Abstract
African trypanosomes cause sleeping sickness in humans and Nagana in cattle. The parasites multiply in the blood and escape the immune response of the infected host by antigenic variation. Antigenic variation is characterized by a periodic change of the parasite protein surface, which consists of a variant glycoprotein known as variant surface glycoprotein (VSG). Using a SELEX (systematic evolution of ligands by exponential enrichment) approach, we report the selection of small, serum-stable RNAs, so-called aptamers, that bind to VSGs with subnanomolar affinity. The RNAs are able to recognize different VSG variants and bind to the surface of live trypanosomes. Aptamers tethered to an antigenic side group are capable of directing antibodies to the surface of the parasite in vitro. In this manner, the RNAs might provide a new strategy for a therapeutic intervention to fight sleeping sickness.
Trypanosoma brucei is the causative agent of sleeping sickness in humans and Nagana in cattle. An estimated 50 million people worldwide are at risk of an infection, and the number of new cases per year likely exceeds the reported 350,000 cases significantly (31). The disease in domestic animals has a severe impact on agricultural development in large parts of Africa (35), and the human form of the disease is fatal if left untreated. The available antiparasitic drugs are highly toxic and difficult to administer. Thus, new experimental strategies for developing novel therapeutics are required (8).
Trypanosomes are extracellular blood parasites. Their cell surface is covered with a dense layer of a single protein termed variant surface glycoprotein (VSG) (4). VSGs have a molecular size of ca. 60 kDa. They form homodimers and are prototypic glycosylphosphatidylinositol-anchored membrane proteins. VSGs induce a T-cell-independent immunoglobulin M (IgM) response and a T-cell-dependent B-cell response that elicits VSG-specific IgG (32). The parasites evade the host immune response by temporarily expressing immunologically unrelated VSG variants (6, 30). This phenomenon, known as antigenic variation, has its molecular basis in the surface presentation of structurally polymorphic N-terminal domains of the different VSGs. Although at any given time point only one VSG variant is expressed and presented on the cell surface, the genome contains a repertoire of hundreds of different vsg genes (39). With a likelihood of 10−2 to 10−7 per cell cycle the parasites switch to the expression of a different VSG variant thereby evading the host's immune response (18). Thus, the VSG surface can be viewed as providing an exclusion barrier for larger molecules, such as antibodies, as well as disarming the infected host's means of clearing the infection through its variable characteristics.
In addition to the variable features, the parasite surface also exhibits constant attributes. Invariant surface glycoproteins, receptor complexes, and transporter molecules are embedded within the VSG layer (24, 27). Even the VSGs show conserved characteristics. Despite a very low identity on the amino acid level, different VSG variants adopt very similar tertiary structures (1). These conserved structural epitopes are not accessible to antibodies but can be accessed by molecules of smaller molecular size such as the protease trypsin (23 kDa). The protease has been shown to be able to penetrate into the molecular cavities between the VSG homodimers (41).
Based on these characteristics, we asked the question whether a SELEX (systematic evolution of ligands by exponential enrichment) protocol (37, 40) could be designed to allow the selection of RNAs that bind with high affinity and specificity (aptamer RNAs) to the structurally conserved parts of VSGs. We further sought to determine whether such RNAs could be tethered to a ligand to indirectly label the otherwise-variable surface of African trypanosomes and, lastly, whether a covalently attached antigenic ligand could be used to direct antibodies to the surface of the parasite.
MATERIALS AND METHODS
Trypanosomes.
The bloodstream life cycle stage of T. brucei subsp. brucei was cultivated at 37°C in HMI-9 medium (13) supplemented with 10% (vol/vol) heat-inactivated fetal calf serum. The following trypanosome strains were used: T. brucei Lister 427-MITat serodeme; variant clones MITat 1.2, and MITat 1.4 (4); AnTat 1.1 (22); and ILTat 1.1 (29). Long slender bloodstream forms of T. brucei AnTat 1.1 and Trypanosoma congolense BeNat 1 were harvested from infected rats.
sVSG and procyclin purification.
Soluble VSG (sVSG) was isolated as described previously (5) and analyzed in discontinuous sodium dodecyl sulfate (SDS)-containing polyacrylamide gels. The formation of sVSG homodimers was verified by size exclusion chromatography, and protein folding was analyzed by circular dichroism (CD) spectroscopy. Deglycosylated sVSG was prepared by treating 30 μg of sVSG with 4 U (160 ng) of N-glycosidase F (Roche) in 50 mM NaxHyPO4 (pH 6.1)-15 mM EDTA. The reaction was performed at 37°C for 2 h in the presence of a cocktail of protease inhibitors (40 μg of bestatin/ml, 60 μg of chymostatin/ml, 10 μg of E-64/ml, 0.7 μg of pepstatin/ml, 300 μg of phosphoramidon/ml, 0.5 μg of leupeptin/ml). N-terminal and C-terminal sVSG fragments were prepared by trypsin hydrolysis (16) at a 1:100 molar ratio of enzyme to protein. Incubation was in 50 mM NaxHyPO4 (pH 8.0) at 25°C for 24 h. The proteolytic fragments were separated by gelfiltration (Superdex 200 HR 10/30 column) in 50 mM NaxHyPO4 (pH 7.0)-150 mM NaCl and analyzed by denaturing polyacrylamide gel electrophoresis (PAGE). Dimer formation was tested by size exclusion chromatography. While the N-terminal fragment showed the characteristics of a homodimer, the C-terminal fragments were monomeric.
Procyclin family proteins were isolated from T. brucei Lister 427 essentially as described by Ferguson et al. (9). The purity of the protein preparations was analyzed in SDS-containing 12.5% (wt/vol) polyacrylamide gels stained with the cationic carbocyanine dye Stains-All (2, 11). Procyclin protein concentrations were calculated from the result of an amino acid analysis after acid hydrolysis (6 M HCl) by using an automated amino acid analyzer system.
Oligodeoxynucleotides and RNA pool synthesis.
The single-stranded starting DNA library was synthesized as described previously (14). The library (0.1 mg) was transcribed into 50 μg of RNA, which was used in the first selection round. Subsequent RNA pools were transcribed from 10 to 50 μg of PCR-generated double-stranded DNA templates in final volumes of 35 to 200 μl. Reactions were performed in the presence of 5 μCi of [α-32P]ATP (3,000 Ci/mmol) in 40 mM Tris-Cl (pH 8.0)-12 mM MgCl2-5 mM dithiothreitol (DTT)-2 mM spermidine-0.002% (vol/vol) Triton X-100, containing 0.8 mM ATP and GTP, 2 mM 2′-F-CTP, and 2′-F-UTP (Europa Bioproducts, Ltd.). Reactions were initiated by adding T7 RNA polymerase (3.5 U/μl). Synthesized RNAs were extracted with phenol, ethanol precipitated, and redissolved in buffer A (20 mM NaxHyPO4 [pH 7.4], 2 mM MgCl2, 130 mM NaCl, 5 mM KCl). Transcripts were purified by size exclusion chromatography and preincubated at 37°C for 30 min before usage.
