Abstract
The xylanase system of the filamentous fungus Hypocrea jecorina (Trichoderma reesei) consists of two specific xylanases, Xyn1 and Xyn2, which are simultaneously expressed during growth on xylan but respond differentially to low-molecular-weight inducers. Using in vivo footprinting analysis of xylan-induced and noninduced mycelia, we detected two adjacent nucleotide sequences (5′-AGAA-3′ on the noncoding strand and 5′-GGGTAAATTGG-3′, referred to as the xylanase-activating element [XAE], on the coding strand, respectively) to bind proteins. Among these, binding to the AGAA-box is only observed under noninduced conditions, whereas binding to XAE is constitutive. Electrophoretic mobility shift assay with heterologously expressed components of the H. jecorina Hap2/3/5 protein complex and the cellulase regulator Ace2 suggests that these two transactivators form the protein complex binding to XAE. H. jecorina transformants, containing correspondingly mutated versions of the xyn2 promoter fused to the Aspergillus niger goxA gene as a reporter, revealed that the elimination of protein binding to the AGAA-box resulted in a threefold increase in both basal and induced transcription, whereas elimination of Ace2 binding to its target in XAE completely eliminated transcription under both conditions. Destruction of the CCAAT-box by insertion of a point mutation prevents binding of the Hap2/3/5 complex in vitro and results in a slight increase in both basal and induced transcription. These data support a model of xyn2 regulation based on the interplay of Hap2/3/5, Ace2 and the AGAA-box binding repressor.
β-1,4-Xylans are heteropolysaccharides that have a backbone of β-1,4-linked xylopyranosyl residues, to which side groups such as d-glucuronic acid, l-arabinose, p-coumaric acid, and ferulic acid are attached and which constitute 20 to 35% of the roughly 830 Gt of annually formed renewable plant biomass (33). Enzymes capable of degrading the xylan backbone are formed by various microorganisms and comprise endoxylanases (EC 3.2.1.8) and β-xylosidase (EC 3.2.1.91) (15). Among these, the xylanases of the ascomycete Hypocrea jecorina (anamorph Trichoderma reesei) have received strong attention because of their application in the pulp and paper and feed industry (4).
Although the biochemistry of xylan degradation by H. jecorina has now been studied in detail (2, 10, 23, 25), the mechanism by which the fungus regulates the formation of its xylanases is still largely unsolved (44). H. jecorina forms two specific endo-β-1,4-xylanases, Xyn1 and Xyn2 (EC 3.2.1.8), and the respective genes (xyn1 and xyn2) have been cloned (27, 34). Expression of xyn1 is induced by d-xylose and is repressed by glucose in a Cre1-dependent manner (18, 44), whereas the expression of xyn2 is partially constitutive and further induced by xylobiose, xylan, cellulose, and sophorose (44). However, the regulatory circuits governing xyn1 and xyn2 gene expression have not yet been elucidated. In Aspergillus niger, expression of the xylanolytic system is regulated by the zinc binuclear cluster type transcriptional regulator XlnR (36), in which it appears to be a central regulator, since it controls the expression of more than 10 genes not only involved in the degradation of xylan but also in xylose metabolism and cellulose degradation (6, 9, 35). The cloning of a xlnR homologue of H. jecorina has not yet been reported, but nucleotide motifs resembling the consensus for XlnR binding (5′-GGCAAA-3′) are present in the xyn1 and xyn2 5′-upstream sequences (unpublished data). On the other hand, Aro et al. (1) recently reported that a H. jecorina mutant, in which the gene encoding the cellulase regulator ace2 had been disrupted, exhibited reduced expression of xyn2.
Using promoter deletion analysis, we previously reported that a 55-bp fragment of the xyn2 promoter contains all of the information necessary for regulating xyn2 gene expression (44). Here we identify the nucleotide sequences within these 55 bp that are essential for binding of proteins and responsible for xyn2 regulation by using both in vitro and in vivo strategies. Furthermore, we show that both basal transcription and induction of xyn2 depends on the binding of the Hap2/3/5 complex (42) and Ace2 to an undecameric motif (5′-GGGTAAATTGG-3′; the xylanase-activating element [XAE]) and that this binding is counteracted by an as-yet-unknown DNA-binding protein (complex) binding to an AGAA-box immediately upstream of XAE.
MATERIALS AND METHODS
Microbial strains and plasmids.
H. jecorina QM9414 (ATCC 26921) and H. jecorina RUT C30 (ATCC 56756) were used throughout the present study. H. jecorina TU-6 (8), a pyr4-null mutant of QM9414, was used as recipient strain for pyr4-mediated cotransformation experiments. The strains were maintained on malt agar which contained 5 mM uridine in the case of TU-6. Escherichia coli JM109 (41) was used for the propagation of vector molecules. Plasmids pFG1 (8), pAT3 (34), and pSJ3 (17) were obtained from our department stock.
Fungal growth, induction of xylanases, and preparation of cell extracts.
H. jecorina was grown on glucose and xylan or induced by sophorose and xylobiose, respectively, as described previously (18, 44). Xylan from oat spelts (Sigma, Steinheim, Germany) was used throughout. Cell extracts were prepared as described previously (30).
Construction of pLW reporter plasmids.
The pLW reporter plasmid series was developed from plasmid pSJ3 by fusing the H. jecorina xyn2 5′ noncoding regions (−1 to −850) to the goxA (glucose oxidase-encoding) structural gene of A. niger as reporter. To construct pLW-WT, primers CKT087 and CKT088 (Table 1) were used to amplify a 1,081-bp fragment from the xyn2 5′ noncoding sequences, thereby also generating an additional NheI terminal site at the 3′ end. Amplification was performed with Taq polymerase (Promega, Madison, Wis.) and pAT3 as the template DNA in a Biometra thermocycler by applying 30 cycles of 1 min at 95°C, 1 min at 56°C, and 1 min at 74°C. The PCR product was cloned into pGEM-T (Promega) to yield pLW1, sequenced, and digested with SalI and NheI. Due to a SalI restriction site within the amplified sequence and the gained NheI site at the 3′ end, a 0.8-kb fragment was released and thereafter fused to goxA of pSJ3 by replacing the nag1 5′-noncoding region (cut with XhoI/XbaI). To yield pLWm1, a four-primer PCR mutagenesis strategy was followed: in a first step, two overlapping fragments containing the intended point mutations were amplified with the primers CKT087/xyn2prm2r and CKT088/xynfprm2f (Table 1), respectively, by using pAT3 as a DNA template and Herculase (Stratagene, La Jolla, Calif.) lacking terminal transferase activity. Derived fragments were thereafter subjected as a template to a second PCR with primers CKT087 and CKT088 by using Taq polymerase. PCR protocols and further processing of the derived amplicon essentially followed the procedure described for constructing pLW-WT.
TABLE 1.
