Abstract
Paracoccidioides brasiliensis, a thermodimorphic fungus, is the causative agent of the prevalent systemic mycosis in Latin America, paracoccidioidomycosis. We present here a survey of expressed genes in the yeast pathogenic phase of P. brasiliensis. We obtained 13,490 expressed sequence tags from both 5′ and 3′ ends. Clustering analysis yielded the partial sequences of 4,692 expressed genes that were functionally classified by similarity to known genes. We have identified several Candida albicans virulence and pathogenicity homologues in P. brasiliensis. Furthermore, we have analyzed the expression of some of these genes during the dimorphic yeast-mycelium-yeast transition by real-time quantitative reverse transcription-PCR. Clustering analysis of the mycelium-yeast transition revealed three groups: (i) RBT, hydrophobin, and isocitrate lyase; (ii) malate dehydrogenase, contigs Pb1067 and Pb1145, GPI, and alternative oxidase; and (iii) ubiquitin, delta-9-desaturase, HSP70, HSP82, and HSP104. The first two groups displayed high mRNA expression in the mycelial phase, whereas the third group showed higher mRNA expression in the yeast phase. Our results suggest the possible conservation of pathogenicity and virulence mechanisms among fungi, expand considerably gene identification in P. brasiliensis, and provide a broader basis for further progress in understanding its biological peculiarities.
Paracoccidioides brasiliensis, a thermodimorphic fungus, is the causative agent of the prevalent systemic mycosis in Latin America, paracoccidioidomycosis (PCM). Epidemiological data indicate a broad geographic distribution of this disease in Central and South America, extending from Mexico to Argentina (56). It is estimated that as many as ten million individuals could be infected with P. brasiliensis, acquired by the inhalation of airborne microconidia, which reach the pulmonary alveolar epithelium and then transform into the parasitic yeast form (52).
Pathogenicity appears to be intimately linked to the dimorphic transition since strains of P. brasiliensis (and also of Histoplasma capsulatum and Blastomyces dermatitidis) that are unable to transform into yeasts are not virulent (4, 37, 54, 56). Signaling pathways that control the morphological changes in P. brasiliensis are poorly understood, but the involvement of both cyclic AMP (cAMP) and mitogen-activated protein kinase (MAPK) signal transduction pathways have been reported in other dimorphic fungi (6, 30). Exogenous cAMP inhibits P. brasiliensis yeast-to-mycelium (Y-M) transition, thus maintaining the pathogenic yeast form (3). This situation differs in Candida albicans, in which the Y-M transition is controlled by cAMP and exogenous cAMP stimulates pseudohyphae that are potentially able to invade mammalian cells (53).
The morphological change in P. brasiliensis is accompanied by documented changes in the cell wall composition, including migration and reorganization of the membrane lipids, especially glycosphingolipids (GLSs) (31, 61, 65). Furthermore, an increase in the chitin content is observed in the cell wall, followed by a change in the glucan polymer predominant anomery from β-1,3- to α-1,3-glucan as the fungus adopts the yeast form (56). The surface α-glucan may have a role as a protective layer against the host defense mechanisms because of the incapacity of phagocytic cells to digest α-1,3-glucan (55, 56).
The development of paraccoccioidomycosis depends on interactions between fungal and host components (22). The main antigenic component described in P. brasiliensis is gp43, an exocellular glycoprotein (57, 59) containing a single oligosaccharide chain (1, 12, 49); it elicits a strong humoral response and can be detected in the sera of PCM patients (39). The gp43 is a potential virulence factor as it binds murine laminin, resulting in increased pathogenicity of yeast cells (64). The fungus can also bind to other components of the extracellular matrix, such as fibronectin and collagen (40). Adhesion to host cells and mucosal surfaces is a crucial step in the establishment of P. brasiliensis infection. There is a correlation between adherence and virulence, as suggested by measuring fungal adhesion to cultured mammalian epithelial cells, with virulent strains having a greater capacity to adhere (22, 40). An extracellular serine-thiol protease has also been characterized in P. brasiliensis; this protease is a probable virulence factor since it can hydrolyze components of the basal membrane, such as fibronectin, laminin, collagen IV, and proteoglycans (9, 50, 51). This enzymatic activity could increase the capacity of P. brasiliensis to disseminate, perhaps acting in collaboration with gp43. It is also possible that sialoglycoconjugates exposed at the surface of P. brasiliensis cells may also modulate the interactions between fungi and the host tissue (58).