In vitro selection (SELEX).
RNA binding to sVSG was performed in 0.45 to 1.8 ml of buffer A containing 0.5 mM DTT, 1 μM 32P-labeled pool RNA, and 1 μM sVSG for 1 h at 37°C. sVSG-bound RNAs were separated from unbound RNAs by filter adsorption by using presoaked nitrocellulose and cellulose acetate filters (19). Yeast tRNA (100 ng/μl) was used as a nonspecific blocking reagent. Samples were slowly filtered, and the filters washed four times with 2 ml of buffer A containing 0.5 mM DTT and 10 ng of yeast tRNA/μl. Bound RNA was recovered by three extractions with 8 M urea and phenol and precipitated with ethanol. Binding to live trypanosomes was performed with cells harvested at a cell density of 1 × 106 to 2 × 106 cells/ml. The parasites were washed in buffer B (buffer A containing 20 mM glucose and 0.5 mM DTT), and RNA binding was performed in final volumes of 0.28 to 0.7 ml. 32P-labeled pool RNA concentrations varied between 0.25 to 1 μM, and the parasite cell density varied between 3 × 107 to 1.2 × 108 cells/ml. After incubation for 30 min at 4°C, unbound and weakly associated RNAs were washed off by four washes with 2 ml of buffer B. Trypanosomes and bound RNAs were collected by centrifugation, and bound RNAs were recovered by two phenol extractions, followed by precipitation with ethanol. Isolated RNAs were reverse transcribed and amplified by PCR, and the resulting DNA templates were transcribed into RNA, which was used in the next round of selection. DNA templates from pool 9 were digested with HindIII and EcoRI and cloned into pUC118. DNA sequences were determined by dideoxy terminator sequencing.
Determination of equilibrium dissociation constants.
The binding of RNA aptamers to sVSG was measured by filter binding. Uniformly 32P-labeled pool RNAs or RNA aptamers were incubated with sVSG in buffer A for 15 min at 37°C. Samples were put on ice (2 min) and slowly filtered (0.5 ml/min) through presoaked nitrocellulose-cellulose acetate mixed bed filters. Filters were washed at 4°C with buffer A (four times with 0.5 ml each time) then air dried, and the percentage of bound and free RNA was determined by scintillation counting. Equilibrium dissociation constants were derived from experiments determining the percentage of bound 32P-labeled RNA (0.02 nM) at increasing concentrations of unlabeled RNA (0.06 to 8 nM) at constant VSG concentrations (5 to 7 nM). Kd values were calculated based on the Scatchard equation: r/[A] = n/Kd − r/Kd, with the slope of the plot representing −1/Kd, where A is the free RNA and r is the bound RNA/VSG.
Runoff transcription, radioactive labeling, and biotinylation of RNA aptamers.
RNA aptamers were transcribed from PCR templates (5 to 15 μg) in 40 mM Tris-Cl (pH 8.0), 12 mM MgCl2, 5 mM DTT, 2 mM spermidine, and 0.002% (vol/vol) Triton X-100 by using T7 RNA polymerase (3.5 U/μl). ATP and GTP were at 0.8 mM, and the concentrations of 2′-F-CTP and 2′-F-UTP were at 2 mM. Full-length transcripts were purified by denaturing PAGE. Aptamer 5′ ends were radioactively labeled with alkaline phosphatase, T4 polynucleotide kinase, and [γ-32P]ATP according to standard procedures. RNA 3′-end labeling was performed with 40 μCi of 5′-[32P]pCp and 18 U of T4 RNA ligase. Uniformly 32P-labeled aptamers were prepared by using [α-32P]ATP (10 to 50 μCi) and 0.01 mM ATP in the transcription reaction. Full-length transcripts were purified by denaturing PAGE and renatured in buffer A. Biotinylation of RNA aptamers was achieved by T7 transcription with mutant T7 RNA polymerase Y639F (25) at a concentration of 3 U/μl and biotin-11-ATP (Perkin-Elmer/Life Sciences) at 0.125 mM.
RNA structure and boundary determination.
Theoretical secondary structures were calculated by using the MFOLD subroutine of the GCG software package based on a free energy minimization algorithm (Wisconsin Package version 9.1; Genetics Computer Group, Madison, Wis.). Calculations were performed at 37°C, which is the optimal growth temperature for bloodstream-stage trypanosomes.
Binding boundaries were determined by using 32P-end-labeled RNA preparations, which were subjected to alkaline hydrolysis in 0.5 M NaHCO3 (12 min at 95°C). Samples were cooled on ice and desalted by size exclusion chromatography. Hydrolyzed RNAs (0.6 nM, ∼0.1 μCi) were renatured (30 min at 37°C) and incubated with sVSG (0, 5, 15, 50, and 100 nM) in buffer A containing 300 nM bovine serum albumin (BSA; 15 min at 37°C). sVSG-bound RNA was captured on nitrocellulose-cellulose acetate filters as described above. Eluted RNA was analyzed on denaturing 8% (wt/vol) polyacrylamide gels.
Fluorescence microscopy and imaging.
Biotinylated aptamer preparations were dissolved to 300 ng/μl in buffer A and incubated at 37°C for 30 min. Trypanosomes were harvested at a cell density of 8 × 105 cells/ml by centrifugation at 1,400 × g and washed twice in buffer B. Cells were resuspended to 108 cells/ml in buffer B, and 15 μl of the cell suspension was incubated with 60 to 80 ng of biotinylated RNA/μl for 20 min at 37°C. Unbound RNA was removed by two washes with 2 ml of TDB (20 mM Na2HPO4, 2 mM NaH2PO4 [pH 7.4], 1 mM MgSO4, 80 mM NaCl, 5 mM KCl, 20 mM glucose)-0.5 mM DTT at room temperature. The cell pellet was resuspended in 100 μl of TDB and fixed with 1 vol. of 8% (wt/vol) fresh paraformaldehyde-0.1% (vol/vol) glutaraldehyde in TDB for 30 min at room temperature. For the detection of the surface-bound biotinylated RNAs, the fixed cells were washed twice with TDB-0.5 mM DTT and incubated with 0.1 μg of Alexa Fluor 594-conjugated streptavidin/ml in TDB-0.5 mM DTT-2% (wt/vol) BSA for 10 min at room temperature. After removal of unbound streptavidin by two washes with TDB-0.5 mM DTT, the cells were resuspended in a small volume (∼10 μl) of the same buffer. Immunodetection of surface-bound biotinylated aptamers was performed by using a 1:100 dilution of a monoclonal Cy3-conjugated anti-biotin antibody (Dianova). For the immunodetection of VSGs, fixed cells were incubated with rabbit anti-VSG antisera (1:2,000), followed by incubation with an Alexa Fluor 594-conjugated anti-rabbit second antibody (1:2,000) for 1 h at room temperature. Visualization of the nucleus and kinetoplast was achieved by DAPI (4′,6′-diamidino-2-phenylindole) staining.