Oligonucleotides used throughout this studya
| Name | Sequence | Nucleotide range (reference) |
|---|---|---|
| CKT087 | 5′-ATCGGAGTCGACACTCGCATCCG-3′ | −1081 to 1059 (43) |
| CKT088 | 5′-ATGCTAGCGTTGATGTCTTCTTGCTTCAGC-3′ | −22 to −1 (43) |
| Prxyn2f | 5′-TGATGAAAGGAGAACAACTTCTAGACTGGGTAAATTGGTCAAT-3′ | −249 to −207 |
| Prxyn2f | 5′-AGCGGCTGGCCATTGACCAATTTACCCAGTCTAGAAGTTGTTCTCC-3′ | −196 to −241 |
| Prxyn2M2f | 5′-TGATGAAAGGAGAACAACGGAGAGACTGGGTAAATTGGTCAAT-3′ | −249 to −207 |
| Prxyn2M2r | 5′-AGCGGCTGGCCATTGACCAATTTACCCAGTCTCTCCGTTGTTCTCC-3′ | −196 to −241 |
| Prxyn2bf | 5′-CTAGACTGGGTAAATTGGTCAATGGCCAGCCGCTCGGCCGTGCGGAGACGAGGCA-3′ | −229 to −175 |
| Prxyn2br | 5′-AGCTTGCCTCGTCTCCGCACGGCCGAGCGGCTGGCCATTGACCAATTTACCCAGT-3′ | −171 to −225 |
| Prxyn2bM3f | 5′-CTAGACTGGGTAAAAAGGTCAATGGCCAGCCGCTCGGCCGTGCGGAGACGAGGCA-3′ | −229 to −175 |
| Prxyn2bM3r | 5′-AGCTTGCCTCGTCTCCGCACGGCCGAGCGGCTGGCCATTGACCTTTTTACCCAGT-3′ | −171 to −225 |
| Prxyn2bM4f | 5′-CTAGACTGGGTTTATTGGTCAATGGCCAGCCGCTCGGCCGTGCGGAGACGAGGCA-3′ | −229 to −175 |
| Prxyn2bM4r | 5′-AGCTTGCCTCGTCTCCGCACGGCCGAGCGGCTGGCCATTGACCAATAAACCCAGT-3′ | −171 to −225 |
| Prxyn2bM5f | 5′-CTAGACTGTGTAAATTGGTCAATGGCCAGCCGCTCGGCCGTGCGGAGACGAGGCA-3′ | −229 to −175 |
| Prxyn2bM5r | 5′-AGCTTGCCTCGTCTCCGCACGGCCGAGCGGCTGGCCATTGACCAATTTACACAGT-3′ | −171 to −225 |
| xyn2P1n | 5′-CTTCTCACATGTATCGGAGCAGAAG-3′ | −357 to −333 |
| xyn2P2n | 5′-ATCTATGTGAGCTGACTCGAGACGGC-3′ | −326 to −301 |
| xyn2P3n: | 5′-AGCTGACTCGAGACGGCTGAGACAGCA-3′ | −317 to −291 |
| xyn2P1c | 5′-CTAGGAGTTGTTGTGTCTTTTG-3′ | −25 to −46 |
| xyn2P2c | 5′-GTTGTTCAGGTTTGGCTTCTTG-3′ | −83 to −104 |
| xyn2P3c | 5′-CAGGTTTGGCTTCTTGAGTCTTTAGG-3′ | −89 to −114 |
| Prxyn2af | 5′-TGATGAAAGGAGAACAACTTCTAGACTG-3′ | −249 to −222 |
| Prxyn2ar | 5′-TGACCAGTCTAGAAGTTGTTCTCCTTTC-3′ | −222 to −245 |
| Prxyn2aM2f | 5′-TGATGAAAGGAGAACAACGGAGAGACTG-3′ | −249 to −222 |
| Prxyn2aM2r | 5′-TGACCAGTCTCTCCGTTGTTCTCCTTTC-3′ | −222 to −245 |
| Prxyn1.1f | 5′-TTGGCAGGCTAAATGCGACATCTTAGCCGGA-3′ | −430 to −400 |
| Prxyn1.1r | 5′-TGCATCCGGCTAAGATGTCGCATTTAGCCTG-3′ | −396 to −421 |
Oligonucleotides already used in earlier studies are referenced. Positions of the oligonucleotides in the respective promoter are given. Underlined letters indicate bases added for labeling or generating restriction enzyme sites, and double underlined bases indicate introduced point mutations.
All further mutations of the xyn2 5′-noncoding region were performed in pLW1 as a template and thereafter transferred into pSJ3 as described above. To obtain pLWM2, pLW1 was cleaved with XbaI and HindIII; protruding ends were blunted with the mung bean exonuclease (Promega) by applying 0.12 U (37°C, 30 min)/μg of DNA and thereafter religated. To construct pLWM3, pLWM4, and pLWM5, the synthetic oligonucleotides Prxyn2bM3f and Prxyn2bM3r; Prxyn2bM4f and Prxyn2bM4r, and Prxyn2bM5f and Prxyn2bM5r (Table 1) containing the respective point mutations were annealed, thereby producing appropriate 5′-protruding ends for an insertion into the XbaI- and HindIII-cleaved plasmid pLW1.
Isolation and manipulation of nucleic acids.
Genomic DNA was isolated as described previously (8). After electrophoretic separation, DNA was blotted onto Hybond-N membranes (Amersham Pharmacia Biotech, Little Chalfont, United Kingdom) and hybridized at 64°C for 20 h according to standard protocols (28). Washing was performed with 0.2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% (wt/vol) sodium dodecyl sulfate (SDS) at 68°C (2 times for 15 min each time). Standard methods were used for plasmid isolation, restriction enzyme digestion, and random priming (28).
DNA transformations.
E. coli transformations were carried out according to standard techniques (28). Transformation of H. jecorina TU-6 was carried out according to an optimized protocol for particle bombardment (11), applying cotransformation of pFG1 with the respective reporter constructs. For each construct, at least 30 uridine prototrophs were purified to mitotic stability, and their DNA was isolated and examined for integration and copy number by Southern analysis. Cotransformation frequency varied from 90 to 100%, yielding copy numbers between 1 and 5, integrated into ectopic loci.
Determination of gene copy number and integration locus in transformants.
Southern hybridization was carried out as described by Sambrook et al. (28). Chromosomal DNA was digested with PstI and hybridized with a [α-32P]dCTP-labeled 440-bp fragment bearing 312 bp of the xyn2 promoter and 128 bp of the goxA gene. The endogenous xyn2 gene and the integrated vector copies were quantified by imagizer analysis, and the values obtained were normalized to the length of the labeled probe.
Glucose oxidase assay.
Glucose oxidase activity was assayed as described previously (17). One unit of activity is defined as the amount of enzyme which oxidizes 1 μmol of glucose per min at pH 5.8 and 25°C.
EMSA.
Oligonucleotides used for electrophoretic mobility shift assay (EMSA) were annealed with their complementary oligonucleotides (Table 1) and end labeled with [α-32P]dCTP by using Sequenase version 2.0 (Amersham Pharmacia Biotech). The resulting double-stranded oligonucleotides were purified by nondenaturing polyacrylamide gel electrophoresis (PAGE). The binding assay and PAGE were performed essentially as described previously (30). Binding was achieved by incubating 100 μg of protein of the cell extract with 5 ng of labeled fragment (15 min, 0°C). For competition experiments, unlabeled synthetic oligonucleotides were used in a 50- or 100-fold molar excess. Unlabeled oligonucleotides were annealed with the complementary synthetic oligonucleotide as described by Strauss et al. (31). After annealing, double strands were filled in by using Sequenase version 2.0 (Amersham Pharmacia Biotech). Fragments bearing DNA-binding regions of A. niger XlnR and H. jecorina Hap2, Hap3, Hap5 and Ace2 were expressed as glutathione S-transferase (GST) fusion proteins in strain BL21 as described by van Peij et al. (36), Zeilinger et al. (42), and Aro et al. (1), respectively. The resulting GST fusion proteins and the proteins after thrombin cleavage were used in EMSAs at the following concentrations: GST-XlnR (0.1, 0.2, 0.5, and 1 μg), XlnR (0.05, 0.1, 0.2, and 0.5 μg); Hap2, Hap3, and Hap5 (0.5 μg each); GST-Ace2 (1 μg); and Ace2 (0.5 μg). Oligonucleotide PRxyn1.1 (Table 1) was used as a positive control for EMSA with XlnR.
