Abstract
The Tec1 and Tec2 transposons of the ciliate Euplotes crassus carry a gene for a tyrosine-type site-specific recombinase. The expression of the Tec2 gene apparently uses a programmed +1 frameshift. To test this hypothesis, we first examined whether this gene has evolved under purifying selection in Tec1 and Tec2. Each element carries three genes, and each has evolved under purifying selection for the function of its encoded protein, as evidenced by a dearth of nonsynonymous changes. This distortion of divergence is apparent in codons both 5′ and 3′ of the frameshift site. Thus, Tec2 transposons have diverged from each other while using a programmed +1 frameshift to produce recombinase, the function of which is under purifying selection. What might this function be? Tyrosine-type site-specific recombinases are extremely rare in eukaryotes, and Tec elements are the first known eukaryotic type II transposons to encode a site-specific recombinase. Tec elements also encode a widespread transposase. The Tec recombinase might function in transposition, resolve products of transposition (bacterial replicative transposons use recombinase or resolvase to separate joined replicons), or provide a function that benefits the ciliate host. Transposons in ciliated protozoa are removed from the macronucleus, and it has been proposed that the transposons provide this “excisase” activity.
We have studied the evolution of ciliate Tec transposon sequences to compare their evolution in Euplotes crassus to that of TBE1 transposons in Oxytricha ciliates (49) as well as to examine the appearance of a frameshift site in a Tec2 gene (22). We have confirmed that this frameshift has been a functional mechanism, and we demonstrate that this gene and its homolog in Tec1 elements encode a tyrosine-type site-specific recombinase.
The Tec and TBE elements encode a DDE transposase (conserved Asp, Asp, Gln residues in the active site) (10) and appear to be type II elements (“cut-and-paste” or “DNA-mediated” elements) (5). The evolution of eukaryotic type II transposons has proved difficult to understand: purifying selection for transposase function is not expected to act on the transposase genes of eukaryotic cut-and-paste transposons within a host population and is generally not found (36, 37, 48, 49). However, TBE1 transposons are a notable exception to this expectation. TBE1 transposon genes have evolved under strong selection for the function of their encoded proteins within their host ciliates (49): that is, nonsynonymous—or missense—mutations have been selected against and so removed from the pool of transposons in the host genome, while synonymous mutations have accumulated unabated. The reason for this selection is at present unknown, but it has been proposed that TBE1 transposons are under selection for a specific host function (28, 46, 49).
Ciliated protozoa are unique transposon hosts because they carry two types of nuclei, a micronucleus and a macronucleus. The macronucleus is a terminally differentiated version of the micronucleus and provides all of the transcripts necessary for cell growth. When a new macronucleus differentiates, all ∼4,000 TBE1 insertions are removed, at least some by precise excision (for a review, see reference 28). This process regenerates many genes that were inactivated in the micronucleus due to TBE1 insertions. Developmental excision is, in essence, “wholesale” somatic reversion of germ line insertion mutations. Our proposal was that the three TBE1 genes are selected for function because they are responsible for the excision of TBE1 insertions during differentiation. This proposal is no longer tenable, because these genes are not detectably expressed during macronuclear development (see below).
The Tec1 and Tec2 elements of E. crassus are ciliate transposons comparable to TBE1 in their general features. They are present in high copy numbers and are precisely excised during macronuclear development. Tec genes are not expressed at levels sufficient to excise the ∼12,000 Tec1 elements and ∼12,000 Tec2 elements (24). Both Tec1 and Tec2 elements are 5.7-kb elements and have identical organization of their three genes and 690-bp inverted terminal repeats (ITRs). The encoded protein sequences of Tec1 and Tec2 are easily aligned but are only ∼40% identical. The noncoding sequences inside the ITRs have diverged to the point at which they cannot be aligned; thus, Tec1 and Tec2 diverged from each other a very long time ago and have since evolved into two quite independent families of elements.
The Tec and TBE elements are not demonstrably related except by the inclusion of their transposases in the IS630/Tc1/Mariner subfamily of the D,D35E transposase superfamily (10). That is, Tec and TBE transposases do not form a ciliate clade of transposases: they are as like Tc1 or IS630 as they are like each other. Nonetheless, both Tec and TBE1 elements carry three genes (11, 22), whereas eukaryotic type II transposons typically carry only a transposase gene (5). Other than the transposase genes, the other Tec1 or Tec2 and TBE genes are unrelated.
Oddly, in all Tec1 elements, open reading frame (ORF) 2 is a single ORF, while in all Tec2 elements, ORF 2 is divided by a frameshifting single-base-pair insertion into ORF 2A and ORF 2B. Jahn et al. (22) proposed that the complete Tec2 gene product is made by a programmed frameshift. Here we demonstrate that the two sections of ORF 2 have been under selection for protein function since the appearance of the frameshifting insertion, indicating that a programmed +1 frameshift has been used to produce the protein. We also show that ORF2 encodes a tyrosine family site-specific recombinase (34). Site-specific recombinases, of either the serine or the tyrosine families, are extremely rare in eukaryotes, and Tec elements are, to our knowledge, the first eukaryotic type II transposons found to encode a site-specific recombinase. Jacobs et al. (21a) report that the newly described Tec3 elements also encode such an enzyme. In prokaryotes, recombinases are associated with transposons that transpose replicatively, forming cointegrates that the recombinase subsequently resolves. That Tec elements possess a recombinase suggests that Tec elements might transpose replicatively, a complicated process in organisms with linear chromosomes.