Image acquisition was performed with a motorized Zeiss Axiophot2 widefield microscope equipped with a Zeiss ×63/1.4 NA oil differential interference contrast (DIC) objective lens, a ×2.5 optovar, and a Princeton Instruments Micromax cooled (−15°C) slow-scan charge-coupled device camera (Kodak KAF-1400 charge-coupled device chip). For acquisition of three-dimensional images a PIFOC objective z-stepper was driven by the piezo-amplifier E662 LVPZT (Physik Instrumente, Karlsruhe, Germany). The microscopic setup was integrated and controlled by using the scripting feature of the IPLab for Macintosh software (v3.2; Scanalytics, Fairfax, Va.). Three-dimensional images were acquired by using the optimal sampling density derived from the optical setup and the Alexa Fluor fluorophors (z-step size, ≥50 by 150 nm). The point spread function of the microscope was measured by using fluorescent 0.17-μm microspheres (PS-Speck Microscope Point Source Kit; Molecular Probes). After image acquisition, the raw data were exported to the Huygens System software (v2.1.8; Scientific Volume Imaging, B.V., Hilversum, the Netherlands), and digital deconvolution was performed by using the maximum-likelihood estimation algorithm (100 iterations). The restored image data set was visualized and verified by using the Imaris software package (version 2.6.8 or 3.0 for IRIX; Bitplane AG, Zurich, Switzerland).
RESULTS
Selection of VSG-specific RNA aptamers.
For the in vitro selection, we used a combinatorial library of 2 × 1014 unique RNA sequences (14). The library had a size of 40 nucleotides (nt), flanked by primer binding sites of 24 and 21 nt, to restrict the molecular mass of the selected aptamers to a value of <30 kDa. RNA transcription was performed in the presence of 2′ F-modified pyrimidine nucleotides to select serum-stable aptamers (28). The selection strategy was based on the following three characteristics. First, to specifically enrich for VSG-interacting aptamers, we incubated the starting RNA pool with a homogeneous sVSG preparation. Second, the selected RNAs were then challenged with live trypanosomes that stably express the same VSG variant to select for RNAs that can bind to the protein in the context of living cells. Third, to select for VSG variant-independent binding characteristics, later selection rounds were performed with a second VSG variant. Again, the protein was first presented as purified sVSG and then in the form of a parasite cell line stably expressing this VSG variant. The following T. brucei variant antigen clones were chosen: T. brucei MITat 1.4, which expresses VSG 117, and T. brucei MITat 1.2, expressing VSG 221 (4). sVSGs were isolated as described previously (5) and purified by ion-exchange chromatography. CD spectra confirmed the presence of a high proportion of α-helical structure in agreement with the X-ray structure of VSG 221 (10). The dimerization of the soluble protein preparations was confirmed by size exclusion chromatography (data not shown). Selection rounds with live trypanosomes were performed at 4°C in order to inhibit endocytosis (15), whereas selections with sVSGs were done at 37°C (the optimal growth temperature for the parasite).
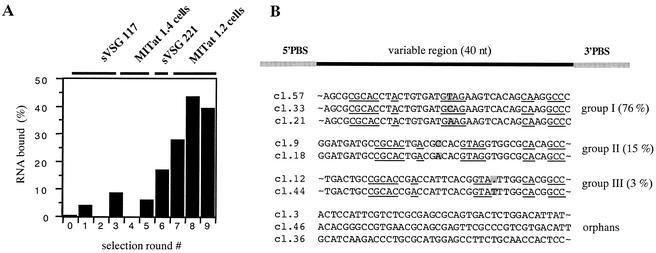
Nine rounds of selection and amplification were performed. The course of the experiment was monitored by determining the percentage of bound RNA in every SELEX round (Fig. 1A). Maximal binding was achieved in round 8 with ca. 40% of the RNAs capable of interacting with sVSG 221. DNA templates from round 9 were cloned, and the nucleotide sequences of 60 individual clones were determined. Based on shared sequence motifs, each aptamer can be placed into one of three groups. Group I, contains 76% of all of the sequences; group II contains 15%, and group III contains 3%. Six percent of the sequences are orphan RNAs (Fig. 1B).
FIG. 1.
Course of the SELEX experiment and primary structures of selected aptamers. (A) Graphical representation of the percentage of bound RNA in each round of the selection. Rounds 1 to 3 were performed with sVSG 117 as the binding target. Rounds 4 and 5 used live MITat 1.4 trypanosomes. Selection round 6 was performed with sVSG 221, followed by rounds 7 to 9 with MITat 1.2 parasites. (B) DNA from round 9 was cloned and the sequences of 60 clones were determined. The derived sequences were put into three groups (I, II, and III) plus three orphan RNAs. Identical sequence regions are underlined. Primer-binding sites (PBS) are shaded gray, and single variable nucleotides are also shaded gray. The name of each aptamer is given in the margin on the left. The percent values represent the relative abundance of the aptamer group within the total number of clones. The primary sequences of the PBS are as follows: 5′PBS-GGTCGAGAATTCAGTCGGACAGCG and 3′PBS-CTACCTGCTTATAGCAGAGGGATATCACTCAGCAATTTCGAAGCG.
Structural features of the selected aptamers.
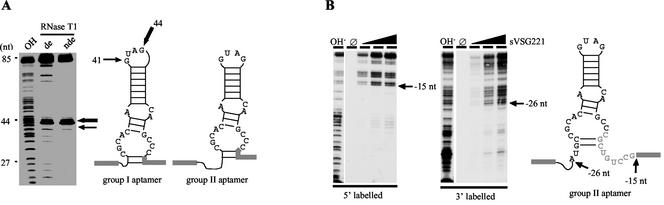
Theoretical RNA secondary (2D) structures were computed (including the primer-binding sequences) based on a minimal free-energy folding algorithm. Figure 2A shows the calculated 2D structures for aptamer clone (cl.) 57 (group I) and aptamer cl. 9 (group II). The two RNAs can be folded into imperfect hairpin loops of similar structure. Conserved nucleotide sequences are located within the single-stranded regions of the hairpins. The two structures differ from each other in (i) the size of the apical loop, which is 2 nt shorter for the group II RNAs, and (ii) in the length of the upper stem, being 1 bp shorter in the case of the group II aptamers. The single-stranded nature of the two G residues within the apical loop was experimentally verified by RNase T1 footprinting (Fig. 2A). As expected from the theoretical structures, both G nucleotides are highly accessible and thus are probably localized on the surface of the RNAs. Although the group III aptamers share some of the sequence motifs of group I and group II RNAs, their computed minimal free-energy structures are different. The same was found for the various orphan aptamers.
FIG. 2.