In vivo genomic footprinting via ligation-mediated PCR.
Methylation of genomic DNA was performed at 30°C in a shaking water bath by transferring 18-ml aliquots of H. jecorina cultures grown on glucose, glycerol, or xylan for the period indicated to 100-ml Erlenmeyer flasks and pulsing them with 40 μl of DMS in 2 ml of 200 mM methyl ethanesulfonate MES buffer (pH 5.5) for 2 min. Methylation was stopped by addition of 50 ml of ice-cold TLEβ buffer (10 mM Tris [pH 8], 1 mM EDTA, 300 mM LiCl, 2% [vol/vol] β-mercaptoethanol). Mycelial samples were filtered and washed twice with 50 ml of TLEβ buffer, and genomic DNA was extracted according to a standard protocol (8). The extracted methylated DNA was cleaved at methylated guanine and adenine residues by incubation of 20 μl of DNA with 1.25 μl of 0.5 M HCl for 1.5 h on ice. After precipitation with ethanol, the DNA was disolved in 48 μl of bidistilled water and incubated at 90°C for 30 min with 2 μl of 1 M NaOH, followed by a second precipitation with ethanol. Finally, DNA was dissolved in 20 μl of Tris-EDTA. In vitro methylation and cleavage of genomic DNA was performed as described by Mueller and Wold (22). Methylated and cleaved DNA was analyzed by ligation-mediated PCR as described by Garrity and Wold (5), and as modified by Wolschek et al. (38), by using Vent polymerase (NEB, Beverly, Mass.). To visualize the noncoding strand, the primers xyn2P1n, xyn2P2n, and xyn2P3n were used, and to visualize the coding strand, the primers xyn2P1c, xyn2P2c, and xyn2P3c were used.
Chromatin analysis.
Micrococcal nuclease (MNase)-based mapping of chromatin organization was carried out as described previously (7). Mycelia were harvested by filtration, pressed dry with filter paper, frozen in liquid nitrogen, and ground to a fine powder. Then, 200-mg portions of the mycelial powder were suspended in 2 ml of nuclease digestion buffer (15 mM Tris-HCl [pH 7.5], 250 mM sucrose, 60 mM KCl, 15 mM NaCl, 0.05 mM CaCl2, 3 mM MgCl2, 0.5 mM dithiothreitol). Next, 200-μl aliquots of the digestion mixture were incubated with MNase for 5 min at 37°C by using various MNase concentrations of between 2 U (100 U/g of mycelium) and 0.01 U (0.5 U/g of mycelium). The reaction was terminated by adding 200 μl of 40 mM EDTA-2% SDS, followed by two rounds of phenol-chloroform extraction and ethanol precipitation. The resuspended DNA was treated with RNase A and passed through a Sephacryl S-200 microspin column (Amersham Pharmacia Biotech). For the preparation of the naked DNA control, 20 μg of purified chromosomal DNA were digested with appropriate concentrations of MNase as described above and then processed as for the chromatin samples.
Indirect end-labeling analysis.
Indirect end labeling was carried out as described previously (39). After secondary restriction enzyme digestion with NsiI, the samples were electrophoresed in 1.2% agarose gels in 1× TAE, transferred to nylon membranes (Hybond-N+; Amersham Pharmacia Biotech) in 10× SSC, and hybridized at 42°C for 20 h in a hybridization solution containing 50% formamide, 5× SSPE, 5× Denhardt solution, 0.5% SDS, 10% (wt/vol) dextran sulfate, and 200 μg of denatured herring sperm DNA/ml. Labeling of the specific probe (0.4-kb HindIII/NsiI fragment isolated from plasmid pAT3) was performed by random priming. The autoradiographs were analyzed by calculating the Rf values for each band.
RESULTS
Identification of in cis-acting elements involved in the regulation of xyn2 expression.
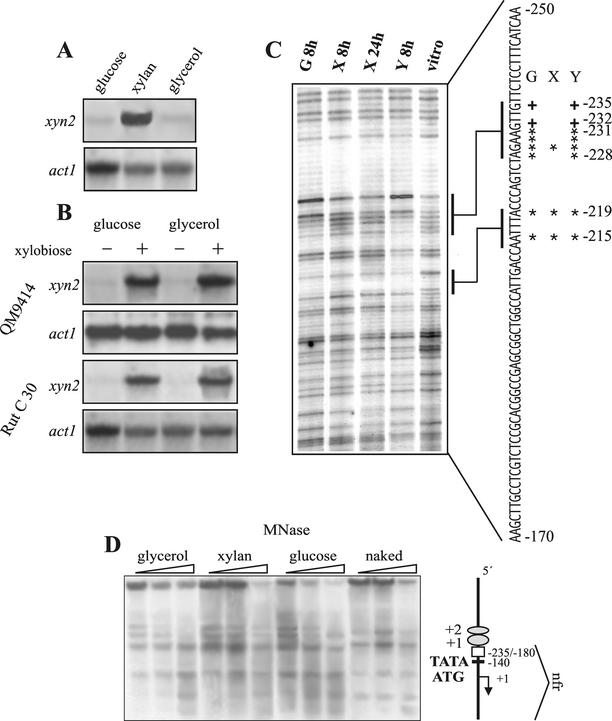
The xyn2 transcript is abundantly accumulating during growth on xylan, but a basal level of expression can also be observed during growth on glycerol and glucose, two carbon catabolite-repressing carbon sources (Fig. 1A). Regulation of xyn2 transcription is not subject to carbon catabolite repression but is strictly dependent on induction since addition of the disaccharide xylobiose triggers its expression similarily on glucose and glycerol in a Cre1 negative background (strain H. jecorina RUT C-30; Fig. 1B). All of this regulation is conferred by a short 55-bp stretch of the xyn2 promoter (44). To identify the in cis-acting regulatory motifs within this 55-bp xyn2 promoter fragment (−235 to −180), we performed in vivo genomic footprinting of the respective nucleotide area by using mycelia pregrown on glycerol and then replaced on medial containing glucose, xylan, or glycerol as carbon source. Under all three conditions, a clear, distinct protection pattern was detected on the noncoding strand (Fig. 1C), whereas no protection was observed on the coding strand (data not shown). Among the protected nucleotides, A−215 within the ATTGG (CCAAT) motif was protected under all conditions, thus corroborating previous in vitro data on the potential involvement of the CCAAT-box in xyn2 regulation (44). In addition, one purine base upstream of the CCAAT-box (−219) was also protected under all conditions. Further upstream at −231 to −228, an AGAA tetranucleotide motif was fully protected under noninducing conditions, whereas only a weak protection of G−229 within this motif was evident under inducing conditions. Furthermore, an increase in the degree of methylation of two guanine bases located immediately upstream of the AGAA-box (G−232 and G−235) was also detected only under noninducing conditions.