MATERIALS AND METHODS
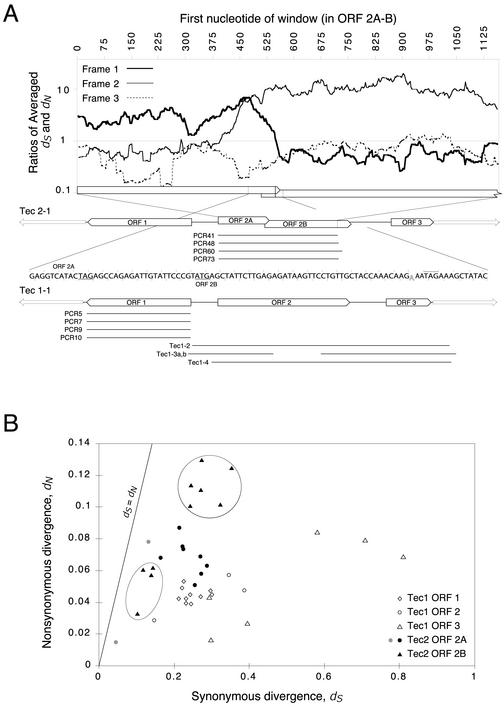
Tec element DNA sequences were determined as described by Jahn et al. (22). The partial ORF 2 sequence from Tec1-3 was not analyzed. Figure 1A indicates the sequences that we have analyzed: a single complete sequence each of a Tec1 element and of a Tec2 element, multiple sequences of all three Tec1 genes, and four sequences of the Tec2 ORF 2 gene that include the proposed frameshift region. Sequence analyses are described in the relevant figure legends.
FIG. 1.
Evidence of the action of purifying selection on the genes of Tec1 and Tec2 elements. (A) Window analysis of divergence in the ORF 2A-ORF 2B region of Tec2. Divergences (dS and dN) were calculated in the three forward reading frames for all 10 pairwise comparisons among five aligned Tec2 sequences by using a 150-bp window and scanning across the alignment in one-codon increments. Ratios of the averaged dS and dN values are plotted (on a logarithmic scale) for each window and each reading frame; frame 1 is the open frame of ORF 2A, and frame 2 is that of ORF 2B. ORF 2A and ORF 2B are drawn to the scale of the plot at its lower boundary. The nucleotide sequence in the immediate vicinity of the putative frameshift is shown, with codons in the two reading frames delimited by tick marks; relevant start and stop codons are underlined (ORF 2B) or overlined (ORF 2A). The subscript “A” denotes the nucleotide whose deletion would fuse the two ORFs (21). The remarkably low dS/dN ratios in out-of-frame intervals is presumably due to selection on coincident in-frame intervals. In sequence maps for Tec2-1 and Tec1-1, sequences are depicted to scale, and ORFs (bars, labeled) and ITRs (arrows) are shown. Regions sequenced from homologous cloned elements are represented by lines aligned beneath their respective “type” elements. Divergence analyses were performed with pairs of sequences drawn from alignments made with ClustalW (43). (B) Scatter plot of synonymous and nonsynonymous divergences (dS and dN) calculated for all pairwise comparisons among sequences within alignments of five ORFs: Tec1 ORFs 1, 2, and 3 and Tec2 ORFs 2A and 2B. Divergences were calculated by the method of Nei and Gojobori (33) as implemented by Ina et al. (21), but with the genetic code of Euplotes (17). Divergence in the absenceof selection should generate points that lie on the line dS = dN. Nearly all comparisons showed dN to be significantly less than dS (P value, < 0.05; one-tailed z test, no correction for nonindependence). The two exceptions, for Tec2 ORF 2A comparisons, are indicated by grey circles. This analysis is influenced by the following considerations. Due to the use of pairwise comparisons, individual estimates are not independent. This analysis tests only for significant evidence of selection in the reading frames of interest, not for significant differences in other patterns or rates of evolution. The stringency of selection on different genes (the proportion of nonsynonymous mutations rejected by selection) and, consequently, the dS/dN ratios may vary between different genes. See the text for an explanation of the two clusters of points that are circled.
Nucleotide sequence accession numbers.
The GenBank accession numbers for the sequences determined here are AF159907 to AF159921 for individual ORF sequences and L03359 to L03360 for the two complete element sequences.
RESULTS
Evolution of Tec genes under purifying selection.
The original counts of synonymous and nonsynonymous differences between Tec gene sequences suggested the action of purifying selection (29). To verify and extend these analyses, we gathered Tec1 and 2 gene sequences and analyzed aligned sections of multiple sequences for each gene (Fig. 1A). The divergence parameters dS and dN were calculated for pairwise comparisons of the aligned sequences; dS and dN represent, respectively, the synonymous and nonsynonymous (silent and missense) divergences of a pair of sequences. If purifying selection has acted on the diverging gene sequences for the encoded protein function, then the accumulation of nonsynonymous (missense) mutations will have been suppressed and the dS/dN ratio will be elevated (33).
Figure 1B shows a significant absence of nonsynonymous mutations during the evolution of each of the three Tec1 genes (all comparisons but two have a significant distortion of the dS/dN ratio away from 1.0). The finding that Tec genes, like TBE genes, have evolved under selection deepens the mystery of ciliate transposons. Why do they show evidence of strong selection, when eukaryotic type II transposons generally do not? The two frameshift-separated sections of the Tec2 ORF 2 gene also have a dS/dN ratio significantly greater than 1, a finding consistent with our suggestion that a programmed frameshift permits the expression of this gene (22).
A programmed +1 frameshift was used to express a single protein from Tec2 ORF 2A and ORF 2B.