Structural features of group I and group II aptamers. (A) Analytical RNase T1 hydrolysis of 5′ 32P-labeled group I aptamer cl. 57. The analysis was performed under denaturing (de) and nondenaturing (nde) conditions, followed by a gel electrophoretic separation of the hydrolysis fragments and visualization by autoradiography. Arrows indicate the primary hydrolysis sites at positions G41 and G44. “OH” represents an alkaline hydrolysis ladder. Fragment sizes are given in nucleotides (nt). (Right panel) Theoretical secondary structures of group I and group II aptamers. Identical sequence motifs are in black; PBSs are shaded in gray. Arrows emphasize the RNase T1-sensitive nucleotides G44 and G41, which are located within the apical loop of the RNAs. (B) Mapping the boundaries of group II aptamer cl. 9 essential for sVSG 221 binding. 32P-radiolabeled aptamer (5′-labeled left panel and 3′-labeled right panel) was partially hydrolyzed at alkaline conditions, and fragments were incubated with sVSG 221 at concentrations of 0, 5, 15, and 50 nM. sVSG-bound RNA fragments were collected by filter binding, eluted from the filter, and analyzed on denaturing polyacrylamide gels. Arrows indicate the boundaries. The corresponding nucleotide positions are indicated in the secondary structure on the right. PBSs are shaded gray.
To identify nucleotide positions that are essential for the aptamer-VSG interaction, RNA boundary experiments were performed. Both 3′-end-labeled and 5′-end-labeled aptamer preparations were subjected to partial alkaline hydrolysis to create populations of random end-labeled RNA fragments. Adsorption to nitrocellulose filters was used to select for those RNA fragments that are competent to bind sVSG protein. Figure 2B shows a representative set of results using group II aptamer cl. 9 as an example. RNAs shortened by 26 nt from the 5′ end and by 15 nt from the 3′ end retain the ability to interact with the protein. Thus, the active binding domain is ca. 44 nt in length and includes the imperfect hairpin loop as the core binding determinant. Though the 3′ boundary includes 9 nt of the primer binding site, the 5′ boundary starts within the variable region. Almost identical results were derived from the analysis of aptamer cl. 57 as a representative group I RNA, thereby confirming that the central hairpin structure is the main binding domain (data not shown). It should be noted that the precise boundary contains an element of uncertainty due to the fact that the aptamers contain 2′ F-modified pyrimidines. The modified nucleotides are inaccessible to alkaline hydrolysis. As a consequence, the true boundaries lie at or in-between the identified boundary and the next purine in the sequence.
Binding affinities of the selected aptamers.
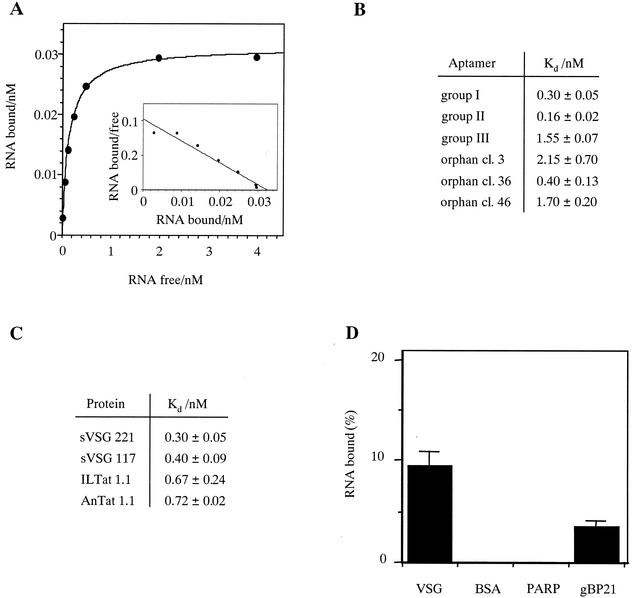
Equilibrium dissociation constants (Kd) for the binding of the three aptamer families to sVSG preparations were determined in nitrocellulose filter-binding assays (19). A representative binding curve and Scatchard analysis is shown in Fig. 3A. The derived Kd values for all three aptamer families are in the nanomolar or even subnanomolar concentration range (Fig. 3B). This is indicative of a high-affinity RNA-protein interaction. The best interactor is aptamer cl. 9 (group II), which binds to sVSG 221 with a Kd of 0.16 ± 0.02 nM. As shown for the group I aptamers (Fig. 3C), the RNAs do not discriminate between sVSG 221 and sVSG 117, the two VSG variants used during the selection experiment: identical Kd values were measured with 0.3 ± 0.05 nM for sVSG 221 and 0.4 ± 0.05 nM for sVSG 117. Strikingly, the aptamers are capable of binding to sVSG preparations not used during the SELEX experiment. As shown in Fig. 3C, group I RNAs bind to sVSG variants from the unrelated T. brucei strains ILTat 1.1 and AnTat 1.1 with Kd values of 0.7 nM. Thus, even sVSG molecules that were never in contact with any of the aptamer pools are recognized by the selected RNAs. This suggests that the RNA molecules might bind to a structural motif of the sVSGs that is present in all tested variants of the protein.
FIG. 3.
Determination of equilibrium dissociation constants (Kd). (A) Representative binding curve with various concentrations of group II aptamer cl. 9 (0.02 to 4 nM) at an sVSG 221 concentration of 4 nM. (Inset) Scatchard plot of the binding data with the slope of the curve representing −1/Kd. (B) Summary of the derived dissociation constants (Kd) for all aptamer families to sVSG 221. (C) Kd values for the binding of group I aptamer cl. 57 to four different VSG variants (sVSG 221, sVSG 117, ILTat 1.1, and AnTat 1.1). Standard deviations were calculated from up to five independent experiments. (D) Relative binding of aptamer cl. 57 to sVSG 221 (50 nM), BSA (50 nM), procyclin (PARP; ≥190 nM), and gBP21 (50 nM). Error bars are standard deviations calculated from three independent experiments.
The specificity of the sVSG-aptamer interaction was analyzed by using three unrelated proteins as controls. BSA was chosen because it represents the dominant protein component in blood serum. As a parasite-specific membrane protein, we used preparations of the procyclin protein family. Although procyclins are only expressed during the insect life cycle stage of the parasite, they are, like VSG, glycosylphosphatidylinositol-anchored surface proteins with the same glycan core structure (20). Lastly, we used gBP21, a mitochondrial RNA/RNA annealing factor from T. brucei. gBP21 has been characterized to bind to irregular RNA hairpin structures (12, 23) and thus should at least in part recognize the hairpin binding core of the selected aptamers. Figure 3D summarizes the results. BSA and the procyclin preparation did not bind aptamer RNA, whereas gBP21, as expected, was able to interact with cl. 57 RNA. However, the binding was significantly less.
Based on the boundary experiments discussed above, the binding affinity of a truncated group II aptamer was determined. The RNA hairpin loop has a length of 47 nt since three G residues were added to the 5′ end to ensure an efficient T7 in vitro transcription. The Kd for this “binding core” aptamer to sVSG 221 is 61 nM. Thus, although the shortened RNA is still able to recognize VSG protein, its binding affinity is significantly weaker than that of the full-length RNA. This indicates that the hairpin loop is able to recognize the surface protein but likely requires additional binding determinants from one or both ends of the RNA. Similar results have been described by Connell et al. (3).
Characterization of the sVSG binding site.
