Abstract
More than 100,000 interstitial segments of DNA (internal eliminated sequences [IESs]) are excised from the genome during the formation of a new macronucleus in Euplotes crassus. IESs include unique sequence DNA as well as two related families of transposable elements, Tec1 and Tec2. Here we describe a new class of E. crassus transposons, Tec3, which is present in 20 to 30 copies in the micronuclear genome. Tec3 elements have long inverted terminal repeats and contain a degenerate open reading frame encoding a tyrosine-type recombinase. One characterized copy of Tec3 (Tec3-1) is 4.48 kbp long, has 1.23-kbp inverted terminal repeats, and resides within the micronuclear copy of the ribosomal protein L29 gene (RPL29). The 23 bp at the extreme ends of this element are very similar to those in other E. crassus IESs and, like these other IESs, Tec3-1 is excised during the polytene chromosome stage of macronuclear development to generate a free circular form with an unusual junction structure. In contrast, a second cloned element, Tec3-2, is quite similar to Tec3-1 but lacks the terminal 258 bp of the inverted repeats, so that its ends do not resemble the other E. crassus IES termini. The Tec3-2 element appears to reside in a large segment of the micronuclear genome that is subject to developmental elimination. Models for the origins of these two types of Tec3 elements are presented, along with a discussion of how some members of this new transposon family may have come to be excised by the same machinery that removes other E. crassus IESs.
Ciliated protozoa undergo a massive reorganization of their genome during the process of sexual reproduction (20, 52). These organisms contain two types of nuclei: nonexpressed micronuclei containing conventional chromosomes and transcriptionally active macronuclei that contain multiple copies of subchromosome-sized DNA molecules. Both types of nuclei are replicated and segregated during asexual reproduction; however, during mating (conjugation), the old macronucleus is destroyed, and a new one is formed from a mitotic copy of the newly formed zygotic micronucleus. Extensive genome remodeling occurs during macronuclear development and includes chromosome fragmentation, de novo telomere formation, and DNA breakage and rejoining.
While all ciliates appear to undergo these various forms of DNA rearrangement, members of the spirotrich group exhibit extreme forms of macronuclear development (17, 20). For the subject of this study, the spirotrich Euplotes crassus (also referred to as Moneuplotes crassus [4]), macronuclear development begins with the endoreplication of micronuclear chromosomes (20 to 45 h after mating is initiated), resulting in the formation of polytene chromosomes. During this period, ∼100,000 interstitial DNA segments (internal eliminated sequences [IESs]) are excised from the chromosomes and flanking sequences are rejoined. The polytene chromosome stage is followed by the vesicle stage, during which chromosomes are fragmented, telomeres are added to chromosome ends, and micronucleus-limited DNA begins to be degraded. The final stage of development includes telomere trimming on macronucleus-destined DNA (51) and additional rounds of DNA replication. The newly generated macronucleus contains linear, highly amplified DNA molecules averaging about 2 kbp in length and usually containing single genes (39).
In E. crassus, there are two classes of IESs that are eliminated during the polytene chromosome stage of development (reviewed in references 17 and 29). The first class is the short IESs, which are noncoding DNA segments ranging in length from approximately 30 to 550 bp. As many as 40,000 of these elements reside within the micronuclear genome. The short IESs have short inverted terminal repeats and are flanked by 5′-TA-3′ direct repeats. The second class of IESs includes members of the closely related Tec1 and Tec2 transposon families (2, 18, 22, 34). Approximately 10,000 to 14,000 copies of each element reside within the micronuclear genome of E. crassus. Both Tec1 and Tec2 are 5.3 kbp in length, have ∼700-bp inverted terminal repeats and, like the short IESs, are flanked by TA direct repeats. The Tec elements contain three degenerate open reading frames (ORFs), one of which encodes a “DDE” transposase most similar to those encoded by the Tc1/Mariner family of transposons (ORF1 [7]), whereas another (ORF2) is predicted to encode a tyrosine-type recombinase (7a). However, there is no evidence that any of these genes is currently highly transcribed or that functional proteins are produced (23).
Two lines of evidence indicate that the same machinery excises the E. crassus short IESs and the Tec1 and Tec2 transposon IESs. First, all of these elements share a short terminal consensus sequence that is similar to the terminal sequences of members of the Tc1/Mariner family of transposons (17, 23). Paramecium IESs have a similar terminal sequence (28), and mutations in the terminal sequence abolish IES excision in this organism; these characteristics argue that the terminal sequence is a key cis-acting element for specifying excision (reviewed in reference 14). Second, both types of E. crassus IESs generate the same excision products. For each one, excision is precise in that the macronucleus-destined DNA retains a single copy of the terminal 5′-TA-3′ direct repeat (48) and the IES assumes a free circular form with an unusual heteroduplex junction structure (21, 24, 32, 34, 46).
In this report, we describe a new type of long-inverted-repeat transposon in E. crassus, Tec3, which also undergoes developmental elimination. The Tec3 elements are distinct from the Tec1 and Tec2 elements, but at least one family member (Tec3-1) has termini that are quite similar to those of the Tec1 and Tec2 elements. Following excision, this element forms a free circle with an unusual junction structure that appears to be identical to those of other characterized E. crassus IESs. A second Tec3 element (Tec3-2) has truncated inverted repeats and appears to reside in a large block of developmentally eliminated DNA. Models for the origins of these two types of Tec3 elements are discussed.
MATERIALS AND METHODS
E. crassus cell culture, mating, and DNA isolation.
E. crassus cells were grown and maintained in artificial seawater by using the alga Dunaliella salina as the food source as described previously (41), with the exception that Reef Crystals (Aquarium Systems, Mentor, Ohio) served as the base for the artificial seawater and vitamin B12 was not included. E. crassus strains X1 (mating type III), X2 (mating type unknown, but not mating type I or III) (11), CT5 (mating type III), and CT27 (mating type unknown, but not mating type I or III) were used in the analyses. Matings of strains X1 and X2 as well as of strains CT5 and CT27 were carried out and the resulting strains were harvested as described previously (41, 47). Total cellular DNA (31), micronuclear DNA (35), and developing macronuclear DNA (27) were purified as described previously.
Molecular biological techniques.
The recombinant LEMIC micronuclear library, constructed from E. crassus strain G1 (3), was screened by hybridization with radiolabeled probes (10, 42). DNA was isolated from recombinant bacteriophage as described previously (42). Plasmid DNAs were isolated from bacterial cells by using a Magic Miniprep DNA purification system (Promega Corp., Madison, Wis.) according to the manufacturer's instructions. PCR products were isolated from low-melting-point agarose (GIBCO BRL Life Technologies, Inc., Rockville, Md.) gels as described previously (40) and cloned by using a TOPO TA cloning kit for sequencing (Invitrogen, Carlsbad, Calif.). DNA restriction digestion, dephosphorylation, and ligation were carried out under conditions recommended by the enzyme suppliers (New England Biolabs, Beverly, Mass., and GIBCO BRL Life Technologies) or with commonly used protocols (42). DNAs were analyzed by electrophoresis on agarose or low-melting-point agarose gels prepared and run in a buffer containing 89 mM Trizma, 89 mM H3BO3, and 2 mM disodium EDTA-H2O.
PCR.
All oligonucleotides were purchased from GIBCO BRL Life Technologies, and PCR was carried out with a model PTC-150 MiniCycler (MJ Research, Cambridge, Mass.). For amplification of the micronuclear RPL29 locus, Hercules polymerase (Stratagene, La Jolla, Calif.) was used under conditions recommended by the manufacturer. Twenty-five cycles of amplification were carried out with oligonucleotides LRPL-4 (5′-CGATCTACCTTATTATCACG-3′) and RFr29-1 (5′-TCTTTCCTAGTATTTGGATG-3′) and 0.5 μg of E. crassus strain X1 total cellular DNA as the substrate. A cycle consisted of 92°C for 30 s, 50°C for 30 s, and 68°C for 8 min.
PCR also was carried out with KlenTaq DNA polymerase (Sigma-Aldrich, St. Louis, Mo.) essentially under the conditions described by the manufacturer and 100 ng of substrate DNA. Excision of the Tec3-1 element was assessed with oligonucleotide primers LRPL-4 and RFr29-1. Twenty-five cycles of amplification were carried out with total DNA from vegetative and developing cells, with a cycle consisting of 95°C for 1 min, 49°C for 1 min, and 72°C for 1 min. For detection of the excised Tec3-1 circle junction, PCR amplification was carried out with primers Tec3.1cirR (5′-TCTGAAGGACGGCATAATTA-3′) and Tec3.1cirL (5′-GCTGAAGGATGTCCTAATCG-3′). Thirty cycles of amplification were performed, with a cycle consisting of 95°C for 1 min and 51°C for 1 min.