All characterized Tec2 ORF 2 sequences—unlike those of Tec1 ORF 2—are interrupted by the insertion of a dA nucleotide that separates the ORF into two overlapping ORFs, ORF 2A and ORF 2B (Fig. 1A); this dA shifts ORF 2 into a frame that results in chain termination at the next codon, TAG. It must be true that either (i) Tec2 elements can produce the encoded recombinase (see below) by a programmed +1 frameshift (22) or (ii) the recombinase gene was inactivated—due to this frameshift mutation—in all Tec2 members during their multiplication and divergence from a common ancestor in the E. crassus genome. The latter is a real possibility: eukaryotic transposon families that have multiplied in their host genomes even though all members of the family share ancestral inactivating mutations are now known (D. J. Witherspoon and H. M. Robertson, submitted for publication).
If the putative programmed frameshift has functioned for the expression of the ORF 2 protein, codons both upstream and downstream of the shift site should show evidence of purifying selection against nonsynonymous mutations. To test this prediction, we used scanning window analysis of the dS/dN ratio. If the recombinase were expressed as a fusion protein, then we would expect to see selection in the coding frame upstream of the frameshift and selection in the new coding frame downstream of the frameshift. For each frame, the dS/dN ratio was plotted for a sliding window of 150 bp across ORF 2A and ORF 2B (Fig. 1A). The codons in the frame of ORF 2A have a distorted dS/dN ratio in positions 5′ of the putative +1 frameshift but not 3′ of it; conversely, in the frame of ORF 2B, no selection is evident 5′ of the shift, but selection is evident 3′ of the shift. These results indicate that, during the time of the divergence of the Tec2 ORF 2 sequences from a common ancestor, this programmed +1 frameshift operated to produce a full-length ORF 2 recombinase, the function of which was under selection.
Since this analysis detected only selection that has acted in the evolutionary past, the gene may no longer be under selection now. The divergence analysis of ORF 2B (Fig. 1B) shows two clusters of points representing two sets of divergences (dS), ancient and intermediate (dS, values, ∼0.3 and ∼0.12, respectively). Selection thus has acted during both ancient evolution and more recent evolution of this ORF. Since this data set lacks recently diverged elements, we cannot address whether selection has acted even more recently. Several of the ORFs in the full data set are clearly damaged: of 20 Tec gene sequences analyzed, 6 have stop codons in frame, in either the original frame or following a frameshifting insertion or deletion (other than the programmed frameshift in Tec2 ORF 2). These particular damaged genes are no longer under selection: as soon as a gene suffers a disabling mutation, it ceases to be under selection and will diverge with a subsequent dS/dN ratio of 1.
Homology of Tec ORF 2 proteins to tyrosine-type site-specific recombinases.
We had been unable to identify the Tec ORF 2 protein function on the basis of homology. However, in a PSI-Blast search (2) of public databases with a profile of the Tec1 and Tec2 ORF 2 proteins, the best match was a Bergeyella integrase (E value, 0.006). The Bergeyella integrase is a member of a diverse set of recombinases, named “tyrosine-type” after the tyrosine residue that forms a transient covalent bond with the DNA substrate, described in detail by Nunes-Duby et al. (34). The family includes phage integrases, yeast FLP recombinases, the transposition function (int) of Tn916, and XisA of the heterocyst-excised nifD interruption of Anabaena. A profile representing the Bergeyella protein and the two Tec proteins matched several other recognized family members aligned by Nunes-Duby et al. (34). The Tec sequences were aligned (Fig. 2) with a well-studied member of each functional subfamily defined by Nunes-Duby et al. (34) as well as with best matches from the profile search. We also included the protein sequence from the recently discovered Tec3 element (21a). The alignment clearly shows the homology of the Tec proteins to the recombinase family. For instance, the Tec proteins share the conserved active-site tyrosine and basic residues in boxes I and II. Further, the regions of greatest similarity between the Tec1 and Tec2 proteins correspond to the most conserved regions of the family (the boxes” and “patches” defined by Nunes-Duby et al. [34]) (Fig. 2), a finding supporting the inferred homology of the ORF 2 proteins to the tyrosine recombinases. While the Tec3 protein is not particularly similar to the Tec1 and Tec2 proteins (21a), it is clearly a member of the family overall.
FIG. 2.
Alignment of the sequences of the proteins encoded by the Tec1 and Tec2 ORF 2 genes and the Tec3 ORF 1 gene with sequences of tyrosine-type site-specific recombinases. Only the C-terminal sections of the Tec proteins are aligned, with truncated recombinase sequences limited to their catalytic domains; the position of the first residue is indicated, as are the numbers of residues removed from the C terminus, if any. Known recombinase family members aligned are of two kinds: (i) several well-studied members (Int, Cre, VLF-1, XerD, Tn916, XisC, and FimB) and (ii) several recognized members (34) with particularly strong matches in a PSI-Blast search with a profile representing the proteins of Tec1, Tec2, and Weeksella (Bergeyella) zoohelcum (W.zooTnpA): PL2, phiCTX, Clostridium butyric (C.but.), Methanococcus jannaschii (M.jan.), W.zooTnpA, and Ye24-shuf; two good hits—RP2 and Int-I3—are not shown in the final alignment because they lengthened the alignment unnecessarily). The sequences are named as described by Nunes-Duby et al. (34). Alignment was achieved in consultation with S. Nunes-Duby (personal communication), yielding a set of five aligned sequences (the Tec1 and Tec2 proteins, Int, Cre, and VLF-1); this core alignment was used as a profile to align the remaining sequences by using ClustalX (42). The alignment was manually adjusted to conform in most cases to the alignment of Nunes-Duby et al. (34). The Tec3 sequence was later aligned against this entire alignment by using ClustalX (42). The two boxes and three patches of strong conservation noted by Nunes-Duby et al. (34) are enclosed in boxes; patches I and III were assigned by S. Nunes-Duby (personal communication). The three absolutely conserved positions (including the catalytic Y) are indicated by asterisks. Residue symbols have been colored to indicate similarity between family members. Colors were assigned to residues as follows. First, a consensus was calculated for each position in the alignment (not shown); the consensus was either an amino acid or an amino acid similarity group (31 or 50%, respectively, as described by Nunes-Duby et al. [34]). Second, based on this consensus, residue colors were designated for alignment positions with five or more nongapped residues. The exchange groups of Dayhoff et al. (7) used for the consensus calculation were SPAGT, RKH, FYW, DEQN, and LIMV. Residues were colored according to chemical type when they contributed to the consensus or fell into a similarity group with the consensus residue. Colors of chemical types are as follows (from reference 34): hydrophobic (I, L, orV; F, Y, or W; M; and P), pink; hydrophilic (S or T and Q or N), green; acidic (D or E), magenta; small (G and A), yellow; and basic (K, R, or H), cyan. The residues of the conserved RRY triad are represented by white letters within dark blue (R) and dark red (Y) backgrounds.