Given the relative lack of information about the P. brasiliensis genome and genes expressed in the yeast pathogenic phase, we have undertaken the first global sequence analysis of P. brasiliensis expressed genes. Our results reveal the presence of several C. albicans virulence and pathogenicity homologues in P. brasiliensis. We also analyzed the mRNA expression during the dimorphic transition yeast-mycelium-yeast (Y-M-Y) for some of the identified P. brasiliensis genes by real-time quantitative reverse transcription-PCR (RT-PCR). Our results support the importance of genomic analysis in human pathogenic fungi and point to a possible conservation of pathogenic and virulence mechanisms among them.
MATERIALS AND METHODS
RNA extraction and cDNA library construction.
P. brasiliensis strain 18 (provided by Z. P. Camargo, UNIFESP, Sao Paulo, Brazil) was inoculated (106 viable yeast given intraperitoneally) into BALB/c mice. After 20 to 30 days of infection, the mice were sacrificed, and the spleens of these animals were plated as detailed by Pinto et al. (46) in order to obtain the CFU counts. Isolated colonies were expanded in slants of modified YPD medium (0.5% Bacto yeast extract, 0.5% casein peptone, 1.5% glucose; pH 6.3) for 10 days at 37°C. A liquid preculture (4 days, with shaking) was prepared from this growth in YPD (10 ml), and the fungus was then grown in 100 ml of fresh medium for three extra days. The cells were checked microscopically for homogeneity, precipitated by centrifugation (1,800 × g for 15 min), and immediately mixed with Trizol (Gibco-BRL) for RNA extraction according to the supplier's recommendations. To verify the RNA integrity, 20 μg of RNA from each treatment was fractionated in a 2.2 M formaldehyde-1.2% agarose gel, stained with ethidium bromide, and visualized with UV light. The presence of intact 28S and 18S rRNA bands was used as a criterion to verify if the RNA was intact. RNase-free DNase treatment was done in a final volume of 100 μl containing 40 mM Tris-HCl (pH 7.5), 6 mM MgCl2, 1 μl of RNasin (40 U/μl; Promega), 10 μl of RNase-free DNase (1 U/μl; Promega or Life Technologies), 2.5 μl of dithiothreitol at 200 mM, and 10 μg of total RNA. The reaction was incubated at 37°C for 60 min and stopped by incubation at 70°C for 30 min. The absence of DNA contamination after the RNase-free DNase treatment was verified by PCR amplification of the GP43 gene (12). Polyadenylated RNA was purified by using oligo(dT)-cellulose, and a unidirectional cDNA library was constructed in plasmid pCMVSport 6.0 by using SuperScript plasmid according to the manufacturer's instructions (Gibco-BRL). The number of clones of the library (transformed into Escherichia coli EMDH10B) was ca. 3 × 106 CFU, with an average insert size of 1.6 kb, and 100% of clones had inserts of >200 bp; 10,000 individual colonies were transferred to 110-by-96-well microtiter plates for storage at −80°C.
For the temperature-induced morphological switch in P. brasiliensis, mycelial and yeast cells growing in the early exponential phase were induced to undergo morphological transformation by changing the temperature of incubation from 26 to 37°C. Samples were collected at 0, 5, 10, 24, 48, and 120 h, and from 37 to 26°C samples were collected at 0, 5, 10, 24, and 48 h.
Plasmid DNA extraction and sequencing.
Bacterial clones were inoculated into CircleGrow (Bio 101) in a 96-well microtiter plate and incubated in a rotatory shaker at 37°C for 22 h. Plasmid DNA was prepared by standard alkaline lysis procedures and also by the boiling method of Marra et al. (36).
Single-run sequencing was done by using the dideoxy chain termination method, and dye termination chemistry (Applied Biosystems) and analysis was done on ABI 377 and 3100 fluorescence automated sequencers.