To obtain additional copies of the core regions of Tec3 elements, three separate PCRs were performed with total cellular DNA and sets of primers designed to give overlapping products. The sequences of the primer pairs were based on either the Tec3-1 sequence or a combination of the Tec3-1 and Tec3-2 sequences and were as follows: Int6 (5′-GCTACTGTGTACCATGCAAC-3′) and Int7 (5′-GCCTTGAAGACAAGAATGTC-3′), F4 (5′-TGCCTCTWGTRAACTTTTCR-3′, where W is A or T and R is A or G) and B4 (5′-GTCWAGAGYMGAAGAGGATA-3′, where Y is C or T and M is A or C), and F5 (5′-ATTKSGARRTCTCCTTTCCC-3′, where S is G or C and K is T or G) and B5 (5′-YCCAARTCTYTYCTCTSTRC-3′). Twenty-five cycles of amplification were performed, with a cycle consisting of 95°C for 30 s, 1 min at the annealing temperature, and 72°C for 1 min. The annealing temperatures were 54, 49, and 51°C for the Int6-Int7, F4-B4, and F5-B5 primer combinations, respectively.
DNA sequencing and analysis.
Sequencing was performed at the University of Connecticut Health Center Molecular Core Facility by using a Taq DyeDeoxy termination cycle sequencing kit (Perkin-Elmer Corp., Norwich, Conn.). For sequencing of the Tec3-containing clones (Mic.RPL-29-2 and PhCl.4), restriction fragments were subcloned into the pBluescript SK(+) phagemid (Stratagene). The subclones were then sequenced from their termini with primers that were complementary to vector sequences, followed by sequencing with primers that were complementary to internal regions of the subclones. Both strands of the entire Mic.RPL-29-2 insert and the Tec3-2 transposon in PhCl.4 were sequenced, while only one strand of the PhCl.4 region containing the Tec1 elements was sequenced. Sequences were compiled and analyzed by using SeqEd version 1.0.3 software (Applied Biosystems, Inc., Foster City, Calif.) and MacVector version 4.1.4 software (Accelrys, Princeton, N.J.).
Database searches were performed by using BLAST version 2.2.1 (1). DNA and protein sequences were aligned by using the ClustalW program (16, 50), accessed on the Baylor College of Medicine (Houston, Tex.) Human Genome Sequencing Center Search Launcher site (43), and edited by using SeqVu version 1.1 (Garvan Institute of Medical Research, Sydney, New South Wales, Australia). Default parameters were used in the alignments of tyrosine recombinase proteins, but some manual adjustments were made to conform to the alignments of Nunes-Düby et al. (38). The additional tyrosine recombinase proteins analyzed were obtained from GenBank and were as follows (with designations and accession numbers given in parentheses): E. crassus Tec1 (Ecra Tec1, 397763), E. crassus Tec2 (Ecra Tec2, 397768), bacteriophage lambda integrase (Lambda 95 Int, 138569), P1-like virus recombinase CRE (P1 CRE, 132262), Autographa californica nucleopolyhedrovirus very late expression factor vlf1 (AcN vlf1, 1175103), Acholeplasma phage L2 integrase/recombinase-like protein (PL2 Int, 9626516), bacteriophage phi CTX integrase (PhiCTX Int, 4063818), Escherichia coli integrase XerD (Ecoli XerD, 139819), Clostridium butyricum hypothetical protein (Cbut hypo, 481912), Methanocaldococcus jannaschii integrase/recombinase (Mjan Int, 15668543), Bergeyella zoohelcum integrase (Bzoo Int, 557887), Haemophilus influenzae Rd putative integrase/recombinase (HinfInt, 16273329), Streptococcus pneumoniae Tn1545 integrase (Spne TnInt, 47463), E. coli fimbria regulatory protein (Ecoli FimB, 729489), Methanothermobacter thermautotrophicus integrase/recombinase protein (Mthe Int, 7428936), Lactobacillus leichmannii XerC recombinase (Llei XerC, 1359910), Staphylococcus aureus recombinase XerD (Saur XerD, 3747042), Rickettsia prowazekii integrase/recombinase XerD (Rpro XerD, 7443326), Vibrio cholerae integrase/recombinase XerD (Vcho XerD, 11355590), Nostoc sp. recombinase XisA (Nostoc XisA, 20141864), Anabaena sp. recombinase XisC (Anab XisC, 1094355), E. coli integrase/recombinase XerC (Ecoli XerC, 14917067), Bacillus thuringiensis Tn4430 resolvase TnPI (Bthur TnPI, 135957), S. aureus transposon Tn554 TnPA (Saur TnPA, 135955), S. aureus phage φ11 integrase (φ11 Int, 166159), Enterobacter phage P22 integrase (P22 Int, 138565), bacteriophage P21 integrase (P21 Int, 138558), and Streptomyces ambofaciens integrase (Samb Int, 124698). Patterns of variation were analyzed by using MacClade version 4.0 software (Sinauer Associates, Sunderland, Mass.).
Genealogical analysis of Tec3 clone and PCR product sequences was done with the neighbor-joining (NJ) algorithm in Paup*4.0, with distances estimated by the Tamura-Nei model (45) and variability in rates among sites estimated by a gamma parameter with an α value of 0.5. Genealogies of tyrosine recombinase amino acid sequences were constructed by using several algorithms in order to assess the impact of different evolutionary models on relationships among sequences. These analyses were done with 95 amino acids corresponding to the boxes and patches characterized by Nunes-Düby et al. (38; for the alignment and regions analyzed, see the supplemental figure at www.science.smith.edu/departments/Biology/lkatz/align.html) and included the cloned Tec3-1 element, a consensus sequence (conTec3-2) derived from Tec3-2, and the PCR clones generated from the tyrosine recombinase regions of other Tec3 elements. Genealogies were constructed by using the NJ algorithm with mean character distances and maximum parsimony (MP) analyses with 25 additional random sequences in Paup* 4.0 (Sinauer Associates). Bootstrap support was calculated by using 100 replicates for both models. Maximum-likelihood (ML) analyses were performed through quartet puzzling by using the JTT model (25), allowing for variation in rates among sites, as implemented by Tree-puzzle 5.0 (44). Resulting puzzle quartet (PZ) support values are reported.
Southern and dot blot hybridizations.
DNA restriction fragments and PCR products used as hybridization probes were labeled with [α-32P]dATP by the random hexamer priming method (10). Southern blotting was carried out as described previously (5, 33). Hybridization was performed with a high-efficiency hybridization system (HS 114; Molecular Research Center, Cincinnati, Ohio) essentially as described by the manufacturer. Blots were routinely washed twice at room temperature for 5 min in 1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), twice at 65°C for 30 min in 1× SSC, and twice at 65°C for 30 min in 0.5× SSC or 0.1× SSC. All SSC buffers contained 1% sodium dodecyl sulfate.
For the dot blot analysis to determine the Tec3 copy number, DNA samples were boiled for 5 min before being spotted in triplicate on Nytran membranes (Schleicher & Schuell, Keene, N.H.) by using a Hybri-Dot (GIBCO BRL Life Technologies) filtration manifold as described by the manufacturer. Blots contained 10 μg of E. crassus strain X2 total vegetative-cell DNA, along with 11, 33, and 109 pg of a 2-kbp EcoRI restriction fragment derived from the Tec3-2 element as copy number standards. The amounts of the 2-kbp EcoRI fragment were chosen to be equivalent to 10, 30, and 100 copies of the Tec3 element/cell in a 10-μg sample of total cellular DNA, based on a DNA content of 50 pg for an E. crassus cell (30). Sufficient salmon sperm DNA was added to the standard samples to ensure that DNA gram amounts equal to those of the E. crassus samples were applied to the slots. Following hybridization with the 2-kbp EcoRI fragment as the probe, the filters were washed as described for Southern blots; the final washes were done with 0.5× SSC at 65°C. The amount of probe bound was then determined by using an Instant Imager (Packard BioScience Co., Meriden, Conn.).
Nucleotide sequence accession numbers.
Sequences determined here were deposited in GenBank under accession numbers AY115662 (Mic.RPL-29-2), AY115671 (PhCl.4), and AY115663 to AY115670 (Tec3 PCR product clones).
RESULTS
A Tec3 element interrupts the micronuclear copy of the ribosomal protein L29 gene.
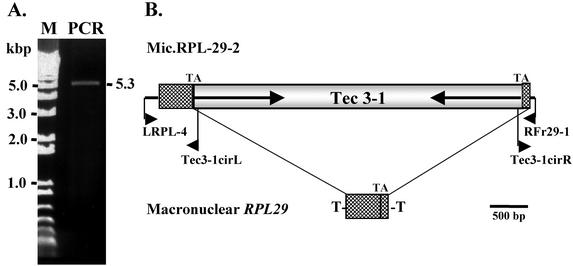
In a previous study (27), inverse PCR was used to isolate and clone the ends and flanking regions of the micronuclear copy of the ribosomal protein L29 gene (RPL29). To obtain the complete micronuclear copy of the RPL29 gene, we attempted to carry out PCR with a pair of oligonucleotide primers complementary to the regions flanking the locus (LRPL-4 and RFr29-1) (Fig. 1). Repeated standard PCRs with this pair of primers, as well as other combinations of primers, failed to give a specific PCR product. Since the macronuclear chromosome containing the RPL29 gene is only 553 bp long (plus telomeres) (19), the failure to obtain a PCR product suggested that the micronuclear copy of the locus contained one or more IESs. To investigate this possibility, we carried out PCR with the above two primers under conditions that would allow for the amplification of large products (see Materials and Methods). A single product of 5.3 kbp was obtained (Fig. 1A) and inserted into the TOPO TA vector to generate clone Mic.RPL-29-2.