Very few eukaryotic tyrosine recombinases are known, making the Tec genes nearly unique. The other known eukaryotic tyrosine recombinases are the FLP enzymes of yeasts, which form a clade very diverged from the rest of the recombinases (34); baculovirus VFL-1 (50); and the recently characterized recombinases of the DIRS retrotransposon family (14). Do eukaryotic tyrosine recombinases form a clade? The Tec recombinases do not match the FLPs at all well and only match VFL-1 strongly in the box II region immediately adjacent to the catalytic tyrosine (Fig. 2). There is also no particular similarity to the DIRS recombinases. These findings fail to support the existence of a unique eukaryotic subfamily of recombinases and suggest that there have been multiple transfers of prokaryotic recombinases into eukaryotic lineages.
DISCUSSION
We find that the three Tec element genes have evolved under selection for the functions of their encoded proteins (Fig. 1B). The Tec2 ORF 2A-ORF 2B protein product is produced by a programmed frameshift (Fig. 1A) and is a tyrosine site-specific recombinase (Fig. 2).
What selection has acted on Tec transposon genes?
It has been difficult to explain the selection that we have detected on Tec genes (Fig. 1B) and the selection on TBE1 genes (11, 29, 49). Genes must be expressed in order for selection to act, but very little, if any, expression is seen for these genes. Jarczewski et al. (24) found only very low levels of Tec gene transcripts in exconjugant cells: ∼60 polyadenylated messages per cell, enough to support Tec transposition in the germ line. TBE1 genes are expressed at undetectably low levels (K. Williams, T. G. Doak, and G. Herrick, unpublished results). Given the lack of expression, it is difficult to formulate a function for the transposon genes that would lead to selection.
We have considered two explanations (not mutually exclusive) for the selection on Tec and TBE1 genes. Either transposon genes are under selection because elements with functional genes transpose more often than do elements with inactivated genes (selection for transposition), or the genes are selected due to a host benefit that they provide (selection for host fitness).
A host fitness explanation for TBE1 selection was proposed by Witherspoon et al. (49). That is, functional TBE1 genes increase the fitness of host cells that carry them, relative to host cells that carry defective TBE1 genes, and so functional TBE1 genes are more likely to be replicated. In particular, Williams et al. proposed that TBE1 transposase catalyzes the precise excision of TBE1 genes during macronuclear development (46). Precise excision of a TBE1 element (or Tec element or most other internal eliminated sequences) removes the element and one flanking target site duplication (for a review, see reference 28) and restores functional genes in the macronucleus. Thus, the selection acting on TBE1 genes would be based on their essential role in precise excision (49). In such a model, the “price” of selection is “paid for” in reduced host fitness. Selection for Tec functions can be imagined in the same fashion (28). However, the low levels of both Tec and TBE1 gene expression seem to rule out this explanation: for both Tec and TBE1, there are thousands of elements to be excised, and it seems implausible that so few transcripts could provide enough transposase to catalyze massive precise excision (24). Furthermore, Tec excision products do not appear to be formed by a transposase-like activity (23, 27), as is also true for the intermediates and products of TBE1 excision (9; K. Williams and T. G. Doak, unpublished data). It is possible that both Tec and TBE1 genes were under selection for excision functions until recently but that now their hosts have independently assumed alternative excision processes and the transcription of thousands of Tec and TBE1 elements has been suppressed (a refinement of the “invade, bloom, abdicate, and fade” model of Klobutcher and Herrick [28]); however, this possibility also seems unlikely.
Alternatively, selection could be directed at transposition functions by one of two mechanisms. First, preferential cis action of transposase on the particular element that encodes it (a known mechanism in prokaryotes; see, e.g., reference 8) would impose selection (26). However, it is difficult to imagine a molecular mechanism in eukaryotes whereby proteins encoded by one of many nearly indistinguishable DNA elements and then translated in the cytoplasm would act preferentially on those particular elements in the nucleus. In contrast, this mechanism appears to function with retroelements, whereby a reverse transcriptase molecule acts in the cytoplasm on the retroelement RNA molecule that encoded it (44). Thus, retrotransposon transposition genes are generally found to be under strong selection (see, e.g., references 19, 25, 31, and 39; D. J. Witherspoon, unpublished results), due to this selection for transposition. While it remains possible that Tec and TBE elements transpose via an RNA intermediate, there is no evidence against the assumption that they are simple type II DNA elements.