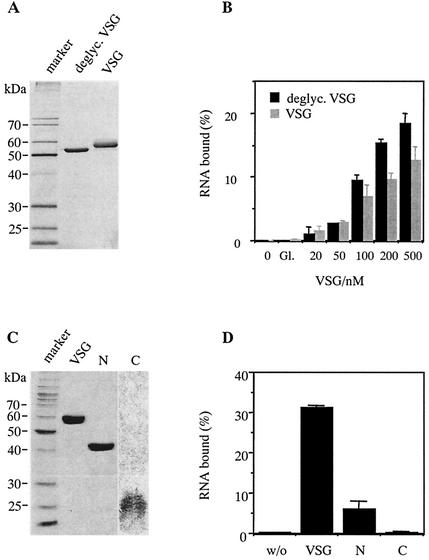
Since VSGs are glycosylated proteins, we tested whether the oligosaccharide side chains (20) might contribute to the binding of the RNAs. Deglycosylated sVSG preparations were prepared by N-glycosidase F treatment (34) (Fig. 4A) and analyzed for their ability to bind aptamer cl. 57 (Fig. 4B). At protein concentrations up to 100 nM deglycosylated sVSG was able to bind RNA indistinguishable from glycosylated sVSG. Thus, the sugar side chains do not contribute to the aptamer-binding domain. At higher protein concentrations (≥200 nM), deglycosylated sVSG bound somewhat better (by ca. 30%), which might suggest a masking effect by the oligosaccharides.
FIG. 4.
Characterization of the aptamer-binding domain on sVSG 221. (A) Silver-stained SDS-containing polyacrylamide gel (12.5% [wt/vol]) of glycosylated and deglycosylated sVSG. (B) Percentage of aptamer cl. 57 bound at different concentrations of glycosylated (VSG) and deglycosylated sVSG (deglyc. VSG). (C) Gel electrophoretic analysis of N-terminal (N) and C-terminal (C) sVSG fragment preparations after digestion with trypsin and fractionation by size exclusion chromatography. Staining was performed by using Coomassie brilliant blue. (D) Percentage of aptamer cl. 57 bound to full-length sVSG and to N-terminal (N) and C-terminal (C) fragments at 4°C. Protein concentrations were at 500 nM. “w/o” refers to a no-protein control. Apparent molecular masses are in kilodaltons, and error bars indicate the standard deviations.
Tryptic digestion of sVSG allows the isolation of an N-terminal fragment of ca. 40 kDa and C-terminal peptides of ca. 25 kDa (16) (Fig. 4C). N-terminal fragments form homodimers similar to full-length sVSG, whereas the C-terminal fragments are in a monomeric state. Using these preparations, we were able to assess whether the aptamer binding site might be restricted to one of two domains. Although the ability of the C-terminal fragments to bind RNA was essentially zero, the N-terminal segment showed 20% of the binding of uncleaved sVSG (Fig. 4D). This indicates that the ability to bind aptamer RNA resides within the full-length protein. However, since the N-terminal fragment was still able to bind, it might at least be part of the RNA interaction domain.
Aptamer stability and importance of the 2′ F-modification for the binding reaction.
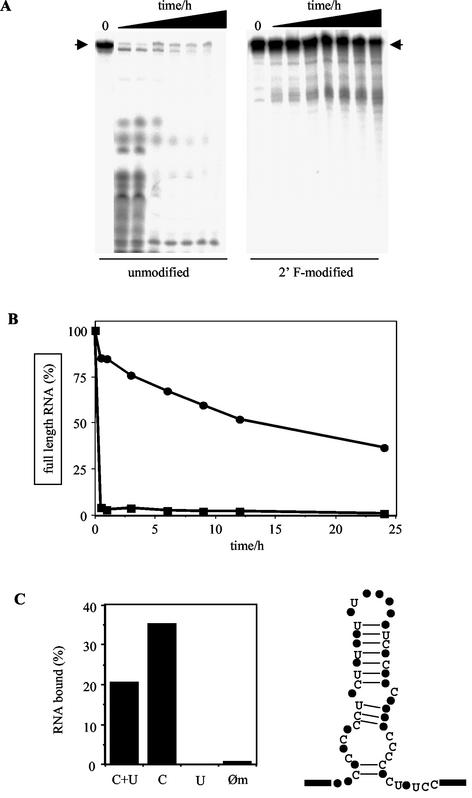
A prerequisite for a potential therapeutic use of the aptamers is a reasonable stability in mammalian serum. This was tested by incubating uniformly 32P-labeled aptamer preparations in blood serum for up to 24 h. As a comparison, we analyzed the 2′ F-modified RNAs side by side to the corresponding unmodified RNAs. Figure 5A shows a representative example with group I aptamer cl. 57. The unmodified RNA has a half-life (t1/2) of ≤10 s, whereas the same RNA containing 2′ F-modified pyrimidines has a t1/2 of ca. 15 h (Fig. 5B). Thus, as expected, the modified aptamers show a significant serum stability.
FIG. 5.
Serum stability of the selected RNA aptamers and influence of the 2′ F-modification on aptamer binding. (A) Stability of unmodified (left) and 2′ F-modified (right) aptamer cl. 57 in mouse serum. Uniformly radiolabeled RNA was incubated in serum for 0, 0.5, 1, 3, 6, 9, 12, and 24 h at 37°C and analyzed in denaturing polyacrylamide gels. Arrows denote the electrophoretic mobility of full-length RNA. (B) Graphical representation of the data in panel A for the determination of half-lives (t1/2). Symbols: •, 2′ F-modified aptamer; ▪, unmodified aptamer. (C) On the left is shown sVSG binding of various aptamer cl. 57 preparations, modified to different degrees. C+U, all C and U nucleotides are modified; C, only C nucleotides are modified; U, only U nucleotides are modified; Øm, no modification. The sVSG concentration was at 150 nM. On the right is the secondary structure of aptamer cl. 57 showing the positions of all C and U nucleotides within the proposed hairpin structure. Boxes represent primer-binding sites.
The importance of the 2′ F-modification for the binding reaction was further tested by comparing the aptamer-sVSG interaction with RNA preparations that were modified to different degrees. Strikingly, unmodified and 2′ F-U-modified aptamers had lost their ability to interact with sVSG, whereas aptamers containing only 2′ F-modified C-nucleotides showed enhanced binding (Fig. 5C). As described before (17, 26), this likely demonstrates that some or all of the 2′ F-C modifications contribute to the higher-order folding of the aptamers which seems to be required for the recognition of the protein.
Aptamer binding to live trypanosomes.