FIG. 1.
Isolation and characterization of Tec3-1. (A) Agarose gel containing PCR products obtained from E. crassus strain X1 total DNA with primers LRPL-4 and RFr29-1 (lane PCR), along with size markers (lane M). (B) Maps of micronuclear clone Mic.RPL-29-2 and the macronuclear chromosome containing the RPL29 gene. Sequences forming macronuclear RPL29 are shown as cross-hatched rectangles, flanking micronuclear DNA is shown as narrow lines, and the Tec3-1 element is shown as a grey rectangle. The arrows inside the grey rectangle indicate the inverted repeats of Tec3-1, and “-T” represents the telomeres of the macronuclear DNA molecule. The positions of oligonucleotides LRPL-4 and RFr29-1, which were used as the PCR primers in panel A, and of primers Tec3-1cirL and Tec3-1cirR (see Fig. 4) are indicated below the micronuclear map, with arrowheads denoting the direction in which they prime DNA synthesis
Complete sequencing of Mic.RPL-29-2 revealed that it contained a perfect match to all of the sequences present in macronuclear RPL29, with the exception of telomeric repeats. However, the micronuclear copy of RPL29 contains a single 4,483-bp IES inserted within the fourth codon prior to the termination codon of the RPL29 ORF. Like all other E. crassus IESs, it is bounded by a 5′-TA-3′ dinucleotide repeat, one copy of which is retained in the RPL29 macronuclear DNA molecule (Fig. 1B). The RPL29 element has large, imperfect (90% identical) inverted repeats of 1.23 kbp at its ends, surrounding a central core region of 2.02 kbp (Fig. 1B). These structural features, as well as other data presented below, indicate that the IES in the micronuclear RPL29 locus is a transposable element. Based on the previous naming of elements in E. crassus, we refer to this family of elements as Tec3 (transposon of E. crassus 3) and to the particular element in the RPL29 locus as Tec3-1. In other analyses, it has been found that there is a perfect duplication of the first 258 bp of the RPL29 locus, along with at least 316 bp of sequence that is a nearly perfect match to left flanking DNA, located ∼6.5 kbp upstream of the micronuclear region represented in clone Mic.RPL-29-2 (A. Sánchez-Blanco and L. A. Klobutcher, unpublished results). However, the remainder of the RPL29 locus, including the region interrupted by the Tec3-1 element, is not present in the intervening DNA.
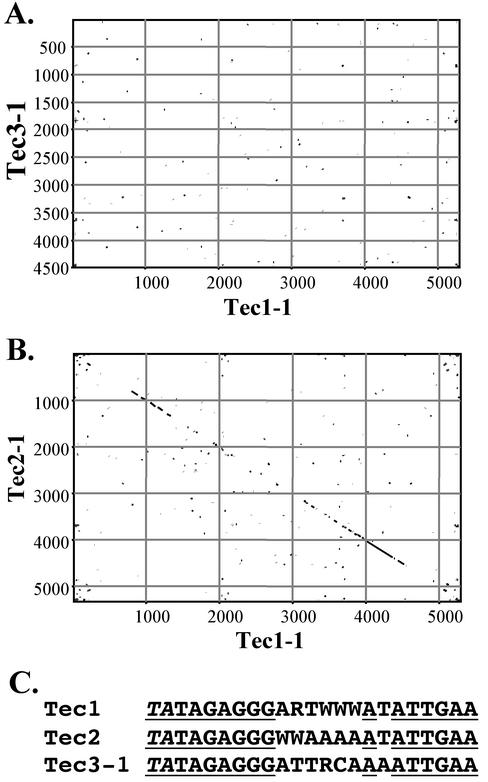
The Tec3-1 element is organizationally similar to the previously characterized Tec1 and Tec2 E. crassus transposon IESs (21, 24, 34), but they appear to be distinct elements on the basis of a number of criteria. First, Tec3-1 is shorter than the Tec1 and Tec2 elements, which are both 5.3 kbp long. Second, the Tec1 and Tec2 elements have shorter inverted repeats of ∼700 bp. Third, there is little primary sequence similarity between Tec3-1 and Tec 1 or Tec2. The latter was assessed by carrying out dot matrix comparisons of the various elements. When Tec3-1 was compared to either Tec1 (Fig. 2A) or Tec2 (data not shown), no sequence similarity was evident. In contrast, as initially shown by Jahn et al. (18), Tec1-1 and Tec2 display significant similarity when compared by using the same parameters (Fig. 2B).
FIG. 2.
Dot matrix comparisons of Tec elements. (A and B) Dot plot matrix comparisons of Tec1-1 (GenBank accession no. L03359) and the Tec3-1 sequence (A) and of Tec1-1 and Tec2-1 (GenBank accession no. L03360) (B). The comparisons were carried out by using MacVector DNA sequence analysis software, with a window size of 30 bases and a minimum score of 65%. (C) Comparison of the terminal sequence of Tec3-1 to the consensus terminal sequences of the Tec1 and Tec2 transposons (24). The terminal 5′-TA-3′ direct repeat is indicated in italic type, and positions that are identical in all elements are underlined.
Despite this lack of overall sequence similarity, the termini of Tec3-1 are quite similar to those of the Tec1 and Tec2 elements (Fig. 2C). Of the first 23 positions at the ends of the Tec1 and Tec2 elements, 16 are universally or nearly universally conserved among members of the two transposon families (Fig. 2C) (24). The ends of Tec3-1 are identical at all of these positions and are similar to the consensus sequences of Tec1 and Tec2 at other, less conserved terminal positions (Fig. 2C). This region includes the first 9 bp (including the flanking 5′-TA-3′ direct repeats), which are identical in all three elements.
Excision of Tec3-1 during macronuclear development.
Previous studies with both E. crassus and Paramecium suggested that the terminal sequences of IESs in these organisms play an important role in specifying their developmental excision (reviewed in references 14 and 20). Thus, the similarity of the Tec3-1 termini to the ends of the Tec1 and Tec2 elements suggested that Tec3-1 might be removed by the same excision machinery during macronuclear development. To test this proposal, two types of analyses were carried out.
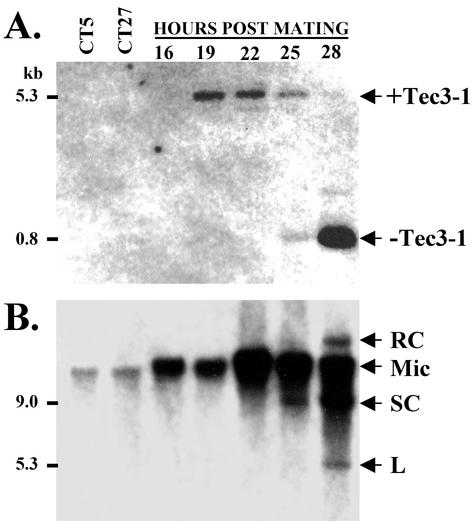
First, we used a PCR procedure to determine whether Tec3-1 is excised from the RPL29 locus during the same developmental period as that during which the Tec1 and Tec2 IESs are removed. PCR was performed with total DNA isolated from cells at different stages of development and with oligonucleotide primers LRPL-4 and RFr29-1, which bind to sequences flanking the micronuclear copy of RPL29 (Fig. 1B). These primers will amplify the micronuclear copy of the RPL29 locus with or without the Tec3-1 element up until the chromosome fragmentation stage of macronuclear development but will not amplify the macronuclear chromosome containing the RPL29 gene. PCR amplification was carried out with vegetative whole-cell DNA and DNA isolated at various times after mixing of cells of complementary mating types to initiate conjugation and macronuclear development (Fig. 3A). Developing-cell DNAs at 19 and 22 h yielded only a 5.3-kbp PCR product representing the micronuclear RPL29 locus containing the Tec3-1 element. While the vegetative-cell and 16-h DNA preparations, which contained fewer copies of the micronuclear RPL29 locus, did not produce detectable amounts of the 5.3-kbp product in this analysis, we have observed the expected products under different experimental conditions (Fig. 1A and data not shown). Beginning at 25 h, an 0.8-kbp band was also detected, representing the micronuclear RPL29 locus lacking the Tec3-1 element (Fig. 3A). The 0.8-kbp signal increased at 28 h, concomitant with the reduction of the larger PCR product representing the Tec3-1-containing forms. These results indicate that Tec3-1 is excised from the RPL29 locus during the early polytene chromosome stage of macronuclear development, the same period during which excision of the Tec1 and Tec2 transposon IESs begins (21, 34). To confirm that Tec3-1 is indeed excised at the same time, Southern hybridization analysis of the same developing-cell DNA preparations was carried out with a Tec1-specific probe. Free, extrachromosomal circular and linear forms of Tec1 also were first detected at 25 h of development (Fig. 3B).