A second situation that leads to selection for transposition functions has been described by Witherspoon (47, 48). The force of the selection is dependent on the variance in the proportions of wild-type elements in individual hosts: simply stated, in hosts with more wild-type elements, transposition is more frequent, so that wild-type elements are preferentially transposed. This population structure model can explain selection on the Tec transposition functions under certain conditions, in particular, if the rate of element turnover (loss, balanced by transposition) is sufficiently high. Conditions exist under which a small number of competent elements would be maintained in a large pool of mutant elements, consistent with the large number of damaged Tec elements that we have found. This selective force is very weak in the presence of lower turnover rates, and in such instances, all elements will become mutant, as has been observed for Mariner elements (see, e.g., reference 37). We have not determined the rate of Tec or TBE1 transposition but plan to do so for TBE1 as a test of this population structure model (Witherspoon, unpublished).
Programmed +1 frameshift in the Tec2 ORF 2 gene.
Jahn et al. (22) proposed that the Tec2 gene was expressed as a single fusion protein, colinear with the homologous Tec1 protein. A +1 frameshift in the overlap of ORF 2A and ORF 2B was proposed that skips one of the A's in a run of three, adjusting the reading frame to that of ORF 2B and avoiding an immediate TAG stop codon (Fig. 1A). The resulting Tec2 fusion protein would have the same amino acids (KN) at the shift site as the Tec1 protein at the homologous position. The Tec2 elements all share the same sequence, AAATAG, with an “extra A,” indicating that this sequence and the need for a frameshift were present in their most recent common ancestor. Descendants of Tec2 ORF 2A and ORF 2B have diverged from that ancestor under selection for the function of the encoded recombinase. Thus, the evolutionary pattern of Tec2 recombinase gene evolution is unique proof of a functional programmed frameshift.
Further evidence of functional frameshifting in the genus Euplotes was recently documented for several genes, those encoding the p43 telomerase subunit (1) and two protein kinases, Eopkar (40) and Eondr2 (41). Most recently, Z. Karamysheva et al. (submitted for publication) identified two functional +1 frameshift sequences in an E. crassus gene for the telomerase protein TERT. In all five examples, amino acid homology between a known protein and the candidate frameshifted protein is evidence for a programmed frameshift. The reasoning is that the similarity would not have persisted in the absence of selection for function of the encoded protein, a notion which in turn implies that the full-length protein was expressed via the programmed frameshift. For p43 and TERT, a protein of the predicted full length is observed, and a predicted p43 peptide has been sequenced.
A common feature of sequences that promote frameshifting is the ability to cause the ribosome to pause while translating a codon paired to a tRNA that also can pair with the overlapping codon in the new frame (for reviews, see references 12 and 13). This codon is often located in a string of identical nucleotides (in the present example, a string of three A's) (Fig. 1A), allowing pairing in the two different frames (with AAA or AAU). A common pause-causing sequence is an in-frame stop codon terminating the first ORF just 3′ of the shift codon (see, e.g., references 6, 16, 30, and 45). The Euplotes +1 frameshift consensus, now AAATAA/G (41; Karamysheva et al., submitted), fits this pattern. Another feature that can cause pausing is a 3′ segment with a strong secondary structure; however, we have not found convincing structures in the ORF 2 gene sequence. A codon that requires a rare tRNA just before the shift can also impede translation and stimulate shifting, but there is no rare codon near the end of ORF 2A (see reference 20 for Euplotes codon usage tables).
Regardless of the details, it is clear that a robust mechanism for frameshifting functions in euplotids. Hence, the appearance of a frameshift mutation in Tec2 ORF 2 might have been effectively silent. It could not have been very deleterious, since the same purifying selection that has maintained the surrounding coding sequence would have eliminated it. Most frameshift sequences represent the independent mutation of nonhomologous sequences to analogous frameshifting signals. Since such signals can be quite short, random mutation is likely to produce them in genes at an appreciable frequency. Thus, a frameshifting sequence may serve no function at all. Note that Tec1 elements, which lack the ORF 2 frameshift, have been as successful as Tec2 elements. Alternatively, the frameshift may have a function such as producing stoichiometrically balanced levels of full-length recombinase and the N-terminal fragment, with the fragment modulating the activity of the recombinase. Many programmed frameshifts used by transposons serve such functions (for a review, see reference 4).
We have considered two alternative explanations for the frameshift and conservation that we have seen in the Tec2 ORF 2A-ORF 2B gene. The first explanation is that the Tec2 ORF 2A-ORF 2B gene is now expressed as a two-peptide recombinase by translational reinitiation in the ORF 2B frame; given the mounting number of frameshift examples in euplotids, we do not favor this explanation.
The second explanation is that gene conversion patched an inactivating frameshift mutation into all modern Tec2 elements, after they had diverged under selection, and that modern Tec2 elements are inactivated by this frameshift. Such a pervasive directional gene conversion seems inherently unlikely, but our data further rule it out. The proposed gene conversion would have erased differences between the Tec2 elements near the frameshift site, but there are base pair differences within 15 nucleotides of the frameshift site on either side, and nucleotide diversity is not reduced near the frameshift site (data not shown). If gene conversion were so frequent, then it should also have acted on the rest of the Tec sequence, but we still saw evidence of selection, which would have had to accumulate after the proposed gene conversion. Therefore, we must still propose an ancestral Tec2 under selection for the function of ORF 2A-ORF 2B with the ancestral frameshift.
Eukaryotic transposons encoding a site-specific recombinase.