Data handling and analysis.
A pipeline was built to analyze and assemble the P. brasiliensis expressed sequence tag (EST) sequences (The P. brasiliensis EST Genome Project is available online [http://143.107.203.68/est/default.html]). Sequences were automatically edited for each EST by using the programs Phred (16, 17), CAP3 (24), Consed (21), and Crossmatch from Phrap (P. Green, http://bozeman.mbt.washington.edu/phrap.html). The sequences were cleaned from the pCMVSport 6.0 vector, and ESTs with a minimum size of 300 bases and a Phred quality value of at least 20 were considered for further analysis. Edited sequences were clustered by using the CAP3 program (24). To identify potential homologues to the P. brasiliensis genes, the BLASTX and TBLASTX algorithms (2) were used to compare the predicted protein sequences with several databases (see below). The values presented for putative homologues derive from results obtained with sequences of known function as well as with those of unknown function. The database sequence matches (putative homologues) were divided into highly significant (P ≤ 10−19), moderately significant (1e-10 to 1e-19), weakly significant (1e-03 to 1e-10), and nonsignificant (>1e-03) classes (data not shown). When P values of >10−3 were obtained, they were considered not statistically significant (no significant match).
The classification of functional groups used here was mainly based on that developed at The Institute for Genomic Research (TIGR; Rockville, Md. [http://www.tigr.org/docs/tiger-scripts/egad_scripts/role_report.spl]) (66). The following annotation strategy was used: the clusters were used as query sequences to search the S. cerevisiae databank (ftp://ftpmips.gsf.de/yeast/ORF_SEQ_15_Oct_2001/), Gproteome (a compilation of 217,319 entries that contain predicted protein sequences from 41 completely sequenced organisms [60]), and the Neurospora crassa and Schizosaccharomyces pombe predicted proteins (http://www-genome.wi.mit.edu/annotation/fungi/neurospora and ftp://ftp.sanger.ac.uk/pub/yeast/pombe/CDS_bases) by using the BLASTX program (2). A databank containing Candida spp., H. capsulatum, and B. dermatitidis (2,592 protein sequences) was compiled from the nonredundant protein database at the National Center for Biotechnology Information (NCBI) and BLASTX (2) was used to translate and compare the P. brasiliensis cluster query sequences. In addition, the clusters were translated and used as query sequences to search the translated Pneumocystis carinii (http://biology.uky.edu/Pc/), Cryptococcus neoformans (http://www.genome.ou.edu/cneo.html), Aspergillus nidulans, and N. crassa EST sequences (http://www.genome.ou.edu/fungal.html) by using the TBLASTX program (2). For all comparisons, BLOSUM62 was used as a matrix, except for GPROTEOME, for which PAM250 was used; for all of the other parameters, such as filtering, BLAST programs with their default settings were used.
PCR and RT-PCRs.
All of the PCR and RT-PCRs were performed by using an ABI Prism 7700 sequence detection system (Perkin-Elmer/Applied Biosystems). TaqMan EZ RT-PCR kits (Applied Biosystems) were used for the RT-PCRs. The thermal cycling conditions comprised an initial step at 50°C for 2 min, followed by 30 min at 60°C for RT, 95°C for 5 min, and 40 cycles at 94°C for 20 s and 60°C for 1 min. TaqMan PCR reagent kits were used for the PCRs. The thermal cycling conditions comprised an initial step at 50°C for 2 min, followed by 10 min at 95°C, and 40 cycles at 95°C for 15 seconds and 60°C for 1 min.