FIG. 3.
Developmental excision of Tec3-1. (A) Timing of Tec3-1 excision from the RPL29 locus. Shown is a Southern blot containing the PCR amplification products obtained from strain CT5 and CT27 vegetative whole-cell DNAs and whole-cell DNAs isolated at various times after mating of the CT5 and CT27 cell lines (lanes representing 16, 19, 22, 25, and 28 h). The blot was hybridized with the 804-bp PCR product generated from the micronuclear RPL29 locus after Tec3-1 excision with primers RFr29-1 and LRPL-4 (Fig. 1B). Hybridization signals at 5.3 kbp represent the RPL29 locus with the Tec3-1 element (+Tec3-1), while the 0.8-kbp signal represents the locus following excision (−Tec3-1). (B) Southern blot of the same undigested, whole-cell DNAs as those used as the substrates for PCR following hybridization with a 1.6-kb HindIII/SalI restriction fragment derived from the Tec1 region of clone PhCl.4 (see Fig. 7B). The Tec1 elements in micronuclear DNA are indicated (Mic), along with the positions of free supercoiled circles (SC), relaxed circles (RC), and linear forms (L).
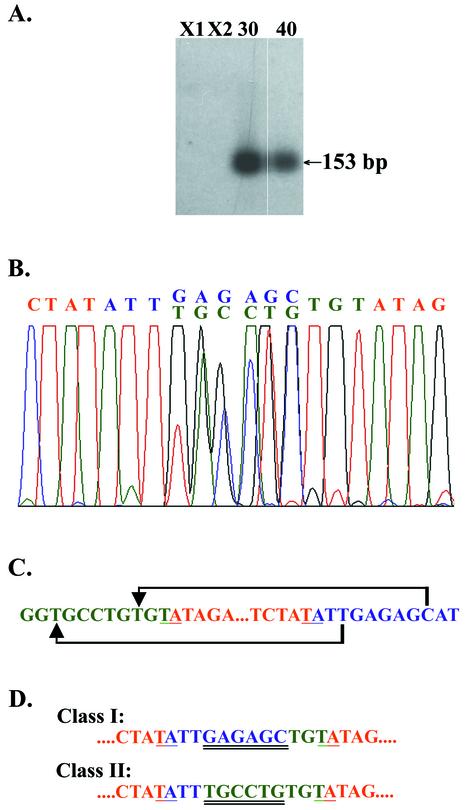
As a second means of assessing whether Tec3-1 excision might be mediated by the same machinery as that which removes the Tec1 and Tec2 transposon IESs, we characterized its excision product. Previous studies indicated that both E. crassus small IESs and Tec1 and Tec2 transposon IESs exist as free circles following their developmental excision (21, 24, 32, 34, 46). The junctions, or joining points, of these circles are unusual: they contain two copies of the terminal 5′-TA-3′ direct repeat separated by 10 bases that are derived from the macronucleus-destined DNA immediately flanking the IES. The central six bases of this junction region appear to be a heteroduplex, with one DNA strand being derived from right flanking sequences and the other being derived from left flanking sequences (for models of the excision process, see references 24 and 32). To determine whether Tec3-1 excision produces circles with this unusual junction structure, PCR was performed with primers that were complementary to regions 72 bp from the ends of the Tec3-1 element and that directed DNA synthesis toward the ends of the element (Tec3-1cirL and Tec3-1cirR; Fig. 1B). These primers should produce a PCR product of ∼150 bp only if the ends of the Tec3-1 element are joined to form a circle following excision. Because Tec3-1 has inverted repeats at its ends, the PCR product produced from the circle junction will be palindromic. As such structures are difficult to amplify by PCR, the two primers were chosen to be complementary to a region of the Tec3-1 inverted repeats that was not perfectly complementary. PCR amplification of DNA isolated from developing macronuclei at 30 and 40 h produced a product of the expected size (Fig. 4A), while a control PCR with vegetative-cell DNA produced no detectable product (Fig. 4A).
FIG. 4.
Characterization of excised Tec3-1 circles. (A) Southern blot containing the PCR amplification products obtained from strain X1 and X2 vegetative-cell DNAs and developing macronuclear DNAs isolated at 30 and 40 h after mating of the two strains with primers Tec3-1cirL and Tec3-1cirR (Fig. 1). The blot was hybridized with a plasmid subclone containing a 0.98-kbp EcoRI restriction fragment from the left end of clone Mic.RPL-29-2, which includes the left end of the Tec3-1 element. (B) DNA sequencing chromatogram from the bulk analysis of the 153-bp PCR product derived from the Tec3-1 circles. The junction region, containing six ambiguous positions, is shown. See the description of panel C for an explanation of colors. (C) Sequences at the ends of the Tec3-1 element. The Tec3-1 sequence is shown in red, and the left flanking and right flanking sequences that will form macronuclear RPL29 are shown in green and blue, respectively. The 5′-TA-3′ repeats flanking the element are underlined, and the brackets illustrate which bases are joined to form the two types of PCR products obtained. (D) Sequences of the two classes of clones obtained from PCR amplification of the Tec3-1 circle junction. Color coding is as described above, with double underlining indicating the proposed heteroduplex region of the circle junction.
To further characterize the circle junction, the ∼150-bp PCR product was gel purified, reamplified by PCR, and directly sequenced with oligonucleotide Tec3-1cirR as the sequencing primer. The resulting sequencing chromatogram confirmed that the PCR product was indeed derived from DNA molecules in which the left and right ends of Tec3-1 were joined and that the predicted type of circle junction was present (Fig. 4B) (note that circle junctions from other Tec3 elements were not observed, presumably because the primers were chosen to match divergent positions of the Tec3-1 inverted repeats). The junction region contained a pair of 5′-TA-3′ direct repeats separated by 10 bases. The central six bases of the junction region were ambiguous in the chromatogram; at each position, two peaks were present, a result that one would expect if the PCR product were derived from a substrate DNA molecule containing a heteroduplex region. The sequences of the six ambiguous positions can be interpreted to indicate that two types of PCR products were generated: one with the six bases derived from left flanking sequences and the other with the six bases derived from right flanking sequences (Fig. 4B and C). To demonstrate that this was the case, the PCR products were inserted into a plasmid vector, and four individual clones were isolated and sequenced. Two classes of clones were obtained. For two clones (Fig. 4C and D, class I), the junction region could be viewed as being formed by the joining of the eighth base distal to the right 5′-TA-3′ direct repeat of Tec3-1 to the second base upstream of the left 5′-TA-3′ repeat. For the other two clones, the junction was such that the second base to the right of the element was joined to the eighth base to the left of the element to form the circle junction (Fig. 4C and D, class II). These results parallel those obtained in similar analyses of the circle junctions of E. crassus small IESs (32). While the results do not prove that a heteroduplex is present at the circle junction, the two observed classes of clones are consistent with a circle junction in which the central six bases are a heteroduplex. We conclude from these analyses that Tec3-1 is excised at the same time of macronuclear development as Tec1 and Tec2 and that it forms a free circular form with a structure that appears to be identical to those of other excised IESs in E. crassus.
Identification and characterization of additional Tec3 elements in the micronuclear genome.
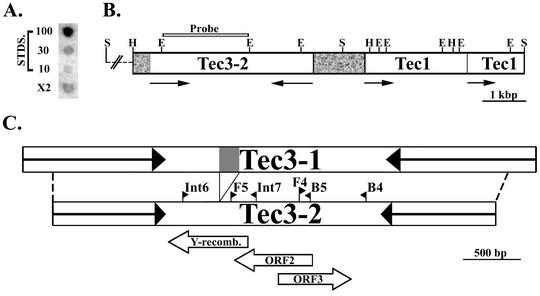
Other studies were carried out to determine whether additional copies of the Tec3 element were present in the micronucleus and to further characterize the element family. To assess the abundance of Tec3 elements, we carried out a dot blot hybridization analysis (Fig. 5A). Hybridization filters were prepared with a known amount of E. crassus total cellular DNA as well as with various amounts of a cloned 2-kbp EcoRI fragment derived from a Tec3 element (Fig. 5B), which served as copy number standards. The filters were then hybridized with the same 2-kbp EcoRI fragment and washed at moderate stringency, and the amount of bound radioactivity was determined. Repeated analyses of this type indicated that there are only 20 to 30 copies of Tec3 residing within the micronuclear genome. This number is significantly smaller that those of the E. crassus Tec1 and Tec2 elements, which are each present at 5,000 to 7,000 copies per haploid micronuclear genome (2, 21, 34). Southern hybridization of total cellular DNA with a variety of Tec3-derived probes indicated that all copies of the element are eliminated during the formation of the macronucleus (data not shown).
FIG. 5.