The ORF 2 gene of Tec elements encodes a site-specific recombinase (Fig. 2), while the ORF 1 gene encodes a conventional DDE transposase (10). Given that the detected purifying selection might indicate that both of these genes are required for Tec transposition, we now consider how such transposition might occur. A tyrosine recombinase performs the transposition reaction (in the absence of a DDE enzyme) for members of the bacterial Tn916 family (35) and may as well for the eukaryotic retroelement DIRS (14). We are unaware of any eukaryotic type II transposon, other than the Tec elements (see also Jacobs et al. [21a]), that encodes a recombinase in addition to a DDE transposase. However, transposons that carry a gene for both a DDE transposase and a site-specific recombinase are common in bacteria. Such elements transpose replicatively, creating a temporary cointegrate structure during interreplicon transposition (3, 38; for a review, see reference 18). When both the donor and the target replicons are circles, as in bacteria, the cointegrate is a composite circle.
The generation of the bacterial cointegrate requires two steps. First, the DDE transposase nicks 3′ of each of the donor element strands, and then each 3′ OH end is covalently linked to the target in two strand transfer reactions. Second, the two displaced flanking target 3′ OH ends prime replication across each strand of the element, generating old and new heteroduplexes. The two old and new element copies lie at the junctions of the fused replicons, oriented in the same direction around the circle. The completion of transposition is accomplished by the resolution of the cointegrate circle back into the two original replicons. This resolution reaction is effected by a site-specific recombinase (usually a serine type; not homologous but analogous to the Tec tyrosine recombinase). The recombinase synapses the resolution site from each copy of the element and catalyzes recombination between them. This process unlinks the two replicons and leaves an element copy at the original location on the donor and an element integrated at the target site on the acceptor. The result is a new copy of the element at the target site location.
Do Tec elements transpose replicatively? If they do, they must have evolved solutions to problems arising from transposition within a genome of linear eukaryotic chromosomes and arising from the large number of Tec elements in the genome. During bacterial interreplicon transposition, the two replicons are temporarily joined in a single structure, the cointegrate. This circle serves to keep the two elements physically linked and in the proper orientation, so that they can participate in the subsequent resolution step. Transposition immunity would ensure that only two elements are present in this structure—the two that must participate to unlink the two replicons and restore them to their initial structures.
Neither condition pertains to Tec elements. First, like other eukaryotic genomes, the E. crassus germ line genome consists of multiple linear mitotic chromosomes. Second, there are hundreds of Tec elements within each chromosome. Replicative transposition by itself, without resolution, would cause chromosome rearrangements. Intrachromosomal transposition without resolution would cause one of two rearrangements: a deletion would result if the new element was inserted into the target in the same orientation as the donor element, or an inversion would result if the new insertion was in the opposite orientation. Similarly, without resolution, transposition between linear chromosomes would result in two possible structures, depending on the orientation of the two elements with respect to centromeres and proximal telomeres of their chromosomes. The two outcomes are a reciprocal (balanced) translocation, if the elements have the same relative orientations, or an acentric fragment and a dicentric fusion resulting from two elements with opposite orientations.
These cointegrate intermediate analogs would not present a problem if resolved before host DNA replication or mitosis. However, with thousands of transposon copies in the genome, the two cointegrate elements must be held together until resolution is complete: otherwise, they could not be correctly rejoined. These two element copies are generated side by side during the local DNA replication that completes cointegrate formation: if a mechanism to hold them together initiates at that point, then they could be held together for subsequent resolution. A precedent for such a replication-dependent mechanism is sister chromatid cohesion (32).
If the ORF 2 protein acts as a site-specific recombinase in transposition, a recognition or recombination site should reside on the element; resolution sites of serine-type recombinases in general consist of an 11- to 13-bp inverted repeat, split by 6 to 8 bp of a central sequence (for a review, see reference 15). We have not found a credible resolution site in the Tec1 or Tec2 sequences.
Summary.
By examining patterns of sequence divergence, we have shown that the E. crassus Tec families of transposons have evolved under purifying selection for their protein products, and we have identified a functional programmed frameshift site in the Tec2 transposons. This site joins a number of other, similar identified shift sites in Euplotes. The Euplotes protein produced as a result of this frameshift is a member of the tyrosine site-specific recombinase family. This finding is surprising, given that tyrosine site-specific recombinases are extremely rare in eukaryotes and that no other known eukaryotic cut-and-paste transposon carries a tyrosine recombinase gene. The selection that we have characterized insists that the protein is needed for either element transposition or host fitness. We consider that Tec elements may transpose replicatively, the recombinase reverting the chromosomal rearrangements that replicative transposition creates.
Acknowledgments
This work was supported by Public Health Service grants GM25203 to G.H. and GM37661 to C.L.J. from the National Institutes of Health.
We thank Mark Krikau, John Jaraczewski, and John Frels for determination of Tec gene sequences. We also thank Dorothy Shippen, John Atkins, Simone Nunes-Duby, Dominic Esposito, Makuni Jayaram, Pat Higgins, Nigel Grindley, Martin Pato, David Stillman, Tim Formosa, and especially Larry Klobutcher for stimulating discussions, constructive criticism, and encouragement.