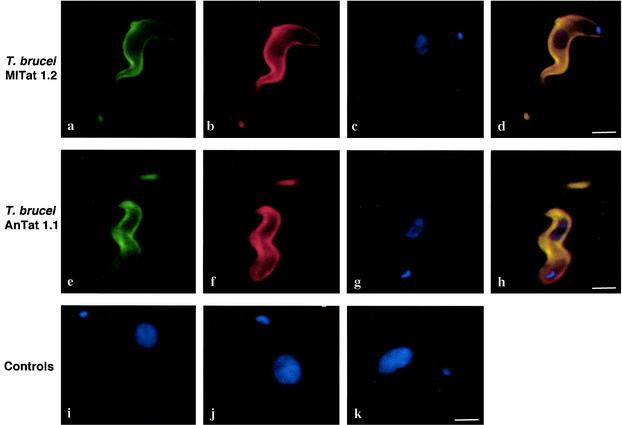
In order to verify the surface binding of the aptamers to live trypanosomes, we performed indirect in situ fluorescence labeling experiments with internally biotinylated aptamer preparations. As a biotinylation substrate, we used the tethered biotin-11-ATP nucleotide (Perkin-Elmer/Life Sciences). The modified ATP was cotranscriptionally incorporated into the aptamer RNAs with a stoichiometry of one to two biotin residues per RNA. After they bound to live trypanosomes, surface-bound, biotin-conjugated aptamers were detected by incubation with Alexa Fluor 488-conjugated streptavidin. Figure 6 summarizes the results. Incubation of T. brucei MITat 1.2 cells (which express VSG 221) with biotinylated group I aptamer cl. 57 resulted in staining of the entire parasite cell surface (Fig. 6a). An identical staining signal was obtained when the parasites were decorated with a rabbit anti-VSG221 antiserum, followed by a fluorophore-coupled anti-rabbit secondary antibody (Fig. 6b). The colocalization of the two signals was verified by the merging of both pictures, along with the localization signals of the nucleus and kinetoplast DNA (Fig. 6c and d). The same results were obtained with T. brucei AnTat 1.1 cells a parasite strain not used during the selection procedure. As controls, we tested the staining of a biotinylated pool 0 RNA preparation (Fig. 6i), a biotinylated tRNA preparation (Fig. 6j), and a nonbiotinylated aptamer preparation (Fig. 6k). In all three cases only background signals were recorded. Taken together, these results indicate that the aptamer RNAs bind to the surface of live trypanosomes. The results show that the RNAs show no VSG-variant specific features and that they are capable of binding different VSG surfaces. Lastly, the results demonstrate that the tethered biotin ligand is accessible on the cell surface. Digitally deconvoluted three-dimensional image data (21) were used to further verify the surface distribution of the aptamers. The data are provided online as supplementary material in the form of two QuickTime animations (http://biosun.bio.tu-darmstadt.de/goringer/index.html).
FIG. 6.
Surface localization of trypanosome-bound, biotin-substituted RNA aptamers. T. brucei MITat 1.2 parasites were incubated with aptamer cl. 57 modified with a tethered biotin side group. (a) Identification of the biotin moiety with Alexa Fluor 488-conjugated streptavidin. (b) Immunodetection of VSG 221 with a rabbit anti-VSG antiserum, followed by an Alexa Fluor 594-conjugated anti-rabbit secondary antibody. (c) DAPI staining of the kinetoplast and nucleus. (d) Merge picture of panels a to c indicating colocalization (yellow). (e to h) Same as panels a to d but with T. brucei AnTat 1.1, a parasite strain not used during the SELEX experiment. Controls: (i) incubation of a MITat 1.2 parasite cell with biotin-conjugated pool 0 RNA followed by incubation with Alexa Fluor 488-conjugated SAV. (j) Staining pattern of a biotin-conjugated tRNA preparation. (k) Staining pattern of nonbiotinylated aptamer cl. 9. The position of the kinetoplast and nucleus is indicated by blue DAPI fluorescence.
Directing antibodies to the trypanosome surface.
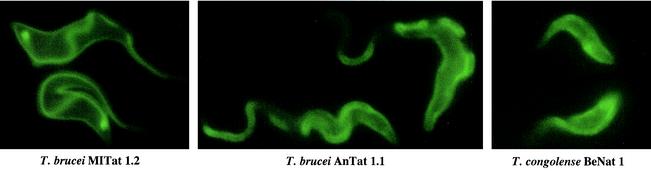
Lastly, as proof-of-concept, we wanted to test whether antibodies could be directed to the surface of the parasite by using an antigen-coupled aptamer preparation. As described above, we used aptamer RNAs that contained a tethered biotin ligand as a model antigen. As shown in Fig. 7, fluorophore-coupled anti-biotin antibodies were able to recognize the biotin-modified RNAs on the surface of T. brucei MITat 1.2 cells, on AnTat 1.1 trypanosomes, and even on the surface of another species: T. congolense (strain BeNat 1). Thus, at the in vitro incubation conditions, antigen-coupled aptamer preparations can be used to direct antibodies to the surface of trypanosome cells.
FIG. 7.
Antibody binding to surface-bound RNA aptamers. T. brucei MITat 1.2 cells, AnTat 1.1, and T. congolense strain BeNat 1 (from left to right) were incubated with biotinylated aptamer cl. 57, followed by incubation with a monoclonal Cy3-conjugated anti-biotin antibody.
DISCUSSION
African trypanosomes escape the immune response of the infected host by antigenic variation. Here we demonstrate the selection of high-affinity RNA ligands to the VSG of the parasite. Three aptamer families were isolated which, on the primary structure level, share a significant sequence identity (37%). The two main aptamer families (groups I and II) likely adopt irregular hairpin secondary structures with invariant sequence motifs in single-stranded domains. This suggests a common higher-order folding of the selected RNAs and further implicates that specific contacts to the VSG binding target might be mediated by the single-stranded sequence motifs. RNA structure probing experiments with RNase T1 experimentally confirmed the basic features of the aptamer hairpin structure. However, more sophisticated structural details likely exist. Evidence for an intricate higher-order folding was derived from experiments determining the binding core of the two aptamer groups. Although the hairpin structure was clearly identified as the main binding determinant to the VSG target, its binding affinity as an isolated RNA molecule was reduced. Therefore, sequences outside of the hairpin element must contribute to the high-affinity interaction, and these sequences can be presented in trans (i.e., out of the continuous sequence context of the aptamer). Similar phenomena have been described before (3) and argue for a detailed structural analysis of the selected RNAs.
Regardless of the precise three-dimensional folding of the selected aptamers, RNAs lacking the 2′ F-modified pyrimidine nucleotides were found to be highly unstable in mouse serum. The determined half lives are in the range of only a few seconds. In contrast, in the presence of the ribose modification the aptamers show serum stabilities of many hours. Thus, as described by Pieken et al. (28), a significant serum stability was achieved by using a ribose-substituted RNA library in the SELEX experiment. Furthermore, we identified the 2′ F-modified C-nucleotides as being essential for the aptamer-VSG interaction. Although we cannot rule out that some or all of the 2′ F-atoms are in direct contact with the protein, this seems rather unlikely. As a more realistic scenario, we suggest that the 2′ F-modifications contribute to the higher-order folding of the RNA molecules that might be one of the determining factors for the binding to the protein. We have no satisfactory explanation as to why only the modifications of the C-nucleotides showed the described effect, aside from the fact that C-nucleotides are more abundant than Us in aptamer cl. 57 (see Fig. 5C).
All three aptamer families bind to sVSG with affinities in the nanomolar or subnanomolar concentration range and thus are of high affinity. Importantly, as implemented in the selection strategy, none of the RNAs is capable of distinguishing between the two sVSG variants used during the selection process. Moreover, all aptamers bind with high affinity to sVSG variants that were not used during the SELEX experiment. This demonstrates that the same or an almost-identical aptamer binding site is present in all tested sVSGs and further suggests that the RNA-protein interaction likely takes place within a structurally conserved domain of the different VSG molecules. Lastly, it is important to note that the RNAs have no significant affinity for serum albumin, which is the most abundant protein component in blood.