Additional Tec3 elements in the micronuclear genome. (A) Determination of Tec3 copy number. An autoradiograph of a dot blot hybridized with a 2-kbp EcoRI restriction fragment from Tec3-2 (see panel B) is shown. The blot contains 10 μg of E. crassus strain X2 total cellular DNA and amounts of the 2-kbp EcoRI fragment from Tec3-2 that were calculated to be equivalent to 10, 30, and 100 copies of the element per cell (STDS.) (see Materials and Methods). (B) Map of a portion of the insert in recombinant clone PhCl.4. The Tec3-2 and Tec1 elements are shown as open rectangles, with arrows indicating inverted repeats. Grey regions represent other micronucleus-limited sequences flanking the elements. E, EcoRI; H, HindIII; S, SalI. (C) Comparison of the Tec3-1 and Tec3-2 elements, along with the positions of the three pairs of PCR primers (Int6-Int7, F5-B5, and F4-B4) that were used to amplify additional segments of Tec3 from genomic DNA. The positions of the defective tyrosine recombinase ORF (Y-recomb.) and two other long ORFs are indicated as arrows below the map. The 171-bp insertion within the tyrosine recombinase ORF of the Tec3-1 element is indicated in grey.
To obtain additional copies of Tec3, the E. crassus LEMIC micronuclear library (2) was screened with a hybridization probe derived from the core region of Tec3 elements. This screening resulted in the identification of clone PhCl.4, which contains an ∼15.8-kbp insert harboring a Tec3 element. A 9.2-kbp region of the clone containing the Tec3 element (Tec3-2) was subcloned and completely sequenced (Fig. 5B). Tec3-2 is quite similar to Tec3-1 but also displays two significant differences. The core region of Tec3-2 lacks a block of 171 bp that is present in Tec3-1 (Fig. 5C) but is otherwise 68% identical in sequence. The second significant difference in the elements concerns their termini. Tec3-2 is bounded by 5′-TA-3′ direct repeats, but its inverted repeats are only 980 bp long. The Tec3-2 inverted repeats are quite similar to the innermost regions of the Tec3-1 inverted repeats, but the distal 258 bp of the 1.23-kbp Tec3-1 inverted repeats are missing from Tec3-2 (Fig. 5C). As a result of these terminal truncations, the ends of the Tec3-2 element show no significant similarity to the Tec1 or Tec2 terminal consensus sequence.
The Tec3-2 element also appears to differ from Tec3-1 in that it does not reside within macronucleus-destined DNA. Southern hybridization analysis with the flanking regions of Tec3-2 as probes against total cellular DNA showed no strong hybridization to macronucleus-sized DNA (data not shown). Thus, Tec3-2 appears to reside within a segment of the micronuclear genome that is eliminated during development. This conclusion is further supported by the sequence analysis, which identified two incomplete copies of the Tec1 element adjacent to Tec3-2 (Fig. 5B). One copy of the Tec1 element appears to be inserted within another (in inverse orientation), a phenomenon previously observed for the E. crassus Tec2 element (34). Thus, the entire insert of clone PhCl.4 may represent part of a large region of the micronuclear genome that is eliminated during development. Frels et al. (12) previously found that most Tec1 and Tec2 transposons in non-macronucleus-destined regions of the genome are both replicated and eliminated later in macronuclear development than are elements in macronucleus-destined DNA. They suggested that such elements may not be excised as IESs but instead are removed in the context of other eliminated DNA. We suspect that this is the case for Tec3-2, as its truncated inverted terminal repeats lack the conserved termini that appear to be necessary for excision as an IES.
Initial searches of the GenBank database with the core regions of the Tec3-1 and Tec3-2 elements indicated that they contain degenerate ORFs encoding proteins similar to the catalytic domain of a number of prokaryotic tyrosine recombinases (38). However, the analysis was complicated because the putative ORF regions encompassed the 171-bp region in Tec3-1 that is absent in Tec3-2 (Fig. 5C) and by the presence of multiple stop codons and possible frameshifts. Thus, to more thoroughly address the potential coding functions of Tec3 elements, we sought to determine a consensus sequence for the core region. Based on the sequences of the core regions in Tec3-1 and Tec3-2, three pairs of oligonucleotide primers were designed to amplify by PCR three overlapping segments of the core region (Fig. 5C). For each pair of primers, two to four cloned PCR products were generated and sequenced. Each of the PCR products showed some differences in sequence from the Tec3-1 and Tec3-2 elements and from each other, indicating that they were derived from independent elements.
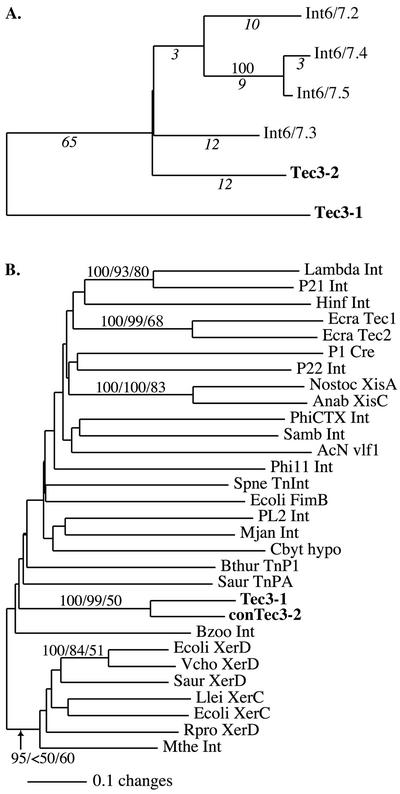
The PCR-generated cloned sequences are more similar to Tec3-2 than to Tec3-1. For example, Fig. 6A shows a genealogical analysis of the PCR clones generated with primers Int6 and Int7 and the corresponding regions of Tec3-1 and Tec3-2. The average pairwise difference between Tec3-1 and the remaining sequences, calculated from 465 overlapping base pairs (excluding gaps) by using the Tamura-Nei model (45) to correct for multiple hits, is 0.193 ± 0.004. This level of divergence is reflected by the 65 fixed differences between Tec3-1- and Tec3-2-like sequences (Fig. 6A). In contrast, the average pairwise difference between the Int6-Int7 PCR-generated clones and Tec3-2 is 0.054 ± 0.018. All of the available sequence data were used to derive a consensus sequence for the core region of the Tec3 elements, which was used in subsequent database searches described below. This analysis also indicates that the extra 171 bp in Tec3-1 relative to Tec3-2 likely represent an insertion, as all of the PCR-generated clones lacked this region.
FIG. 6.
Genealogical analyses. (A) Genealogy of Tec3-1, Tec3-2, and four cloned PCR products generated with primers Int6 and Int7 (Int6/Int7.2-Int6/Int7.5), determined by NJ analysis of nucleotide sequences, excluding gaps. Numbers above branches represent NJ bootstrap support values; branches with less than 50% bootstrap support are not labeled. Branch lengths are proportional to the numbers of changes, and the numbers of unambiguous changes are indicated below branches in italic type. (B) Genealogy of tyrosine recombinases, determined by NJ analysis of amino acid sequences. Numbers above branches represent bootstrap support values estimated by NJ and MP analyses and PZ support values, in that order. Branches with less than 50% bootstrap support are not labeled. Branch lengths are proportional to the numbers of changes in each branch. GenBank numbers and further details on algorithms and models are described in Materials and Methods. For the alignment of the proteins and regions included in the analyses, see the supplemental figure at www.science.smith.edu/departments/Biology/lkatz/align.html.
Tec3 elements encode a tyrosine recombinase.
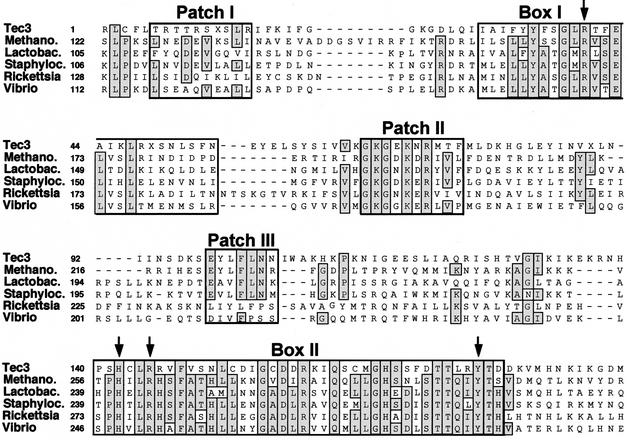
BLAST searches of databases were performed with conceptual translations of the core region consensus sequence to identify potential proteins encoded by the Tec3 elements. Only the left end of the core region produced significant matches in the BLAST search, detecting numerous tyrosine recombinases (38). The best matches (e values of <10−4) were to homologs of the XerC and XerD proteins, which are involved in resolving the products of DNA replication in a number of bacterial species. An alignment of the putative consensus Tec3 protein with the best matches in the databases is shown in Fig. 7. The putative Tec3 protein displays significant sequence similarity to the 180-amino-acid region which has been defined as the tyrosine recombinase domain (amino acids 175 through 355 of the bacteriophage λ Int protein [38]). The sequence similarity is primarily limited to the highly conserved box I and II regions and the three more poorly conserved patch regions that are signatures of the tyrosine recombinase family (Fig. 7). In addition, the putative Tec3 protein contains the four box I and II residues (R-H-R-Y) that are almost universally conserved among tyrosine recombinases and that participate in catalysis (38). While these results provide a strong indication that Tec3 elements contain a tyrosine recombinase gene, it appears that the gene is no longer functional in a significant fraction of the elements, as even the Tec3 consensus sequence has three stop codons (Fig. 7).