FIG. 1.
(A and B) Northern analysis of xyn2 transcript accumulation after replacement of H. jecorina (QM9414 and RUT C-30) on various carbon sources. (A) Replacement on glucose, xylan, and glycerol. (B) Replacement on media containing glucose or glycerol alone (−) or when supplemented with 2 mM xylobiose after 8 h (+). A total of 20 μg of RNA were loaded, and hybridizations were performed with a 0.4-kb HindIII/NsiI fragment of the xyn2 gene and a 1.9-kb KpnI fragment of the act1 (actin-encoding) gene of H. jecorina. (C) Identification of nucleotides contacted by DNA-binding proteins by using in vivo footprinting techniques via ligation-mediated PCR of the noncoding strand previously identified to contain all necessary elements for the regulation of xyn2 transcription (44). Lanes 1 to 4, DNA of H. jecorina cultures grown on glucose (G) for 8 h, xylan (X) for 8 h and 24 h, and glycerol (Y) for 8 h, respectively, was subjected to in vivo methylation, treated with HCl, and cleaved with NaOH. Bases involved in protein-DNA contact are indicated by an asterisk, whereas bases showing increased methylation are indicated by a “+” symbol. Lane 5, control DNA methylated in vitro. (D) Chromatin organization of the xyn2 gene under xylanase noninducing (glycerol and glucose) and inducing (xylan) conditions in H. jecorina. Mycelial samples were treated for 5 min at 37°C with MNase as described in Materials and Methods. The vertical map on the right shows the relative positions of ATG, TATA, and the previously identified upstream regulatory sequences. The naked DNA control is genomic DNA treated with MNase and processed similarly to the chromatin samples. The deduced nucleosome phasing is shown on the right side of the gel. Nucleosomes are given as ellipses and are numbered divergently from the nucleosome-free region.
These data revealed that a part of the xyn2 promoter is constitutively bound by DNA-binding proteins. This would imply that the xyn2 promoter is permanently accessible to its in trans-acting factors. To prove this directly, we investigated the chromatin structure of xyn2 by MNase treatment by using indirect end labeling with an 0.4-kb HindIII/NsiI fragment of the xyn2 gene as a probe (Fig. 1D). MNase treatment yielded a ladder of nucleosome-specific bands with a nucleosomal repetition length of 170 (±9) bp upstream of the previously identified regulatory sequences (above −235). This array of at least two positioned nucleosomes is followed by a nucleosome-free region, emphasized by identical MNase patterns as in the naked DNA control, which spans the detected regulatory nucleotide motifs and the TATA-box and reaches even into the structural gene. No differences were evident between induced and noninduced conditions, thus implying that the TATA-box is permanently accessible, which is in accordance with the constitutive transcription of xyn2.
In vitro characterization of protein binding to the AGAA-box.
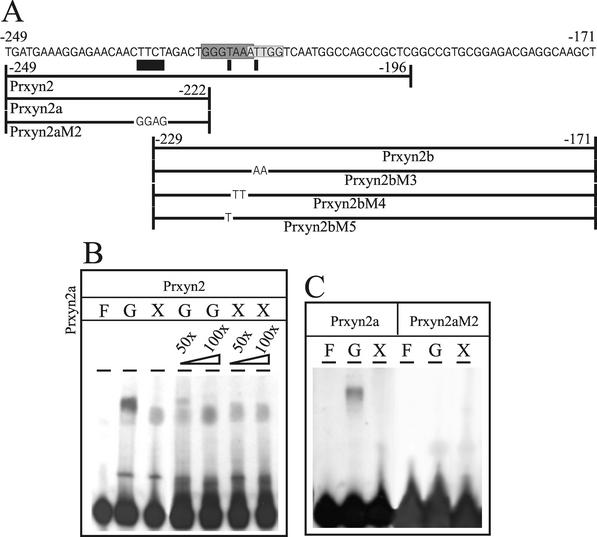
To verify the binding of regulatory proteins to the identified nucleotide motifs, EMSAs were carried out by using cell extracts prepared from H. jecorina mycelia grown on glucose or xylan. Synthetic oligonucleotides (Table 1 and Fig. 2A), covering the whole promoter area for which methylation protection had been observed were used as probes. The 54-bp oligonucleotide Prxyn2, resembling the wild-type xyn2 promoter area between −196 to −249, yielded one major protein-DNA complex with increased mobility under inducing conditions. This is in accordance with the findings that the AGAA-box and two adjacent 5′ guanines are only bound by proteins under noninducing conditions (Fig. 2B, lanes 1 to 3). More direct evidence was obtained by EMSAs with oligonucleotide Prxyn2a, which represents a xyn2 promoter fragment spanning from −222 to −249 and thus only bearing this motif (Fig. 2C, lanes 1 to 3). Only extracts from glucose-grown mycelia resulted in complex formation. To provide evidence that binding to the AGAA-box is sufficient to account for the differences in mobility observed in the binding of cell extracts from induced- and noninduced conditions to the wild-type promoter fragment Prxyn2, competition experiments with 50- and 100-fold molar excesses of unlabeled Prxyn2a over labeled Prxyn2 were performed; these experiments showed that the competing oligonucleotide titrates away the protein(s) binding to the AGAA-box from the labeled wild-type promoter fragment, causing the complexes from induced- and noninduced conditions to migrate with similar mobility in an EMSA (Fig. 2B).
FIG. 2.
(A) Schematic drawing of the oligonucleotides used in EMSAs. The given sequence spans the whole area of the previously identified xyn2 5′-noncoding region responsible for regulation of transcription. Potential regulatory elements are shaded, whereas the new motif identified by methylation protection in vivo footprinting is underlined. (B and C) EMSA analysis with 100 μg of cell extract derived from H. jecorina mycelia replaced on either 1% (wt/vol) glucose (G, repressing conditions) or 1% (wt/vol) xylan (X, inducing conditions) and 5 ng of labeled oligonucleotides as probes: Prxyn2 (B) and Prxyn2a and Prxyn2aM2 (C). “F” indicates free probe; unlabeled Prxyn2a was used as a competitor in 50- or 100-fold molar excess (B, lanes 4 to 7).
In order to investigate whether the AGAA-box is sufficient and essential for this protein binding, EMSAs were performed with a mutated version of Prxyn2a, in which the AGAA-box was mutated to CTCC, whereas the 5′-upstream Gs had been left intact (oligonucleotide Prxyn2aM2; Table 1 and Fig. 2A). These data show that a replacement of the AGAA sequence in the noncoding strand by CTCC completely impaired protein binding (Fig. 2C, lanes 4 to 6). We conclude from these data that the AGAA-box is the essential core motif responsible for protein binding in this area of the xyn2 promoter and that binding under induced and noninduced conditions differs only in the binding of protein(s) to the AGAA-box under noninducing conditions.
The xyn2 promoter contains a regulatory element that is bound by Hap2/3/5 and Ace2.
The permanent protection of A−215 within the CCAAT-box of the xyn2 promoter is consistent with previous findings of constitutive formation of a protein-DNA complex with this motif (44). Interestingly, the inverted CCAAT-box (ATTGG) in the xyn2 promoter is joined 5′ by a GGGTAA motif, which closely resembles the binding site for the xylanase activator XlnR (36) and the cellulase regulator Ace2 (1). Both motifs together (5′-GGGTAAATTGG-3′) also bear a striking resemblance to an inverted version of the cellulase-activating element (CAE) in the H. jecorina cbh2 promoter (5′-ATTGGGTAATA-3′), which is responsible for the induction of cbh2 gene transcription by cellulose and sophorose (43).