A comparison of the aptamer-binding ability of full-length sVSG with that of isolated N-terminal and C-terminal fragments demonstrated that the full-length protein is the preferred binding partner. C-terminal fragments do not bind added aptamer, and the N-terminal polypeptide has only 20% of the RNA-binding ability of uncleaved sVSG. Nonetheless, this might suggest that the aptamer binding site is localized on the N-terminal domain of sVSG. The reduced RNA binding of this fragment could be the result of a conformational change induced by the proteolytic cleavage. However, we cannot exclude that the aptamer binding site is located near or at the trypsin cleavage site which separates the N-terminal peptide from the C-terminal fragments and that the C-terminal polypeptides lose their ability to bind RNA because of their inability to form dimers. Additional support for an N-terminal aptamer-binding domain can be drawn from the result that aptamer cl. 57 is able to recognize the surface of T. congolense. Although most of the known T. congolense VSGs resemble the N-terminal cysteine residue domain type B of T. brucei, they lack corresponding C termini (38). Thus, the N-terminal region of sVSG is the most probable interaction domain for the RNAs, although further evidence is required to substantiate this observation.
Lastly, we verified that the N-Asn linked sugar side chain modifications of the sVSG protein do not contribute to the binding of the RNAs. Deglycosylated sVSG interacts with the aptamer RNAs indistinguishable from glycosylated protein. On the contrary, at higher sVSG concentrations, the sugar modification seems to have a negative effect on RNA binding since deglycosylated sVSG binds moderately better. This can be interpreted as a masking effect probably as a result of the conformational flexibility of the extended sugar structures. This is supported by the crystal structure of the N-terminal domain of sVSG 221 (10). The glycosylation site is located at Asn-263 in the lower half of the N-terminal fragment. Most of the core oligosaccharides attached to Asn-263 pack against the surface of the protein in a defined pocket between two helices. However, the remainder of the oligosaccharide residues could not be located likely due to a high degree of structural flexibility.
Importantly, the aptamer-VSG interaction is not restricted to sVSG preparations. Aptamers substituted with a tethered biotin residue bind to the VSG surface of live trypanosomes. Since fluorescently labeled streptavidin was used for the detection, this implies that the tethered biotin side group is accessible on the surface. The N-terminal VSG domain is a rod-like structure with a length of ca. 100 Å (10). For the bulged hairpin of aptamer cl. 57 plus the tethered biotin ligand, we calculated a minimal length of roughly 60 Å. Thus, the aptamer might penetrate into the VSG layer to some degree, which might explain the observed shielding effect of the oligosaccharide side chains at higher sVSG concentrations (Fig. 4B).
The fluorescence signals from both the streptavidin and the anti-biotin antibody are distributed over the entire surface of the parasite and colocalize with the position of the VSG molecules. This is consistent with the aptamer-sVSG in vitro binding data and demonstrates that anti-biotin antibodies can reach the aptamer-bound biotin ligand. This further illustrates that substituted aptamer preparations can be used to direct antibodies to the otherwise-variable surface of the parasite. Whether this can stimulate an agglutination reaction in vivo (33) remains to be tested. However, other scenarios can be envisaged as well, e.g., the presentation of nonvariant, aptamer-coupled T-cell epitopes on the surface of the parasite in order to activate complement by the alternative pathway (7, 36).
Collectively, the described results open the possibility of using the VSG-specific aptamers as components to side-step antigenic variation. The RNAs have the potential to function as carrier-type delivery molecules for the development of new therapeutic and diagnostic strategies against trypanosome infections (within this context, see also reference 15). Finally, it should be noted that the experimental approach might have a much broader application spectrum, especially in the case of other extracellular pathogens.
Supplementary data.
Supplementary data for the present study are available online (http://biosun.bio.tu-darmstadt.de/goringer/index.html).
Acknowledgments
We thank A. S. Paul for comments on the manuscript. M.E. acknowledges the support of M. Boshart.
The work was supported by an International Research Scholar grant from the Howard Hughes Medical Institute to H.U.G. and the DFG-Schwerpunkt grant En305 to M.E.
REFERENCES
- 1.Blum, M. L., J. A. Down, A. M. Gurnett, M. Carrington, M. J. Turner, and D. C. Wiley. 1993. A structural motif in the variant surface glycoproteins of Trypanosoma brucei. Nature 362:603-609. [DOI] [PubMed] [Google Scholar]
- 2.Clayton, E. C., and M. R. Mowatt. 1989. The procyclic acidic repetitive proteins of Trypanosoma brucei. J. Biol. Chem. 264:15088-15093. [PubMed] [Google Scholar]
- 3.Connell, G. J., M. Illangesekare, and M. Yarus. 1993. Three small ribooligonucleotides with specific arginine sites. Biochem. J. 32:5497-5502. [DOI] [PubMed] [Google Scholar]
- 4.Cross, G. A. M. 1975. Identification, purification and properties of clone-specific glycoprotein antigens constituting the surface coat of Trypanosoma brucei. Parasitology 71:393-417. [DOI] [PubMed] [Google Scholar]
- 5.Cross, G. A. M. 1984. Release and purification of Trypanosoma brucei variant surface glycoprotein. J. Cell. Biochem. 24:79-90. [DOI] [PubMed] [Google Scholar]
- 6.Cross, G. A. M. 1996. Antigenic variation in trypanosomes: secrets surface slowly. Bioessays 18:283-291. [DOI] [PubMed] [Google Scholar]
- 7.De Almeida, M. L. C., and M. J. Turner. 1983. The membrane form of variant surface glycoproteins of Trypanosoma brucei. Exp. Nat. 302:349-352. [DOI] [PubMed] [Google Scholar]
- 8.Ferguson, M. A. J. 2000. Glycosylphosphatidylinositol biosynthesis validated as a drug target for African sleeping sickness. Proc. Natl. Acad. Sci. USA 97:10673-10675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferguson, M. A. J., P. Murray, H. Rutherford, and M. J. McConville. 1993. A simple purification of procyclic acidic repetitive protein and demonstration of a sialylated glycosyl.phosphatidylinositol membrane anchor. Biochem. J. 291:51-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Freymann, D., J. Down, M. Carrington, I. Roditi, M. Turner, and D. Wiley. 1990. 2.9 Å resolution structure of the N-terminal domain of a variant surface glycoprotein from Trypanosoma brucei. J. Mol. Biol. 216:141-160. [DOI] [PubMed] [Google Scholar]
- 11.Green, M. R., J. N. Pastewka, and A. C. Peacock. 1973. Differential staining of phosphoproteins on polyacrylamide gels with a cationic carbocyanine dye. Anal. Biochem. 56:43-51. [DOI] [PubMed] [Google Scholar]
- 12.Hermann, T., B. Schmid, H. Heumann, and H. U. Göringer. 1997. A three-dimensional working model for a guide RNA from Trypanosoma brucei. Nucleic Acids Res. 25:2311-2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hirumi, H., and K. Hirumi. 1994. Axenic culture of African trypanosome bloodstream forms. Parasitol. Today 10:80-84. [DOI] [PubMed] [Google Scholar]
- 14.Homann, M., and H. U. Göringer. 1999. Combinatorial selection of high-affinity RNA ligands to live African trypanosomes. Nucleic Acids Res. 27:2006-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Homann, M., and H. U. Göringer. 2001. Uptake and intracellular transport of RNA aptamers in African trypanosomes suggest therapeutic “piggy-back” approach. Bioorg. Med. Chem. 9:2571-2580. [DOI] [PubMed] [Google Scholar]
- 16.Johnson, J. G., and G. A. Cross. 1979. Selective cleavage of variant surface glycoproteins from Trypanosoma brucei. Biochem. J. 178:689-697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kubik, M. F., C. Bell, T. Fitzwater, S. R. Watson, and D. M. Tasset. 1997. Isolation and characterization of 2′-fluoro-, 2′-amino-, and 2′-fluoro-/amino-modified RNA ligands to human IFN-γ that inhibit receptor binding. J. Immunol. 159:259-267. [PubMed] [Google Scholar]
- 18.Lamont, G. S., R. S. Tucker, and G. A. Cross. M. 1986. Analysis of antigen switching rates in Trypanosoma brucei. Parasitology 92:355-367. [DOI] [PubMed] [Google Scholar]
- 19.Lever, J. E. 1972. Quantitative assay of the binding of small molecules to protein: comparison of dialysis and membrane filter assays. Anal. Biochem. 50:73-83. [DOI] [PubMed] [Google Scholar]
- 20.McConville, M. J. 1996. Glycosyl-phosphatidylinositols and the surface architecture of parasitic protozoa, p. 205-228. In Molecular biology of parasitic protozoa. Oxford University Press, Oxford, United Kingdom.