FIG. 7.
Comparison of the Tec3 tyrosine recombinase consensus sequence with the XerC- or XerD-like proteins of Methanothermobacterium thermoautotrophicum (Methano.; GenBank accession no. 7428936), L. leichmannii (Lactobac.), S. aureus (Staphyloc.), R. prowazekii (Rickettsia), and V. cholerae (Vibrio). The alignment was obtained by using ClustalW, but with some manual adjustments. Positions which are identical in the majority of the proteins are shaded. The conserved box and patch regions characteristic of the tyrosine recombinase protein family (38) are boxed, and the nearly universally conserved residues involved in catalysis are indicated by arrows.
Tyrosine recombinase genes are typically not found in eukaryotic transposons. Nonetheless, a recent analysis indicated that the ORF2 regions of the Tec1 and Tec2 transposons also encode tyrosine recombinases (7a). As we have noted, there is little shared sequence similarity between Tec3 and the Tec1 or Tec2 element at the DNA level. The Tec3 tyrosine recombinase also appears to be distantly related to that of Tec1 or Tec2, as the Tec1 and Tec2 proteins were not among the 50 best matches (e values, <0.14) in the BLAST searches. To further explore this issue, we carried out a genealogical analysis of 26 proteins that represent the major subclasses of the tyrosine recombinase family and the tyrosine recombinases of Tec1, Tec2, and Tec3-1. Also included in the analysis were a consensus sequence (conTec3-2) derived from the Tec3-2 tyrosine recombinase region and the four related Int6-Int7 PCR-generated clones (Fig. 6A). The resulting genealogy reveals only limited phylogenetic information from tyrosine recombinases from eukaryotic, bacterial, viral, and archaeal genomes, as evidenced by the short length of branches at deep nodes and the relatively low bootstrap support at most nodes (Fig. 6B). There is strong support, as indicated by bootstrap values of ≥99%, for the sister relationship of the E. crassus Tec1 and Tec2 ORF2 proteins and for the E. crassus Tec3-1 and conTec3-2 tyrosine recombinases in the NJ and MP analyses (Fig. 6B). In contrast, the ML analyses provide limited support for the sister status of Tec1 and Tec2 (PZ value, 68%) and of Tec3-1 and conTec3-2 (PZ value, 50%).
Although there is little support at deep nodes, there is no evidence that the diverse E. crassus Tec sequences form a monophyletic group. The NJ analysis of aligned box and patch amino acids places the E. crassus Tec1 and Tec2 sequences in a clade with integrase genes from the bacterium Haemophilus influenzae (Fig. 6B, Hinf Int) and the lambda and P21 phages (Fig. 6B, Lambda Int and P21 Int), although NJ bootstrap values are less than 50% for these relationships and these nodes are unresolved in both the MP and the ML (puzzle) analyses. In contrast, the Tec3-1 and conTec3-2 sequences do not appear to be closely related to any known tyrosine recombinase (Fig. 6B). The results of a Kishino-Hasegawa test (26) that compared the most parsimonious tree with one constraining all of the Tec elements to be monophyletic were not significant, indicating that we cannot reject a single origin for E. crassus Tec tyrosine recombinases based on these analyses. The only other well-supported nodes in the genealogy, based on bootstrap and PZ support values of >70%, are for two recombinase genes isolated from cyanobacteria (Fig. 6B, Nostoc XisA and Anab XisC), two phage integrase genes (Fig. 6B, Lambda Int and P21 Int), E. coli and V. cholerae XerD sequences, and a large clade containing diverse XerC, XerD, and XerC- or XerD-like proteins from bacteria and archaea. Analyses of the highly conserved box regions alone or of alternative alignments had no significant impact on the resulting genealogy (data not shown). There is a relatively high divergence (∼19%, excluding gaps) between the Tec3-1 and conTec3-2 tyrosine recombinase sequences, indicating that these two classes of sequences have coexisted within the genome of E. crassus for quite some time (Fig. 6B).
While the remainder of the Tec3 core region generated no significant BLAST matches, other coding regions are likely to be present. Two relatively long regions that are uninterrupted by stop codons are present in the core region consensus sequence (ORF2 and ORF3 in Fig. 5C; bases 3310 to 2641 and 3034 to 3657 in Mic.RPL-29-2, respectively). ORF3 would be transcribed in the orientation opposite that of the tyrosine recombinase-coding region and so might encode a second protein. ORF2 would be transcribed in the same direction as the tyrosine recombinase-coding region, but it is in a different reading frame. As a result, ORF2 might also encode a separate protein. However, many tyrosine recombinases have N-terminal regions that are not conserved among family members, and it is possible that ORF2 represents such an N-terminal region that has been separated from the Tec3 tyrosine recombinase by a frameshift mutation or by the presence of an intron.
DISCUSSION
In this study, we have identified a small family of transposons in E. crassus that differ significantly from the previously identified Tec1 and Tec2 elements. One copy of the Tec3 family (Tec3-1) interrupts the RPL29 locus. Tec3-1 is 4.48 kbp long and has 1.23-kbp terminal inverted repeats. A second Tec3 family member (Tec3-2) resides in a region of the genome that contains Tec1 elements. Tec3-2 shares a ∼1.9-kbp core region with Tec3-1 but lacks 258 bp of the inverted terminal repeats. Which of these two end structures is generally characteristic of the Tec3 element family is unclear at present, as we have been unable to isolate additional copies of element termini by using a number of PCR-based procedures. All copies of the element family are eliminated during the process of macronuclear development.
The Tec3-1 element residing within the micronuclear RPL29 locus behaves as an IES. It is excised during the early polytene chromosome stage of macronuclear development, the same period during which removal of the Tec1 and Tec2 IESs begins (21, 34). In addition, the results indicate that Tec3-1 excision products are likely identical to those of other Euplotes IESs. Tec3-1 is precisely excised, resulting in an RPL29 macronuclear DNA molecule that retains one copy of the terminal 5′-TA-3′ direct repeat of the element and forms a circle following excision. Moreover, the data indicate that the Tec3-1 element forms a circle following excision which appears to have the same type of heteroduplex junction structure as other E. crassus IESs. These results are significant in that Tec3-1 shares little sequence similarity with the Tec1 and Tec2 elements, except for its termini, where 16 of 23 bp are identical to Tec1 and Tec2. These terminal sequences, which are also similar to the consensus sequences for the E. crassus short IESs, Paramecium short IESs, and the Tc1/Mariner transposons (17, 28), have been suggested to play a major role in specifying IES excision. As the termini are the only discernible feature shared among the E. crassus IESs, the similarity in the timing and products of Tec3-1 excision provides additional evidence that these sequences play a major role in specifying IES removal.
The results also indicate that the IES terminal sequence may be a bipartite structure. In a comparison of the ends of the three E. crassus transposons, a terminal conserved block of 9 bp is separated from a second conserved block of 6 bp by an AT-rich 8-bp region (Fig. 2C). The two shared blocks represent nearly universally conserved regions of members of the Tec1 and Tec2 element families (24), arguing that they play a role in some function important for the elements. This bipartite structure is reminiscent of the V(D)J recombination signal sequences (RSS) that specify gene rearrangement in the vertebrate immune system. The RSS are comprised of a conserved heptamer and nonamer that are separated by either 12 or 23 bp (reviewed in reference 13). Intriguingly, a number of lines of evidence suggest that the V(D)J recombination system evolved from a transposable element (reviewed in reference 13), and some similarity between the RSS heptamer and the termini of the Tc1/Mariner family of transposons has been noted (8).
Tec3 tyrosine recombinase gene.
The Tec3 elements contain a defective ORF that appears to have encoded a tyrosine recombinase protein. Tyrosine recombinases are a broad family of proteins that carry out a variety of functions, including viral integration, DNA inversion, transposition, and the resolution of catenated DNA circles (see, e.g., reference 38). All of the analyzed Tec3 ORFs are defective by virtue of the presence of indels and/or stop codons, making it unlikely that the predicted tyrosine recombinase protein plays a role in the IES excision process at present. It seems more likely that it was involved in the transposition of Tec3 elements. Resolvase genes are found in prokaryotic elements that transpose via a replicative mechanism (reviewed in reference 15). The resolvases are usually members of the distinct family of serine recombinases, but some elements encode a tyrosine recombinase that functions as a resolvase (see, e.g., reference 36). During replicative transposition, a cointegrate structure forms that links the transposon donor DNA with the target DNA. The resolvase carries out site-specific recombination between sites within the two transposon copies to resolve the donor and target DNAs. Resolvase genes have not been identified yet in eukaryotic transposons. As discussed by Doak et al. (7a), the resolution of replicative transposition products in eukaryotic genomes with multiple linear chromosomes and multiple transposon copies would likely give rise to a variety of deleterious chromosome rearrangements, unless a mechanism existed that restricts recombination to the donor and target copies of the transposon. Thus, while it is possible that the Tec3 tyrosine recombinase served as a resolvase, a replicative transposition mechanism does pose some problems.