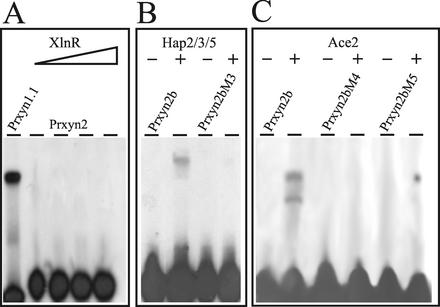
To investigate whether XlnR or Ace2 bind to the 5′-GGTAAATTGG-3′ motif, EMSAs with a heterologously expressed fragment of A. niger XlnR (36) containing the zinc-finger domain were performed with oligonucleotide Prxyn2. However, in contrast to a positive control experiment with oligonucleotide Prxyn1.1 (spanning a fragment of the xyn1 upstream regulatory region bound by XlnR [C. Wacenovsky and R. L. Mach, unpublished data]), complex formation was not observed with the GST fusion protein (Fig. 3A) or with the XlnR zinc-finger domain alone, irrespective of the amount of protein used. In contrast, by using the overexpressed zinc-finger domain of Ace2 (1), both the GST-Ace2 fusion (Fig. 3C, lanes 1 and 2) and the zinc-finger region alone showed protein-DNA binding in an EMSA. Oligonucleotides bearing mutations previously identified to decrease protein binding to the CAE (AA−217/−218 to TT; PRxyn2M4 [43]) and a G−221-to-T mutation (essential for Ace2 binding to the cbh1 promoter; Prxyn2M5 [1]) led to a complete loss of Ace2 binding. We thus conclude that Ace2 binds to the GGTAAA motif 5′-adjacent of the inverted CCAAT-box in the xyn2 promoter.
FIG. 3.
(A) Analysis of the involvement of a XlnR-like factor in binding to the xyn2 5′-noncoding region. EMSA analysis with labeled oligonucleotide Prxyn2 as a probe and increasing amounts of recombinant GST-XlnR (lanes 2 to 4; 0.05, 0.1, 0.2, and 0.5 μg) is shown. As a positive control, oligonucleotide Prxynn1.1 and 0.1 μg of fusion protein were mixed in the binding reaction (lane 1). (B) Reconstitution of the CCAAT-binding activity on the xyn2 promoter with recombinant GST-Hap2/3/5 (0.5 μg each). EMSA was performed with labeled oligonucleotide Prxyn2b (lanes 1 and 2) and Prxyn2bM3 (lanes 3 and 4). (C) Analysis of the involvement of Ace2 in binding to the xyn2 5′ regulatory region. EMSA analysis with labeled oligonucleotide Prxyn2b (lanes 1 and 2) and with oligonucleotides Prxyn2bM4 and Prxyn2bM5 (lanes 3 to 6) as probes and 1 μg of recombinant GST-Ace2. −, Free probe; +, addition of the respective fusion proteins to the binding reaction.
Recently, Zeilinger et al. (42) demonstrated that the Hap2/3/5 protein complex binds to the inverted CCAAT-box (ATTGG) within the CAE of cbh2. In order to prove that this is also the case in the xyn2 promoter, oligonucleotide PRxyn2b and the H. jecorina Hap subunits 2, 3, and 5 were subjected to EMSA. DNA-protein complex formation was clearly observed and abolished by introduction of a TT−214/−215-to-AA mutation within the CCAAT-box (oligonucleotide Prxyn2M3; Fig. 3B). From these data it can be concluded that the CAE-like motif in the xyn2 promoter is bound by the Hap2/3/5 protein complex and Ace2 but not XlnR.
Mutations impairing protein binding in vitro lead to a loss of distinct regulatory functions in xyn2 gene expression in vivo.
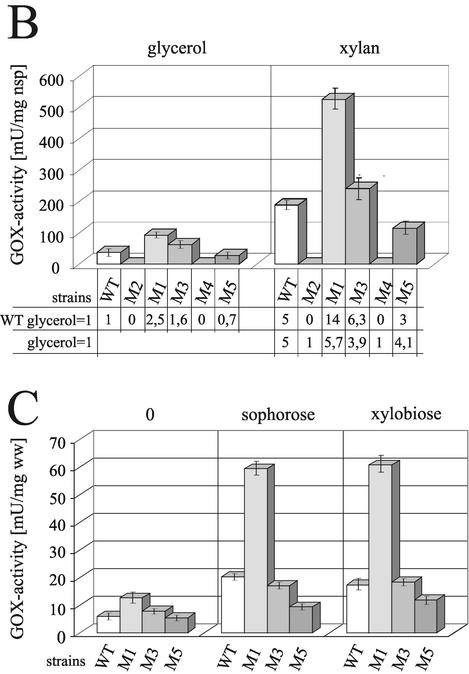
To investigate whether the protein-binding motifs identified above indeed confer regulation of xyn2 transcription in vivo, the promoter mutations studied above were introduced into 850 bp of the 5′-noncoding region of the xyn2 promoter (pLW-WT) and fused to the A. niger goxA (glucose oxidase-encoding) gene as a reporter. Except for a deletion from −171 to −229, which eliminates all putative elements identified (pLW-M2), the mutations inserted essentially resemble those used for the analysis of protein-DNA complex formation in vitro (pLWM1 = PRxyn2aM2; pLWM3 = PRxyn2M3; pLWM4 = PRxyn2M4; pLWM5 = PRxyn2M5; Fig. 4A). The different constructs were introduced into H. jecorina TU-6 by cotransformation with plasmid pFG1 (8) employing biolistic bombardment (11).
FIG. 4.
(A) Schematic representation of the reporter vectors used for transformation and the respective mutations in the xyn2 5′-noncoding region. Point mutations are indicated by arrows; putative motifs are boxed or underlined. (B) Comparison of glucose oxidase (GOX) activity given in milliunits per mg of NaOH-soluble protein measured after cultivation of the strains LW-WT, LWM1, LWM2, LWM3, LWM4, and LWM5 on glycerol (non inducing conditions) and xylan (inducing conditions) for 24 h. The results are the means of two independent experiments performed with three different single-copy integration strains; error bars indicate the standard deviation. Factors representing regulatory switches were either calculated based on the GOX activity obtained from the wild-type reporter strains (WT glycerol = 1) or from the respective reporter strains (glycerol = 1) grown on glycerol. (C) Comparison of GOX activities given in milliunits per milligram (wet weight) of replaced mycelia, measured 12 h after replacement of the strains LW-WT, LWM1, LWM3, and LWM5 on media without a carbon source (0) or supplemented with either sophorose or xylobiose in a final concentration of 2 mM. The results are given as the means derived from an identical experimental set up as in panel B.