- 21.McNally, J. G., T. Karpova, J. Cooper, and J. A. Conchello. 1999. Three-dimensional imaging by deconvolution microscopy. Methods 19:373-385. [DOI] [PubMed] [Google Scholar]
- 22.Michiels, F., G. Matthyssens, P. Kronenberger, E. Pays, B. Dero, S. Van Assel, M. Darville, A. Cravador, M. Steinert, and R. Hamers. 1983. Gene activation and re-expression of a Trypanosoma brucei variant surface glycoprotein. EMBO J. 2:1185-1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Müller, U. F., L. Lambert, and H. U. Göringer. 2001. Annealing of RNA editing substrates facilitated by guide RNA-binding protein gBP21. EMBO J. 20:1394-1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Overath, P., M. Chaudhri, D. Steverding, and K. Ziegelbauer. 1994. Invariant surface proteins in bloodstream forms of Trypanosoma brucei. Parasitol. Today 10:53-58. [DOI] [PubMed] [Google Scholar]
- 25.Padilla, R., and R. Sousa. 1999. Efficient synthesis of nucleic acids heavily modified with non-canonical ribose 2′-groups using a mutant T7 RNA polymerase RNAP. Nucleic Acids Res. 27:1561-1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pan, W., R. C. Craven, Q. Qiu, C. B. Wilson, J. W. Wills, S. Golovine, and J. W. Wang. 1995. Isolation of virus-neutralizing RNAs from a large pool of random sequences. Proc. Natl. Acad. Sci. USA 92:11509-11513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pays, E., and D. P. Nolan. 1998. Expression and function of surface proteins in Trypanosoma brucei. Mol. Biochem. Parasitol. 91:3-36. [DOI] [PubMed] [Google Scholar]
- 28.Pieken, W. A., D. Olsen, F. Benseler, H. Aurup, and F. Eckstein. 1991. Kinetic characterization of ribonuclease-resistant 2′-modified hammerhead ribozymes. Science 253:314-317. [DOI] [PubMed] [Google Scholar]
- 29.Rice-Ficht, A. C., K. K. Chen, and J. E. Donelson. 1982. Point mutations during generation of expression-linked extra copy of trypanosome surface glycoprotein gene. Nature 298:676-679. [DOI] [PubMed] [Google Scholar]
- 30.Rudenko, G., M. Cross, and P. Borst. 1998. Changing the end: antigenic variation orchestrated at the telomeres of African trypanosomes. Trends Microbiol. 6:113-116. [DOI] [PubMed] [Google Scholar]
- 31.Smith, D. H., J. Pepin, and A. H. R. Stich. 1998. Human African trypanosomiasis: an emerging public health crisis. Br. Med. Bull. 54:341-355. [DOI] [PubMed] [Google Scholar]
- 32.Sternberg, J. M. 1998. Immunobiology of African trypanosomiasis. Chem. Immunol. 70:186-199. [DOI] [PubMed] [Google Scholar]
- 33.Takayanagi, T., and G. L. Enriquez. 1973. Effects of the IgG and IgM immunoglobulins in Trypanosoma gambiense infections in mice. J. Parasitol. 59:644-647. [PubMed] [Google Scholar]
- 34.Tarentino, A. L., C. M. Gomez, and T. H. Plummer, Jr. 1985. Deglycosylation of asparagine-linked glycans by peptide N-glycosidase F. Biochemistry 24:4665-4671. [DOI] [PubMed] [Google Scholar]
- 35.Taylor, K. A. 1998. Immune responses of cattle to African trypanosomes: protective or pathogenic? Int. J. Parasitol. 28:219-240. [DOI] [PubMed] [Google Scholar]
- 36.Tetley, L., K. Vickerman, and S. K. Moloo. 1981. Absence of a surface coat from metacyclic Trypanosoma vivax: possible implications for vaccination against vivax trypanosomiasis. Trans. R. Soc. Trop. Med. Hyg. 75:409-414. [DOI] [PubMed] [Google Scholar]
- 37.Tuerk, C., and L. Gold. 1990. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 249:505-510. [DOI] [PubMed] [Google Scholar]
- 38.Urakawa, T., Y. Eshita, and P. A. Majiwa. 1997. The primary structure of Trypanosoma (Nannomonas) congolense variant surface glycoproteins. Exp. Parasitol. 85:215-224. [DOI] [PubMed] [Google Scholar]
- 39.Van der Ploeg, L. H. T., D. Valerio, T. De Lange, A. Bernards, P. Borst, and F. G. Grosveld. 1982. An analysis of cosmid clones of nuclear DNA from Trypanosoma brucei shows that the genes for variant surface glycoproteins are clustured in the genome. Nucleic Acids Res. 10:5905-5923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wilson, D. S., and J. W. Szostak. 1999. In vitro selection of functional nucleic acids. Annu. Rev. Biochem. 68:611-647. [DOI] [PubMed] [Google Scholar]
- 41.Ziegelbauer, K., and P. Overath. 1993. Organization of two invariant surface glycoproteins in the surface coat of Trypanosoma brucei. Infect. Immun. 61:4540-4545. [DOI] [PMC free article] [PubMed] [Google Scholar]