In all experiments, appropriate negative controls containing no DNA template or RNA were subjected to the same procedure to exclude or detect any possible contamination or carryover. Each reaction was analyzed at least three times. All reactions were examined by agarose gel electrophoresis to confirm that only one PCR product was synthesized. The results were normalized by using the CT values obtained for the α-tubulin and/or 18S ribosomal gene RNA amplifications run in the same plate. CT is defined as the first amplification cycle in which fluorescence indicating PCR products becomes detectable. The α-tubulin and the 18S RNA are endogenous controls that were used to normalize the samples for differences in the amount of total RNA added to each cDNA reaction. In real-time PCR analysis, quantification is based on the threshold cycle, which is inversely proportional to the logarithm of the initial copy number (62). Real-time PCR was performed with each primer pair and its TaqMan probe by using known amounts of target obtained by serial dilutions of genomic DNA as a template over several orders of magnitude. The initial template amount used in parallel reactions was estimated by assuming a P. brasiliensis genome size of 30 Mb. A linear relationship was obtained by plotting the threshold cycle against the logarithm of the known amount of the initial template. The equation of the line that best fitted the data was determined by regression analysis. The R2 value was calculated for each data set to estimate the accuracy of the real-time PCR with TaqMan as a quantification method. The amounts of target copies contained in an unknown sample were determined by extrapolation from the linear regression of the standard curve obtained for each primer-TaqMan set (18). For a description of the primers and TaqMan fluorescent probes (Applied Biosystems) used here, see Table 1.
TABLE 1.
List of primers and fluorescent probes used in this study
| Primers and probesa | Sequenceb | P. brasiliensis cluster IDc |
|---|---|---|
| Gpif | 5′-CGATCTCTATGCCCGCTACAC-3′ | PbCC003-D08 |
| Gpir | 5′-AAGCCCAGGACGTTGGAGT-3′ | |
| PGpi | 5′-TET-CCGTCATCGATACCCTCGCCCCT-TAMRA-3′ | |
| H104f | 5′-CGAAATCCATGCTCTGGCTC-3′ | PbCC012-B06 |
| H104r | 5′-TGTTTGGCAGCCTCGAGTC-3′ | |
| PH104 | 5′-TET-AAAAGGATGCGGCGTCGAAGGC′-TAMRA-3′ | |
| H70f | 5′-GACCACACCCTCGTTCGTTG-3′ | PbCC006-G05 |
| H70r | 5′-ACTTGATTTTTGGCGGCATC-3′ | |
| PH70 | 5′-6FAM-CTTCACAGACACCGAGCGTCTGATCGT-TAMRA-3′ | |
| H82f | 5′-ACCGCTGGTGCCGATATTT-3′ | PbCC001-B11 |
| H82r | 5′-GGCCACGAGGTAGGCAGAG-3′ | |
| PH82 | 5′-TET-TGATTGGTCAGTTTGGTGTCGGCTTCT-TAMRA-3′ | |
| Hydf | 5′-GTCCTCGGCACTGTCATCG-3′ | PbCC004-F01 |
| Hydr | 5′-AGGCTGAGTTTGGAGCACTGA-3′ | |
| Phyd | 5′-TET-CAAGGGCGGCAGCGTCGGT-TAMRA-3′ | |
| Isof | 5′-TGAAGTCCAGGCCGTCAAA-3′ | PbCC005-H06 |
| Isof | 5′-TGAACGGGCGCTTGGTATA-3′ | |
| Piso | 5′-TET-AATGGTGGCAGGACTCAAGATGGCG-TAMRA-3′ | |
| Malf | 5′-CCGAGGGAGCCTGCTAGACT-3′ | PbCC002-G09 |
| Malr | 5′-CCCTTCCACCCATCGTTTT-3′ | |
| PMal | 5′-VIC-CCTCCCAGAGACCAAGCACATCCG-TAMRA-3′ | |
| Olef | 5′-CTTCCACCACGAGTTCCCA-3′ | PbCC002-E05 |
| Oler | 5′-ATTTGGTCGGGTCGTACTGG-3′ | |