Three transgenic strains bearing ectopic single-copy integrations of the wild-type construct (LW-WT) or each mutated construct (LWM1-5) were investigated (Fig. 4B). As expected, deletion of the complete nucleotide area between −171 to −229 resulted in a loss of transcription both on noninducing (glycerol), as well as on inducing (xylan), carbon sources. However, the same effect was obtained when mutation 4 (pLWM4), which interfered with the binding of Ace2, was introduced, indicating that AA−217/−218 is essential for transcription. Mutation of the AGAA-box (pLWM1) exhibited a striking effect, since it not only increased the basal transcriptional level on glycerol but drastically increased the inducibility by xylan. The mutation within the CCAAT-box (pLWm3) also increased the basal level of transcription on glycerol but had a less-pronounced effect on the inducibility by xylan. Mutation 5 (pLWM5), which in vitro impairs the binding of Ace2 to the xyn2 promoter, resulted in a moderate (20 to 40%) decrease of xyn2 gene transcription under both noninducing and inducing conditions.
Sophorose- and xylobiose-mediated induction involve the same nucleotide motifs.
We have previously shown that the expression of xyn2 can be induced by both xylan and cellulose and their respective soluble degradation products, xylobiose and sophorose (44). Since transcription of the cellulase genes (cbh1 and cbh2), which are also induced by cellulose and sophorose, is not triggered by xylan or xylobiose, we investigated whether induction of xyn2 expression by cellulose and xylan works via the same promoter motifs. To this end, we studied the inducibility of xyn2 expression by xylobiose and sophorose in the various promoter mutants described above. As shown in Fig. 4C, both sophorose and xylobiose resulted in roughly similar levels of xyn2 induction, which was 2.5-fold over the control (glycerol). A mutation of the AGAA-box (LWM1) led to a dramatic and comparable increase in xyn2 expression with both xylobiose and sophorose. Similar to the behavior on xylan, a mutation in the CCAAT-box (LWM3) had only a small stimulatory effect on the inducibility of xyn2 by xylobiose or sophorose. Also, in accordance with the behavior on xylan, a mutation in the Ace2-binding site (LWM5) reduced xyn2 gene expression by about half. These data show that induction of xyn2 by sophorose and xylobiose strictly follows the pattern observed on xylan, and the nucleotide motifs are therefore responsible for the induction by both xylan and cellulose.
DISCUSSION
By a combination of in vitro and in vivo techniques, two adjacent in cis-acting motifs on the noncoding strand of the xyn2 promoter (5-AGAA-3′ and 5′-GGGTAAATTGG-3′, respectively) were identified as responsible for the regulation of xyn2 gene expression. Although the latter motif in analogy to the CAE in the cbh2 promoter (43), named XAE, is essential for both the basal and induced expression of xyn2, the former is only bound under noninducing conditions and is therefore a repressor of xyn2 gene expression.
Results from in vitro protein-binding studies, in vivo footprinting, and chromatin analysis consistently showed that XAE within the xyn2 promoter is constitutively bound by its in trans-acting factors. In addition, results from EMSA analysis with heterologously expressed and purified DNA-binding proteins suggest that these in trans-acting factors are the Hap2/3/5 protein complex (42) and Ace2 (1). Although a role for the Hap2/3/5 complex has been assumed from EMSA analysis with cell extracts (44), the nature of the other binding partner was unknown. The binding site [(GG)GTAATA] does not exactly match the consensus of any of the known DNA-binding proteins but has high similarity to those proposed for the xylanase regulator XlnR (36) and the cellulase regulator Ace2 (1).
Here we now provide evidence that Ace2 but not XlnR binds to this sequence and that the effect of two mutations in the Ace2-binding motif within XAE (a G-to-T mutation in the second guanine and a AA-to-TT mutation) on binding in vitro are perfectly reflected by the impact of these mutations on xyn2 gene expression in vivo. The importance of individual nucleotides within XAE for binding of Ace2 noted here thereby strongly contrasts with the findings of Aro et al. (1), who reported that a simultaneous mutation in the TAA triplet only partially reduced binding, whereas mutating all three nucleotides of GGC eliminated DNA binding. Thus, the specific nucleotide exchanges performed in the present study produced essentially the opposite effect. The fact that the AA-to-TT mutation completely abolished both basal and induced xyn2 expression indicates that Ace2 acts as a general rather than as a specific transactivator in H. jecorina. The use of cell extracts of H. jecorina in EMSA analysis revealed that impairment of Ace2 binding also affected binding of the Hap2/3/5 protein complex, whereas impairment of binding of the latter had no effect on Ace2 binding (E. Würleitner and R. L. Mach, unpublished data).
The Hap2/3/5-binding CCAAT motif in XAE is a common cis-acting element found in the promoter and enhancer regions of a large number of eukaryotic genes (20), including filamentous fungi (3), and has been shown to be involved in maintaining a transcriptionally active chromatin structure (24). We have recently shown that the CCAAT-box within the CAE of H. jecorina cbh2 is involved in nucleosome assembly (S. Zeilinger, M. Leonhartsberger, M. Pail, R. L. Mach, and C. P. Kubicek, unpublished data) and that a mutation of CCAAT to CCCTT within CAE results in a 30% decrease in gene transcription (43). However, in the xyn2 promoter, a similar mutation in the CCAAT-box of XAE resulted in a small but reproducible increase both in basal and in induced xyn2 gene transcription, indicating that the Hap2/3/5 complex partially mediates xyn2 repression. Negative regulation by the human Hap homologue NF-Y has been observed in some cases (e.g., topoisomerase IIα, the type II transforming growth factor β receptor, and the varicella-zoster virus ORF62 [12, 13, 21]) and, more recently, also for the lysF promoter of the fungus A. nidulans (37). In hematopoietic cells, repression of the protein-tyrosine phosphatase gene by NF-Y was shown to be dependent on its interaction with the histones (40). A similar mechanism does not apply for the Hap2/3/5-mediated repression of xyn2, since chromatin analysis exhibited that the xyn2 promoter is permanently accessible. An alternative mechanism has been found in the SHP-1 gene promoter of MCF7 cells, where activating and repressing effects of the CCAAT-box are due to position-dependent competition with binding of the transactivator COUP-TFII (16). However, we also consider this explanation less likely since the binding sites for Hap2/3/5 and Ace2, albeit partially overlapping, are located on opposite strands of the DNA. Rather, we consider it more likely that the Hap2/3/5 complex is involved in positioning the 5′ nucleosomes, which in its absence therefore would move downstream and partially interfere with the binding of the AGAA-binding repressor protein.
The major difference in nucleotide protection between induced and noninduced conditions was the binding of an as-yet-unidentified protein to the AGAA-box. To the best of our knowledge, no such consensus sequence has so far been reported for other fungal promoters, although these four nucleotides form the core of the glucocorticoid receptor element (5′-AGAACA-3′ [26]), which occurs in two inverted copies, separated by 3 bp (29). Interestingly, the AGAA-box of the xyn2 promoter was also accompanied by an inverted copy (5′-AGAAcaacTTCT-3′), leading us to assume that the respective factor may actually bind to both boxes. However, mutation of the 3′ TTCT-box had no effect on the binding to AGAA on the noncoding strand (Würleitner and Mach, unpublished), and this hypothesis was thus refuted.
The fact that the mutation in the AGAA element resulted in a >3-fold increase in xyn2 gene transcription under both induced and noninduced conditions renders the xyn2 promoter a potentially useful alternative for protein expression in H. jecorina. Currently, the cbh1 promoter is almost exclusively used for these purposes, since the corresponding protein Cel7A (CBH I) accounts for >60% of the total protein secreted by the fungus (14). Average secreted amounts of Xyn2 are approximately one-fifth of the amount of Cel7A (32), implying that a xyn2 promoter with a mutated AGAA motif may lead to protein yields almost as high as from the cbh1 promoter.