An alternative is that the Tec3 tyrosine recombinase was involved in the initial steps of transposition. A tyrosine recombinase protein mediates the excision and transposition of the Tn916 and Tn1545 conjugative transposons (49), so that it is conceivable that the Tec3 tyrosine recombinase played a similar role in the multiplication of this element. Intriguingly, the excised circular form of Tn916, which is believed to be a transposition intermediate, has a heteroduplex region at its junction that is derived from the sequences originally flanking the element (6). Although the structure of the Tn916 circular junction differs somewhat from that of the E. crassus circular small IESs and Tec transposons, these are the only examples that we are aware of in which excised circular forms of elements have heteroduplex junctions.
Our genealogical analysis indicates that the Tec3 tyrosine recombinase is not closely related to those of the Tec1 and Tec2 transposons. Moreover, the phylogenetic analyses failed to provide strong support for the association of the Tec3 tyrosine recombinase with any of the functionally distinct subfamilies of tyrosine recombinases. The possibility of elevated mutation rates in E. crassus Tec3 sequences is likely part of the explanation for the lack of resolution in our genealogies. Although these analyses did not provide insight into the function of the Tec3 protein, it is clear that it is more closely related to prokaryotic members of the group than to eukaryotic members (e.g., the yeast FLP recombinase proteins), which differ significantly in sequence (9, 38). In fact, the yeast sequences are so divergent that they could not be unambiguously aligned with the prokaryotic, viral, and ciliate sequences and were not included in our analyses. The high level of divergence between ciliate Tec tyrosine recombinases and other eukaryotic tyrosine recombinases may be an indication of lateral transfer of Tec elements from a prokaryote or virus to Euplotes. In nature, ciliates meet their nutritional requirements by engulfing other microorganisms, including prokaryotes, and it has been suggested that such food organisms might be the source of at least some ciliate transposons (29).
Possible origins of multiple developmentally excised transposon families.
Our studies establish the Tec3 element as another family of sequences that undergo developmental elimination in E. crassus, with at least one family member (Tec3-1) behaving as an IES. The previously described Tec1 and Tec2 elements behave in a similar manner, raising the question of how these various elements are related. The Tec1 and Tec2 elements are very similar elements, so that these families may have diverged from one another following the invasion of the micronuclear genome by an ancestral element. However, this putative ancestral element is unlikely to have also given rise to the Tec3 element. The Tec3 element shares little sequence similarity at the DNA level with the Tec1 and Tec2 elements and appears to lack a gene encoding a Tc1/Mariner-like transposase, which both Tec1 and Tec2 possess (7). Furthermore, even though all of the elements have a tyrosine recombinase ORF, our analyses indicate that the Tec1 or Tec2 protein is distantly related to the Tec3 protein. Thus, we suggest that the Tec3 element independently invaded the E. crassus micronuclear genome.
If so, how have members of these different element families come to be subject to the same developmental excision system? It was recently suggested that ciliate IES removal represents a means of ridding the genome of repetitive DNA elements, possibly serving as a defense against the deleterious events of transposable elements (20, 37, 52). It was also proposed that transposable elements were responsible for establishing the excision system, an event which would serve to enhance their survival in the host (29). Whatever the origin and purpose of the excision system, it appears to be a sequence-specific process in E. crassus, as the majority of IESs share similar termini.
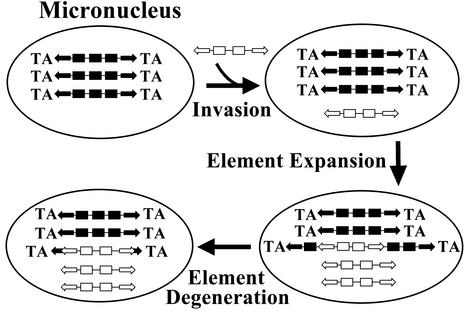
Scenarios for the origin of the Tec3 element in the E. crassus micronucleus need to explain the two types of terminal structures observed for the Tec3-1 and Tec3-2 family members. While there are a number of possible explanations, one interesting possibility is that some Tec3 elements acquired their terminal sequences from Tec1 or Tec2 by an “end-swapping” process (Fig. 8). In this scheme, the original Tec3 element that entered the micronuclear genome possessed the shorter inverted terminal repeats of Tec3-2. This original element would have been under the selective constraints typical for transposons in most organisms, as it could not be developmentally excised. However, during limited expansion in the micronuclear genome, a Tec3 element might have inserted within a Tec1 or Tec2 element. Insertions of transposons within transposons are found in many organisms. Indeed, in the micronuclear clone harboring Tec3-2, an insertion of one Tec1 element into another was found (Fig. 5B), and Krikau and Jahn (34) previously described a Tec2 transposon residing within another. Once such an insertion of a Tec3 element into Tec1 or Tec2 occurred, sequence deletions may have resulted in a composite element primarily composed of Tec3 sequences but with Tec1 or Tec2 termini. By virtue of acquiring Tec1 or Tec2 termini, this composite element would now be subject to the IES excision process, enhancing its chances of survival in the host genome. In addition, since terminal sequences are typically the cis-acting sequences required for transposition, end-swapping may also have allowed the Tec3 element to transpose by using Tec1 or Tec2 transposase in trans. This transposon end-swapping hypothesis is attractive in that it provides a straightforward means for disparate transposons to fall under the same system of developmental removal.
FIG. 8.
End-swapping model for the origins of multiple transposon IESs. The micronucleus initially harbors one transposon family with termini compatible with developmental excision. A second transposon invades the micronuclear genome, occasionally inserting within an element of the original family. Deletions result in the formation of a composite element that can also be excised. Within the oval diagrams, rectangles represent coding regions, arrows indicate inverted repeats, and “TA” denotes the conserved terminal sequence required for excision.
Acknowledgments
We thank Tom Doak and Glenn Herrick for assistance in the alignment of tyrosine recombinase proteins and for helpful discussions.
This work was supported by National Science Foundation grant MCB-9816765 to L. A. Klobutcher and a National Science Foundation CAREER award to L. A. Katz.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baird, S. E., G. M. Fino, S. L. Tausta, and L. A. Klobutcher. 1989. Micronuclear genome organization in Euplotes crassus: a transposonlike element is removed during macronuclear development. Mol. Cell. Biol. 9:3793-3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baird, S. E., and L. A. Klobutcher. 1989. Characterization of chromosome fragmentation in two protozoans and identification of a candidate fragmentation sequence in Euplotes crassus. Genes Dev. 3:585-597. [DOI] [PubMed] [Google Scholar]
- 4.Bernhard, D., A. Stechmann, W. Foissner, D. Ammermann, M. Hehn, and M. Schlegel. 2001. Phylogenetic relationships within the class Spirotrichea (Ciliophora) inferred from small subunit rRNA gene sequences. Mol. Phylogenet. Evol. 21:86-92. [DOI] [PubMed] [Google Scholar]
- 5.Boswell, R. E., L. A. Klobutcher, and D. M. Prescott. 1982. Inverted terminal repeats are added to genes during macronuclear development in Oxytricha nova. Proc. Natl. Acad. Sci. USA 79:3255-3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caparon, M. G., and J. R. Scott. 1989. Excision and insertion of the conjugative transposon Tn916 involves a novel recombination mechanism. Cell 59:1027-1034. [DOI] [PubMed] [Google Scholar]
- 7.Doak, T. G., F. P. Doerder, C. L. Jahn, and G. Herrick. 1994. A proposed superfamily of transposase genes: transposon-like elements in ciliated protozoa and a common “D35E”motif. Proc. Natl. Acad. Sci. USA 91:942-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7a.Doak, T. G., D. J. Witherspoon, C. L. Jahn, and G. Herrick. 2003. Selection on the genes of Euplotes crassus Tec1 and Tec2 transposons: evolutionary appearance of a programmed frameshift in a Tec2 gene encoding a tyrosine family site-specific recombinase. Eukaryot. Cell 2:95-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dreyfus, D. H. 1992. Evidence suggesting an evolutionary relationship between transposable elements and the immune system recombination sequences. Mol. Immunol. 29:807-810. [DOI] [PubMed] [Google Scholar]
