D., T. Ishitani, J. C. Carter, N. Hisamoto, J. Ninomiya-Tsuji, C. J. Thorpe, D. R. Hamill, K. Matsumoto, and B. Bowerman. 1999. MAP kinase and Wnt pathways converge to down-regulate an HMG domain repressor in C. elegans. Nature 399:793-797. [DOI] [PubMed] [Google Scholar]
- 17.Miller, J. R., and R. T. Moon. 1996. Signal transduction through β-catenin and specification of cell fate during embryogenesis. Genes Dev. 10:2527-2539. [DOI] [PubMed] [Google Scholar]
- 18.Molenaar, M., M. van de Wetering, M. Oosterwegel, J. Peterson-Maduro, S. Godsave, V. Korinek, J. Roose, O. Destree, and H. Clevers. 1996. XTcf-3 transcription factor mediates β-catenin-induced axis formation in Xenopus embryos. Cell 86:391-399. [DOI] [PubMed] [Google Scholar]
- 19.Natarajan, L., N. E. Witwer, and D. M. Eisenmann. 2001. The divergent Caenorhabditis elegans β-catenin proteins BAR-1, WRM-1 and HMP-2 make distinct protein interactions but retain functional redundancy in vivo. Genetics 159:159-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peifer, M., and P. Polakis. 2000. Wnt signaling in oncogenesis and embryogenesis: a look outside the nucleus. Science 287:1606-1609. [DOI] [PubMed] [Google Scholar]
- 21.Rocheleau, C. E., W. D. Downs, R. Lin, C. Wittmann, Y. Bei, Y.-H. Cha, M. Ali, J. R. Priess, and C. C. Mello. 1997. Wnt signaling and an APC-related gene specify endoderm in early C. elegans embryos. Cell 90:707-716. [DOI] [PubMed] [Google Scholar]
- 22.Rocheleau, C. E., J. Yasuda, T. H. Shin, R. Lin, H. Sawa, H. Okano, J. R. Priess, R. J. Davis, and C. C. Mello. 1999. WRM-1 activates the LIT-1 protein kinase to transduce anterior/posterior polarity signals in C. elegans. Cell 97:717-726. [DOI] [PubMed] [Google Scholar]
- 23.Roose, J., M. Molenaar, J. Peterson, J. Hurenkamp, H. Brantjes, P. Moerer, M. van de Wetering, O. Destree, and H. Clevers. 1998. The Xenopus Wnt effector XTcf-3 interacts with Groucho-related transcriptional repressors. Nature 395:608-612. [DOI] [PubMed] [Google Scholar]
- 24.Sachdev, S., L. Bruhn, H. Sieber, A. Pichler, F. Melchior, and R. Grosschedl. 2001. PIASy, a nuclear matrix-associated SUMO E3 ligase, represses LEF1 activity by sequestration into nuclear bodies. Genes Dev. 15:3088-3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sampson, E. M., Z. K. Haque, M. C. Ku, S. G. Tevosian, C. Albanese, R. G. Pestell, K. E. Paulson, and A. S. Yee. 2001. Negative regulation of the Wnt-β-catenin pathway by the transcriptional repressor HBP1. EMBO J. 20:4500-4511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sawa, H., L. Lobel, and H. R. Horvitz. 1996. The Caenorhabditis elegans gene lin-17, which is required for certain asymmetric cell divisions, encodes a putative seven-transmembrane protein similar to the Drosophila Frizzled protein. Genes Dev. 10:2189-2197. [DOI] [PubMed] [Google Scholar]
- 27.Shin, T. H., J. Yasuda, C. E. Rocheleau, R. Lin, M. Soto, Y. Bei, R. J. Davis, and C. C. Mello. 1999. MOM-4, a MAP kinase kinase kinase related protein, activates WRM-1/LIT-1 kinase to transduce anterior/posterior polarity signals in C. elegans. Mol. Cell 4:275-280. [DOI] [PubMed] [Google Scholar]
- 28.Snider, L., H. Thirlwell, J. R. Miller, R. T. Moon, M. Groudine, and S. J. Tapscott. 2001. Inhibition of Tcf3 binding by I-mfa domain proteins. Mol. Cell. Biol. 21:1866-1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tago, K., T. Nakamura, M. Nishita, J. Hyodo, S. Nagai, Y. Murata, S. Adachi, S. Ohwada, Y. Morishita, H. Shibuya, and T. Akiyama. 2000. Inhibition of Wnt signaling by ICAT, a novel β-catenin-interacting protein. Genes Dev. 14:1741-1749. [PMC free article] [PubMed] [Google Scholar]
- 30.Thorpe, C. J., A. Schlesinger, J. C. Carter, and B. Bowerman. 1997. Wnt signaling polarizes an early C. elegans blastomere to distinguish endoderm from mesoderm. Cell 90:695-705. [DOI] [PubMed] [Google Scholar]
- 31.Treisman, R. 1996. Regulation of transcription by MAP kinase cascades. Curr. Opin. Cell Biol. 8:205-215. [DOI] [PubMed] [Google Scholar]
- 32.van de Wetering, M., M. Oosterwegel, D. Dooijes, and H. Clevers. 1991. Identification and cloning of TCF-1, a T lymphocyte-specific transcription factor containing a sequence-specific HMG box. EMBO J. 10:123-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van de Wetering, M., J. Castrop, V. Korinek, and H. Clevers. 1996. Extensive alternative splicing and dual promoter usage generate Tcf-1 protein isoforms with differential transcription control properties. Mol. Cell. Biol. 16:745-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van de Wetering, M., R. Cavallo, D. Dooijes, M. van Beest, J. van Es, J. Loureiro, A. Ypma, D. Hursh, T. Jones, A. Bejsovec, M. Peifer, M. Mortin, and H. Clevers. 1997. Armadillo coactivates transcription driven by the product of the Drosophila segment polarity gene dTCF. Cell 88:789-799. [DOI] [PubMed] [Google Scholar]
- 35.Waltzer, L., and M. Bienz. 1998. Drosophila CBP represses the transcription factor TCF to antagonize Wingless signalling. Nature 395:521-525. [DOI] [PubMed] [Google Scholar]
- 36.Zorn, A. M., G. D. Barish, B. O. Williams, P. Lavender, M. W. Klymkowsky, and H. E. Varmus. 1999. Regulation of Wnt signaling by Sox proteins: XSox17 α/β and XSox3 physically interact with β-catenin. Mol. Cell 4:487-498. [DOI] [PubMed] [Google Scholar]