We have previously reported that xyn2 is subject to induction both by cellulose and by xylan and some of its corresponding degradation products (i.e., sophorose and xylobiose, respectively [44]). The results from the present study show that induction by both systems acts via the same promoter motif, which is also constitutively bound by its in trans-acting factors, thus implying that specificity of induction does not involve a specific DNA-binding protein and rather must be due to signaling mechanisms. The relief from repression by the AGAA-binding protein alone is not sufficient to explain induction because both the induced and the basal transcription of xyn2 are increased in the AGAA-to-GAGG mutant. Furthermore, an EMSA with cell extracts showed that the protein complex binding XAE exhibits the same migration property under induced and noninduced conditions and thus most likely consists of the same proteins. This argues against the possibility that a further protein would bind to the Hap2/3/5-Ace2-XAE complex under inducing conditions and thereby stimulate transcription. Rather, our working hypothesis is that the induction by xylan and/or cellulose may be mediated by covalent modification of Ace2, e.g., by phosphorylation. The identical migration of the XAE-protein complex from induced and noninduced conditions would not contradict this assumption, since the excess of oligonucleotide used likely masks the introduction of a phosphate group into Ace2 and thus will not give rise to a difference in the electrophoretic mobility. Our previous findings that xyn2 gene transcription can be inhibited by calmodulin antagonists, which led us to propose the involvement of a Ca2+-calmodulin-dependent protein kinase in its induction (19), and the abundance of protein kinase targets in Ace2, including those for calmodulin-dependent protein kinase II (predicted by “PhosphoBase” [15]; http://www.cbs.dtu.dk/databases/PhosphoBase/predict/predict.html) would be in accordance with this speculation.
Acknowledgments
This study was supported by a grant from Fonds zur Förderung Wissenschaftlicher Forschung to R.L.M. (P-12918 MOB).
REFERENCES
- 1.Aro, N., A. Saloheimo, M. Ilmen, and M. Penttila. 2001. ACEII, a novel transcriptional activator involved in regulation of cellulase and xylanase genes of Trichoderma reesei. J. Biol. Chem. 276:24309-24314. [DOI] [PubMed] [Google Scholar]
- 2.Biely, P., M. Vrsanska, M. Tenkanen, and D. Kluepfel. 1997. Endo-β-1,4-xylanase families: differences in catalytic properties. J. Biotechnol. 57:151-166. [DOI] [PubMed] [Google Scholar]
- 3.Brakhage, A. A., A. Andrianopoulos, M. Kato, S. Steidl, M. A. Davis, N. Tsukagoshi, and M. J. Hynes. 1999. HAP-Like CCAAT-binding complexes in filamentous fungi: implications for biotechnology. Fungal Genet. Biol. 27:243-252. [DOI] [PubMed] [Google Scholar]
- 4.Buchert, J., T. Oksanen, J. Pere, M. Siika-aho, A. Suurnäkki, and L. Viikari. 1998. Application of Trichoderma reesei enzymes in pulp and paper industry, p. 343-357. In G. E. Harman and C. P. Kubicek (ed.), Trichoderma and Gliocladium, vol. 2. Taylor & Francis, Ltd., London, United Kingdom.
- 5.Garrity, P. A., and B. J. Wold. 1992. Effects of different DNA polymerases in ligation-mediated PCR: enhanced genomic sequencing and in vivo footprinting. Proc. Natl. Acad. Sci. USA 89:1021-1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gielkens, M. M., E. Dekkers, J. Visser, and L. H. de Graaff. 1999. Two cellobiohydrolase-encoding genes from Aspergillus niger require d-xylose and the xylanolytic transcriptional activator XlnR for their expression. Appl. Environ. Microbiol. 65:4340-4345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gonzalez, R., and C. Scazzocchio. 1997. A rapid method for chromatin structure analysis in the filamentous fungus Aspergillus nidulans. Nucleic Acids Res. 25:3955-3956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gruber, F., J. Visser, C. P. Kubicek, and L. H. de Graaff. 1990. The development of a heterologous transformation system for the cellulolytic fungus Trichoderma reesei based on a pyrG-negative mutant strain. Curr. Genet. 18:71-76. [DOI] [PubMed] [Google Scholar]
- 9.Hasper, A. A., J. Visser, and L. H. de Graaff. 2000. The Aspergillus niger transcriptional activator XlnR, which is involved in the degradation of the polysaccharides xylan and cellulose, also regulates d-xylose reductase gene expression. Mol. Microbiol. 36:193-200. [DOI] [PubMed] [Google Scholar]
- 10.Havukainen, R., A. Torronen, T. Laitinen, and J. Rouvinen. 1996. Covalent binding of three epoxyalkyl xylosides to the active site of endo-1,4-xylanase II from Trichoderma reesei. Biochemistry 35:9617-9624. [DOI] [PubMed] [Google Scholar]
- 11.Hazell, B. W., V. S. Te'o, J. R. Bradner, P. L. Bergquist, and K. M. Nevalainen. 2000. Rapid transformation of high cellulase-producing mutant strains of Trichoderma reesei by microprojectile bombardment. Lett. Appl. Microbiol. 30:282-286. [DOI] [PubMed] [Google Scholar]
- 12.Isaacs, R. J., A. L. Harris, and I. D. Hickson. 1996. Regulation of the human topoisomerase IIα gene promoter in confluence-arrested cells. J. Biol. Chem. 271:16741-16747. [DOI] [PubMed] [Google Scholar]
- 13.Kelly, D., S. J. Kim, and A. Rizzino. 1998. Transcriptional activation of the type II transforming growth factor-beta receptor gene upon differentiation of embryonal carcinoma cells. J. Biol. Chem. 273:21115-21124. [DOI] [PubMed] [Google Scholar]
- 14.Kubicek, C. P., R. Messner, F. Gruber, M. Mandels, and E. M. Kubicek-Pranz. 1993. Triggering of cellulase biosynthesis by cellulose in Trichoderma reesei. Involvement of a constitutive, sophorose-inducible, glucose-inhibited β-diglucoside permease. J. Biol. Chem. 268:19364-19368. [PubMed] [Google Scholar]
- 15.Kulkarni, N., A. Shendye, and M. Rao. 1999. Molecular and biotechnological aspects of xylanases. FEMS Microbiol. Rev. 23:411-456. [DOI] [PubMed] [Google Scholar]
- 16.Liberati, C., M. R. Cera, P. Secco, C. Santoro, R. Mantovani, S. Ottolenghi, and A. Ronchi. 2001. Cooperation and competition between the binding of COUP-TFII and NF-Y on human epsilon- and gamma-globin gene promoters. J. Biol. Chem. 276:41700-41709. [DOI] [PubMed] [Google Scholar]