- 9.Esposito, D., and J. J. Scocca. 1997. The integrase family of tyrosine recombinases: evolution of a conserved active site domain. Nucleic Acids Res. 25:3605-3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feinberg, A. P., and B. Vogelstein. 1983. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal. Biochem. 132:6-13. [DOI] [PubMed] [Google Scholar]
- 11.Frels, J. S., and C. L. Jahn. 1995. DNA rearrangements in Euplotes crassus coincide with discrete periods of DNA replication during the polytene chromosome stage of macronuclear development. Mol. Cell. Biol. 15:6488-6495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frels, J. S., C. M. Tebeau, S. Z. Doktor, and C. L. Jahn. 1996. Differential replication and DNA elimination in the polytene chromosomes of Euplotes crassus. Mol. Biol. Cell 7:755-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fugmann, S. D., A. I. Lee, P. E. Shockett, I. J. Villey, and D. G. Schatz. 2000. The RAG proteins and V(D)J recombination: complexes, ends, and transposition. Annu. Rev. Immunol. 18:495-527. [DOI] [PubMed] [Google Scholar]
- 14.Gratias, A., and M. Bétermier. 2001. Developmentally programmed excision of internal DNA sequences in Paramecium aurelia. Biochimie 83:1009-1022. [DOI] [PubMed] [Google Scholar]
- 15.Grindley, N. D. F. 1994. Resolvase-mediated site-specific recombination. Nucleic Acids Mol. Biol. 8:236-267. [Google Scholar]
- 16.Higgins, D. G., and P. M. Sharp. 1988. CLUSTAL: a package for performing multiple sequence alignment on a microcomputer. Gene 73:237-244. [DOI] [PubMed] [Google Scholar]
- 17.Jacobs, M. E., and L. A. Klobutcher. 1996. The long and the short of developmental DNA deletion in Euplotes crassus. J. Eukaryot. Microbiol. 43:442-452. [DOI] [PubMed] [Google Scholar]
- 18.Jahn, C. L., S. Z. Doktor, J. S. Frels, J. W. Jaraczewski, and M. F. Krikau. 1993. Structures of the Euplotes crassus Tec1 and Tec2 elements: identification of putative transposase coding regions. Gene 133:71-78. [DOI] [PubMed] [Google Scholar]
- 19.Jahn, C. L., M. Erbeznik, J. W. Jaraczewski, M. Melek, and D. E. Shippen. 1994. Sequence of the macronuclear DNA encoding large subunit ribosomal protein 29 (L29) in Euplotes crassus and cycloheximide sensitivity. Gene 151:231-235. [DOI] [PubMed] [Google Scholar]
- 20.Jahn, C. L., and L. W. Klobutcher. 2002. Genome remodeling in ciliated protozoa. Annu. Rev. Microbiol. 56:489-520. [DOI] [PubMed] [Google Scholar]
- 21.Jahn, C. L., M. F. Krikau, and S. Shyman. 1989. Developmentally coordinated en masse excision of a highly repetitive element in E. crassus. Cell 59:1009-1018. [DOI] [PubMed] [Google Scholar]
- 22.Jahn, C. L., L. A. Nilles, and M. F. Krikau. 1988. Organization of the Euplotes crassus micronuclear genome. J. Protozool. 35:590-601. [DOI] [PubMed] [Google Scholar]
- 23.Jaraczewski, J. W., J. S. Frels, and C. L. Jahn. 1994. Developmentally regulated, low abundance Tec element transcripts in Euplotes crassus—implications for DNA elimination and transposition. Nucleic Acids Res. 22:4535-4542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jaraczewski, J. W., and C. L. Jahn. 1993. Elimination of Tec elements involves a novel excision process. Genes Dev. 7:95-105. [DOI] [PubMed] [Google Scholar]
- 25.Jones, D. T., W. R. Taylor, and J. M. Thornton. 1992. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 8:275-282. [DOI] [PubMed] [Google Scholar]
- 26.Kishino, H., and M. Hasegawa. 1989. Evaluation of the maximum likelihood estimate of the evolutionary tree topologies from DNA sequence data, and the branching order in hominoidea. J. Mol. Evol. 29:170-179. [DOI] [PubMed] [Google Scholar]
- 27.Klobutcher, L. A., S. E. Gygax, J. D. Podoloff, J. R. Vermeesch, C. M. Price, C. M. Tebeau, and C. L. Jahn. 1998. Conserved DNA sequences adjacent to chromosome fragmentation and telomere addition sites in Euplotes crassus. Nucleic Acids Res. 26:4230-4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klobutcher, L. A., and G. Herrick. 1995. Consensus inverted terminal repeat sequence of Paramecium IESs: resemblance to termini of Tc1-related and Euplotes Tec transposons. Nucleic Acids Res. 23:2006-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klobutcher, L. A., and G. Herrick. 1997. Developmental genome reorganization in ciliated protozoa: the transposon link. Prog. Nucleic Acid Res. Mol. Biol. 56:1-62. [DOI] [PubMed] [Google Scholar]
- 30.Klobutcher, L. A., and D. M. Prescott. 1986. The special case of the hypotrichs, p. 111-154. In J. G. Gall (ed.), The molecular biology of ciliated protozoa. Academic Press, Inc., New York, N.Y.
- 31.Klobutcher, L. A., M. T. Swanton, P. Donini, and D. M. Prescott. 1981. All gene-sized DNA molecules in four species of hypotrichs have the same terminal sequence and an unusual 3′ terminus. Proc. Natl. Acad. Sci. USA 78:3015-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klobutcher, L. A., L. R. Turner, and J. LaPlante. 1993. Circular forms of developmentally excised DNA in Euplotes crassus have a heteroduplex junction. Genes Dev. 7:84-94. [DOI] [PubMed] [Google Scholar]
- 33.Knecht, K., and L. A. Klobutcher. 1995. Telomeric repeat sequences are not associated with Tec1 elements in Euplotes crassus. Eur. J. Protistol. 31:201-207. [Google Scholar]
- 34.Krikau, M. F., and C. L. Jahn. 1991. Tec2, a second transposon-like element demonstrating developmentally programmed excision in Euplotes crassus. Mol. Cell. Biol. 11:4751-4759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ling, Z., S. Ghosh, M. E. Jacobs, and L. A. Klobutcher. 1997. Conjugation-specific genes in the ciliate Euplotes crassus: gene expression from the old macronucleus. J. Eukaryot. Microbiol. 44:1-11. [DOI] [PubMed] [Google Scholar]
- 36.Mahillon, J., and D. Lereclus. 1988. Structural and functional analysis of Tn4430: identification of an integrase-like protein involved in the co-integrate-resolution process. EMBO J. 7:1515-1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meyer, E., and O. Garnier. 2002. Non-Mendelian inheritance and homology-dependent effects in ciliates. Adv. Genet. 46:305-337. [DOI] [PubMed] [Google Scholar]
- 38.Nunes-Düby, S. E., H. J. Kwon, R. S. Tirumalai, T. Ellenberger, and A. Landy. 1998. Similarities and differences among 105 members of the Int family of site-specific recombinases. Nucleic Acids Res. 26:391-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Prescott, D. M. 1994. The DNA of ciliated protozoa. Microbiol. Rev. 58:233-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Quan, L., and M. Wilkinson. 1991. DNA fragment purification from LMP agarose. BioTechniques 10:737-738. [PubMed] [Google Scholar]
- 41.Roth, M., M. Lin, and D. M. Prescott. 1985. Large scale synchronous mating and the study of macronuclear development in Euplotes crassus. J. Cell Biol. 101:79-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 43.Smith, R. F., B. A. Wiese, M. K. Wojzynski, D. B. Davison, and K. C. Worley. 1996. BCM Search Launcher—an integrated interface to molecular biology data base search and analysis services available on the World Wide Web. Genome Res. 6:454-462. [DOI] [PubMed] [Google Scholar]
- 44.Strimmer, K., and A. von Haeseler. 1996. Quartet puzzling: a quartet maximum likelihood method for reconstructing tree topologies. Mol. Biol. Evol. 13:964-969. [Google Scholar]
- 45.Tamura, K., and M. Nei. 1993. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 10:512-526. [DOI] [PubMed] [Google Scholar]
- 46.Tausta, S. L., and L. A. Klobutcher. 1989. Detection of circular forms of eliminated DNA during macronuclear development in E. crassus. Cell 59:1019-1026. [DOI] [PubMed] [Google Scholar]
- 47.Tausta, S. L., and L. A. Klobutcher. 1990. Internal eliminated sequences are removed prior to chromosome fragmentation during development in Euplotes crassus. Nucleic Acids Res. 18:845-853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tausta, S. L., L. R. Turner, L. K. Buckley, and L. A. Klobutcher. 1991. High fidelity developmental excision of Tec1 transposons and internal eliminated sequences in Euplotes crassus. Nucleic Acids Res. 19:3229-3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Taylor, K., and G. Churchward. 1997. Specific DNA cleavage mediated by the integrase of conjugative transposon Tn916. J. Bacteriol. 179:1117-1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vermeesch, J. R., and C. M. Price. 1994. Telomeric DNA sequence and structure following de novo telomere synthesis in Euplotes crassus. Mol. Cell. Biol. 14:554-566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yao, M.-C., S. Duharcourt, and D. L. Chalker. 2002. Genome-wide rearrangements of DNA in ciliates, p. 730-758. In N. L. Craig, R. Craigie, M. Gellert, and A. M. Lambowitz (ed.), Mobile DNA II. ASM Press, Washington, D.C.