REFERENCES
- 1.Aigner, S., J. Lingner, K. L. Goodrich, C. A. Grosshans, A. Shevchenko, M. Mann, and T. R. Cech. 2000. Euplotes telomerase contains an La motif protein produced by apparent translational frameshifting. EMBO J. 19:6230-6239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., L. M. Thomas, A. A. Schäffer, J. Zhang, and Z. Zhang. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arthur, A., and S. Sherratt. 1979. Dissection of the transposition process: a transposon-encoded site-specific recombination system. Mol. Gen. Genet. 175:267-274. [DOI] [PubMed] [Google Scholar]
- 4.Chandler, M., and O. Fayet. 1993. Translational frameshifting in the control of transposition in bacteria. Mol. Microbiol. 7:497-503. [DOI] [PubMed] [Google Scholar]
- 5.Craig, N. L., R. Craigie, M. Gellert, and A. M. Lambowitz (ed.). 2002. Mobile DNA II. ASM Press, Washington, D.C.
- 6.Craigen, W. J., and C. T. Caskey. 1986. Expression of peptide chain release factor 2 requires high-efficiency frameshift. Nature 322:273-275. [DOI] [PubMed] [Google Scholar]
- 7.Dayhoff, M. O., R. M. Schwartz, and B. L. Orcott. 1978. A model of evolutionary change in proteins, p. 345-352. In M. O. Dayhoff (ed.), Atlas of protein sequence and structure, vol. 5, suppl. 3. National Biomedical Research Foundation, Washington, D.C. [Google Scholar]
- 8.Derbyshire, K., M. Kramer, and N. Grindley. 1990. Role of instability in the cis action of the insertion sequence IS903 transposase. Proc. Natl. Acad. Sci. USA 87:4048-4052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doak, T. G. 2001. Transposons in ciliated protozoa. Ph.D. dissertation. University of Utah, Salt Lake City.
- 10.Doak, T. G., F. P. Doerder, C. Jahn, and G. Herrick. 1994. A proposed superfamily of transposase genes: transposon-like elements in ciliated protozoa and a common “D35E”motif. Proc. Natl. Acad. Sci. USA 91:942-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doak, T. G., D. J. Witherspoon, F. P. Doerder, K. R. Williams, and G. Herrick. 1997. Conserved features of ciliate TBE1 transposons. Genetica 101:75-86. [DOI] [PubMed] [Google Scholar]
- 12.Farabaugh, P. J. 2000. Translational frameshifting: implications for the mechanism of translational frame maintenance. Prog. Nucleic Acid Res. Mol. Biol. 64:131-170. [DOI] [PubMed] [Google Scholar]
- 13.Gesteland, R. F., and J. F. Atkins. 1996. Recoding dynamic reprogramming of translation. Annu. Rev. Biochem. 65:741-768. [DOI] [PubMed] [Google Scholar]
- 14.Goodwin, T. J., and R. T. Poulter. 2001. The DIRS1 group of retrotransposons. Mol. Biol. Evol. 18:2067-2082. [DOI] [PubMed] [Google Scholar]
- 15.Grainge, I., and M. Jayaram. 1999. The integrase family of recombinases: organization and function of the active site. Mol. Microbiol. 33:449-456. [DOI] [PubMed] [Google Scholar]
- 16.Gramstat, A., D. Prufer, and W. Rohde. 1994. The nucleic acid-binding zinc finger protein of potato virus M is translated by internal initiation as well as by ribosomal frameshifting involving a shifty stop codon and a novel mechanism of P-site slippage. Nucleic Acids Res. 22:3911-3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grimm, M., C. Brunen-Nieweler, V. Junker, K. Heckmann, and H. Beier. 1998. The hypotrichous ciliate Euplotes octocarinatus has only one type of tRNACys with GCA anticodon encoded on a single macronuclear DNA molecule. Nucleic Acids Res. 26:4557-4565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grindley, N. D. F. 2002. The movement of Tn3-like elements: transposition and cointegrate resolution, p. 272-302. In N. L. Craig, R. Craigie, M. Gellert, and A. M. Lambowitz (ed.), Mobile DNA II. ASM Press, Washington, D.C.
- 19.Hardies, S. C., S. L. Martin, C. F. Voliva, C. A. Hutchison, and M. H. Edgell. 1986. An analysis of replacement and synonymous changes in the rodent L1 repeat family. Mol. Biol. Evol. 3:109-125. [DOI] [PubMed] [Google Scholar]
- 20.Hoffman, D. C., R. C. Anderson, M. L. Dubois, and D. M. Prescott. 1995. Macronuclear gene-sized molecules of hypotrichs. Nucleic Acids Res. 23:1279-1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ina, Y., M. Mizokami, K. Ohba, and T. Gojobori. 1994. Reduction of synonymous substitutions in the core protein gene of hepatitis C virus. J. Mol. Evol. 38:50-56. [DOI] [PubMed] [Google Scholar]
- 21a.Jacobs, M. E., A. Sanchez-Blanco, L. A. Katz, and L. A. Klobutcher. 2003. Tec3, a new developmentally eliminated DNA element in Euplotes crassus. Eukaryot. Cell 2:103-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jahn, C. L., S. Z. Doktor, J. S. Frels, J. W. Jaraczeski, and M. F. Krikau. 1993. Structures of the Euplotes crassus Tec1 and Tec2 elements: identification of putative transposase coding regions. Gene 133:71-78. [DOI] [PubMed] [Google Scholar]
- 23.Jaraczewski, J. W., and C. L. Jahn. 1993. Elimination of Tec elements involves a novel excision process. Genes Dev. 7:95-105. [DOI] [PubMed] [Google Scholar]
- 24.Jaraczewski, J. W., J. S. Frels, and C. L. Jahn. 1994. Developmentally regulated, low abundance Tec element transcripts in Euplotes crassus—implication for DNA elimination and transposition. Nucleic Acids Res. 22:4535-4542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jordan, I. K., and J. F. McDonald,. 1999. Tempo and mode of Ty element evolution in Saccharomyces cerevisiae. Genetics 151:1341-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaplan, N., T. Darden, and C. H. Langley. 1985. Evolution and extinction of transposable elements in Mendelian populations. Genetics 109:459-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klobutcher, L. A., L. R. Turner, and J. LaPlante. 1993. Circular forms of developmentally excised DNA in Euplotes crassus have heteroduplex junction. Genes Dev. 7:84-94. [DOI] [PubMed] [Google Scholar]
- 28.Klobutcher, L. A., and G. Herrick. 1997. Developmental genome reorganization in ciliated protozoa: the transposon link. Prog. Nucleic Acid Res. Mol. Biol. 56:1-62. [DOI] [PubMed] [Google Scholar]
- 29.Krikau, M. 1991. Sequence analysis of the tec1 and tec2 repetitive element families in Euplotes crassus. Ph.D. dissertation. University of Illinois, Chicago.