- 17.Mach, R. L., C. K. Peterbauer, K. Payer, S. Jaksits, S. L. Woo, S. Zeilinger, C. M. Kullnig, M. Lorito, and C. P. Kubicek. 1999. Expression of two major chitinase genes of Trichoderma atroviride (T. harzianum P1) is triggered by different regulatory signals. Appl. Environ. Microbiol. 65:1858-1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mach, R. L., J. Strauss, S. Zeilinger, M. Schindler, and C. P. Kubicek. 1996. Carbon catabolite repression of xylanase I (xyn1) gene expression in Trichoderma reesei. Mol. Microbiol. 21:1273-1281. [DOI] [PubMed] [Google Scholar]
- 19.Mach, R. L., S. Zeilinger, D. Kristufek, and C. P. Kubicek. 1998. Ca2+-calmodulin antagonists interfere with xylanase formation and secretion in Trichoderma reesei. Biochim. Biophys. Acta 1403:281-289. [DOI] [PubMed] [Google Scholar]
- 20.Mantovani, R. 1999. The molecular biology of the CCAAT-binding factor NF-Y. Gene 239:15-27. [DOI] [PubMed] [Google Scholar]
- 21.Moriuchi, H., M. Moriuchi, and J. I. Cohen. 1995. The varicella-zoster virus immediate-early 62 promoter contains a negative regulatory element that binds transcriptional factor NF-Y. Virology 214:256-258. [DOI] [PubMed] [Google Scholar]
- 22.Mueller, P. R., and B. Wold. 1989. In vivo footprinting of a muscle specific enhancer by ligation-mediated PCR. Science 246:780-786. [DOI] [PubMed] [Google Scholar]
- 23.Muilu, J., A. Torronen, M. Perakyla, and J. Rouvinen. 1998. Functional conformational changes of endo-1,4-xylanase II from Trichoderma reesei: a molecular dynamics study. Proteins 31:434-444. [PubMed] [Google Scholar]
- 24.Narendja, F. M., M. A. Davis, and M. J. Hynes. 1999. AnCF, the CCAAT binding complex of Aspergillus nidulans, is essential for the formation of a DNase I-hypersensitive site in the 5′ region of the amdS gene. Mol. Cell. Biol. 19:6523-6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ntarima, P., W. Nerinckx, K. Klarskov, B. Devreese, M. K. Bhat, J. Van Beeumen, and M. Claeyssens. 2000. Epoxyalkyl glycosides of d-xylose and xylo-oligosaccharides are active-site markers of xylanases from glycoside hydrolase family 11, not from family 10. Biochem. J. 347(Pt. 3):865-873. [PMC free article] [PubMed] [Google Scholar]
- 26.Pearce, D., W. Matsui, J. N. Miner, and K. R. Yamamoto. 1998. Glucocorticoid receptor transcriptional activity determined by spacing of receptor and nonreceptor DNA sites. J. Biol. Chem. 273:30081-30085. [DOI] [PubMed] [Google Scholar]
- 27.Saarelainen, R., M. Paloheimo, R. Fagerstrom, P. L. Suominen, and K. M. Nevalainen. 1993. Cloning, sequencing, and enhanced expression of the Trichoderma reesei endoxylanase II (pI 9) gene xln2. Mol. Gen. Genet. 241:497-503. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 29.Scheidereit, C., D. von der Ahe, A. C. Cato, M. Wenz, G. Suske, C. Carlson, H. Bosshard, H. M. Westphal, and M. Beato. 1989. Protein-DNA interactions at steroid hormone regulated genes. Endocrinol. Res. 15:417-440. [DOI] [PubMed] [Google Scholar]
- 30.Stangl, H., F. Gruber, and C. P. Kubicek. 1993. Characterization of the Trichoderma reesei cbh2 promoter. Curr. Genet. 23:115-122. [DOI] [PubMed] [Google Scholar]
- 31.Strauss, J., R. L. Mach, S. Zeilinger, G. Hartler, G. Stoffler, M. Wolschek, and C. P. Kubicek. 1995. Cre1, the carbon catabolite repressor protein from Trichoderma reesei. FEBS Lett. 376:103-107. [DOI] [PubMed] [Google Scholar]
- 32.Suominen, P., A. Mäntylä, R. Saarelainen, M. Paloheimo, R. Fagerström, E. Parkkinen, and H. Nevalainen. 1992. Genetic engineering of Trichoderma reesei to produce suitable enzyme combinations for applications in pulp and paper industry, p. 439-446. In M. K. A. M. Shimada (ed.), Bio/technology in the pulp and paper industry. Uni Publishers Co., Ltd., Tokyo, Japan.
- 33.Timell, T. E. 1965. Wood hemicellulose, part I. Adv. Carbohydr. Chem. 20:409-483. [Google Scholar]
- 34.Torronen, A., R. L. Mach, R. Messner, R. Gonzalez, N. Kalkkinen, A. Harkki, and C. P. Kubicek. 1992. The two major xylanases from Trichoderma reesei: characterization of both enzymes and genes. Bio/Technology 10:1461-1465. [DOI] [PubMed] [Google Scholar]
- 35.van Peij, N. N., M. M. Gielkens, R. P. de Vries, J. Visser, and L. H. de Graaff. 1998. The transcriptional activator XlnR regulates both xylanolytic and endoglucanase gene expression in Aspergillus niger. Appl. Environ. Microbiol. 64:3615-3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Peij, N. N., J. Visser, and L. H. de Graaff. 1998. Isolation and analysis of xlnR, encoding a transcriptional activator co-ordinating xylanolytic expression in Aspergillus niger. Mol. Microbiol. 27:131-142. [DOI] [PubMed] [Google Scholar]
- 37.Weidner, G., S. Steidl, and A. A. Brakhage. 2001. The Aspergillus nidulans homoaconitase gene lysF is negatively regulated by the multimeric CCAAT-binding complex AnCF and positively regulated by GATA sites. Arch. Microbiol. 175:122-132. [DOI] [PubMed] [Google Scholar]
- 38.Wolschek, M. F., F. Narendja, J. Karlseder, C. P. Kubicek, C. Scazzocchio, and J. Strauss. 1998. In situ detection of protein-DNA interactions in filamentous fungi by in vivo footprinting. Nucleic Acids Res. 26:3862-3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu, C. 1980. The 5′ ends of Drosophila heat shock genes in chromatin are hypersensitive to DNase I. Nature 286:854-860. [DOI] [PubMed] [Google Scholar]
- 40.Xu, Y., D. Banville, H. F. Zhao, X. Zhao, and S. H. Shen. 2001. Transcriptional activity of the SHP-1 gene in MCF7 cells is differentially regulated by binding of NF-Y factor to two distinct CCAAT-elements. Gene 269:141-153. [DOI] [PubMed] [Google Scholar]
- 41.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 42.Zeilinger, S., A. Ebner, T. Marosits, R. Mach, and C. P. Kubicek. 2001. The Hypocrea jecorina HAP 2/3/5 protein complex binds to the inverted CCAAT-box (ATTGG) within the cbh2 (cellobiohydrolase II-gene) activating element. Mol. Genet. Genomics 266:56-63. [DOI] [PubMed] [Google Scholar]
- 43.Zeilinger, S., R. L. Mach, and C. P. Kubicek. 1998. Two adjacent protein binding motifs in the cbh2 (cellobiohydrolase II-encoding) promoter of the fungus Hypocrea jecorina (Trichoderma reesei) cooperate in the induction by cellulose. J. Biol. Chem. 273:34463-34471. [DOI] [PubMed] [Google Scholar]
- 44.Zeilinger, S., R. L. Mach, M. Schindler, P. Herzog, and C. P. Kubicek. 1996. Different inducibility of expression of the two xylanase genes xyn1 and xyn2 in Trichoderma reesei. J. Biol. Chem. 271:25624-25629. [DOI] [PubMed] [Google Scholar]