- 30.Matsufuji, S., T. Matsufuji, Y. Miyazaki, Y. Murakami, and J. F. Atkins. 1995. Autoregulatory frameshifting in decoding mammalian ornithine decarboxylase antizyme. Cell 80:51-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McAllister, B. F., and J. H. Werren. 1997. Phylogenetic analysis of a retrotransposon with implications for strong evolutionary constraints on reverse transcriptase. Mol. Biol. Evol. 14:69-80. [DOI] [PubMed] [Google Scholar]
- 32.Nasmyth, K., J. M. Peters, and F. Uhlmann. 2000. Splitting the chromosome: cutting the ties that bind sister chromatids. Science 288:1379-1385. [DOI] [PubMed] [Google Scholar]
- 33.Nei, M., and T. Gojobori. 1986. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3:418-426. [DOI] [PubMed] [Google Scholar]
- 34.Nunes-Duby, S. E., H. J. Kwon, R. S. Tirumalai, T. Ellenberger, and A. Landy. 1998. Similarities and differences among 105 members of the Int family of site-specific recombinases. Nucleic Acids Res. 26:391-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Poyart-Salmeron, C., P. Trieu-Cuot, C. Carlier, and P. Courvalin. 1989. Molecular characterization of two proteins involved in the excision of the conjugative transposon Tn1545: homologies with other site-specific recombinases. EMBO J. 8:2425-2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robertson, H. M. 1993. The mariner transposable element is widespread in insects. Nature 362:241-245. [DOI] [PubMed] [Google Scholar]
- 37.Robertson, H. M., and K. L. Zumpano. 1997. Molecular evolution of an ancient mariner transposon, Hsmar1, in the human genome. Gene 205:203-217. [DOI] [PubMed] [Google Scholar]
- 38.Shapiro, J. 1979. Molecular model for the transposition and replication of bacteriophage Mu and other transposable elements. Proc. Natl. Acad. Sci. USA 76:1933-1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Springer, M. S., E. H. Davidson, and R. J. Britten. 1991. Retroviral-like element in a marine invertebrate. Proc. Natl. Acad. Sci. USA 88:8401-8404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tan, M., K. Heckmann, and C. Brunen-Nieweler. 2001. Analysis of micronuclear, macronuclear and cDNA sequences encoding the regulatory subunit of cAMP-dependent protein kinase of Euplotes octocarinatus: evidence for a ribosomal frameshift. J. Eukaryot. Microbiol. 48:80-87. [DOI] [PubMed] [Google Scholar]
- 41.Tan, M., A. Liang, C. Brunen-Nieweler, and K. Heckmann. 2001. Programmed translational frameshifting is likely required for expression of genes encoding putative nuclear protein kinases of the ciliate Euplotes octocarinatus. J. Eukaryot. Microbiol. 48:575-582. [DOI] [PubMed] [Google Scholar]
- 42.Thompson, J., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thompson, J., D. Higgins, and T. Gibson. 1994. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wei, W., N. Gilbert, S. L. Ooi, J. F. Lawler, E. M. Ostertag, H. H. Kazazian, J. D. Boeke, and J. V. Moran. 2001. Human L1 retrotransposition: cis preference versus trans complementation. Mol. Cell. Biol. 21:1429-1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weiss, R. B., D. M. Dunn, J. F. Atkins, and R. F. Gesteland. 1987. Slippery runs, shifty stops, backward steps, and forward hops: −2, −1, +1, +2, +5 and +6 ribosomal frameshifting. Cold Spring Harbor Symp. Quant. Biol. 52:687-693. [DOI] [PubMed] [Google Scholar]
- 46.Williams, K., T. Doak, and G. Herrick. 1993. Precise excision of Oxytricha trifallax telomere-bearing elements and formation of circles closed by a copy of the flanking target duplication. EMBO J. 12:4593-4601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Witherspoon, D. J. 1999. Selective constraints on P-element evolution. Mol. Biol. Evol. 16:472-478. [DOI] [PubMed] [Google Scholar]
- 48.Witherspoon, D. J. 2000. Natural selection on transposable elements of eukaryotes. Ph.D. dissertation. University of Utah, Salt Lake City.
- 49.Witherspoon, D. J., T. Doak, K. R. Williams, A. Seegmiller, and J. Seger. 1997. Selection on the protein-coding genes of the TBE1 family of transposable elements in the ciliates Oxytricha fallax and O. trifallax. Mol. Biol. Evol. 14:696-706. [DOI] [PubMed] [Google Scholar]
- 50.Yang, S., and L. K. Miller. 1998. Expression and mutational analysis of the baculovirus very late factor 1 (vlf-1) gene. Virology 245:99-109. [DOI] [PubMed] [Google Scholar]