| POle | 5′-VIC-CTACCGCAACGCCATCGAATGGC-TAMRA-3′ | |
| Oxif | 5′-CGCACATCGAAATGCTGAGA-3′ | PbCC006-F12 |
| Oxir | 5′-CAGCGCAACATACGAACGG-3′ | |
| Poxi | 5′-VIC-CTGGTCCGATTGGGTTGCCTTGG-TAMRA-3′ | |
| RBf | 5′-GCCGATGTTCAGCGGAGAT-3′ | PbCC008-G03 |
| RBr | 5′-CCTTGCGCTAACAAACACCAA-3′ | |
| PRB | 5′-VIC-ATGCGCGATTCTCCTCGTGGGAT-TAMRA-3′ | |
| Tubf | 5′-TGGCCACTTTCTCTGTCGTTC-3′ | PbCC002-A04 |
| Tubr | 5′-CAGGGTGGCATTGTATGGCT-3′ | |
| Ptub | 5′-VIC-TTCGCCCAAAGTCTCCGACACCG-TAMRA-3′ | |
| Ubif | 5′-TCCTCCTGACCAGCAACGTC-3′ | PbCC003-E02 |
| Ubir | 5′-ATCAGAAAGGGTGCGACCG-3′ | |
| Pubi | 5′-VIC-TTTTCGCCGGCAAGCAGTTGGAG-TAMRA-3′ | |
| Ribf | 5′-CGGAGAGAGGGAGCCTGAGAA-3′ | PbCC012-H11 |
| Ribr | 5′-GGGATTGGGTAATTTGCGC-3′ | |
| PRib | 5′-6FAM-CGGCTACCACATCCAAGGAAGGCAG-TAMRA-3′ | |
| 1067f | 5′-TGTAAATGCCCAATCGACACA-3′ | PbCC012-A11 |
| 1067r | 5′-AATCGAAGGCATCTCGAATGA-3′ | |
| P1067 | 5′-VIC-TGGTCGCAGCACATGACAAGTTGC-TAMRA-3′ | |
| 1145f | 5′-ATCGCCGTTGCTGTCTCG-3′ | PbCC012-H05 |
| 1145r | 5′-GCTGTTCACCGTAATTGCCAG-3′ | |
| P1145 | 5′-TET-CTTGCAGTCTCCGTCAGTGCCGCT-TAMRA-3′ |
Grouped by cluster.
Abbreviations: TET =, 6-carboxy-4,7,2′,7′-tetrachlorofluorescein; 6-FAM, 6-carboxyfluorescein; VIC, trademark product from Applied Biosystems; TAMRA, 6-carboxy-N,N,N′,N′-tetramethylrhodamine.
ID, identification.
RESULTS
General strategy and data clustering.
The EST approach is a rapid and relatively efficient method for quick gene discovery, which may assist studies involving organisms with little or no genetic research history available, as is the case with P. brasiliensis and other human fungal pathogens. A unidirectional cDNA library was constructed for the identification of genes expressed in the yeast phase of P. brasiliensis. M13 reverse and anchored oligo(dT) (see Materials and Methods) primers were used to generate 5′ and 3′ sequence information, respectively. We obtained 13,490 ESTs with a minimum length of 300 bases and a Phred quality value of at least 20 (16, 17). We sequenced from the 5′ end 6,481 ESTs, whereas 7,009 were sequenced from the 3′ end. Clustering by the CAP3 program (24) identified 5,121 singletons and 1,397 contigs. The 913 singleton pairs represented virtual clusters; since they were derived from both ends of the same cDNA clone, the final number of putative P. brasiliensis genes identified in our analyses corresponds to the partial sequences of 4,692 expressed genes.
Gene identification and functional categories.
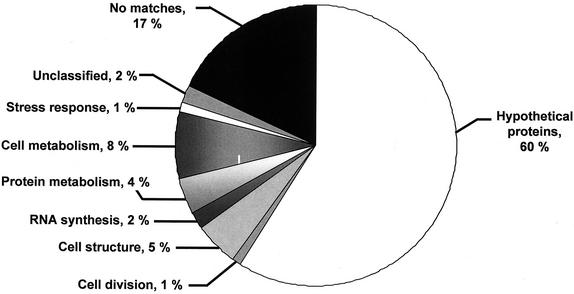
The nucleotide data set of clustered sequences was compared to several databases by using the BLASTX and TBLASTX programs (2) with a threshold E-value of ≤10−3. The clusters encoding putative protein sequences that show similarity to products in our local databanks (see Material and Methods for a description) were classified into functional groups (Fig. 1). During this phase, expert validation was introduced by individual examination of the matches for significance and to assign a functional category (i.e., TIGR classification). The total number of clusters that could be assigned a cellular role was 1,095. Most of the known transcripts belong to housekeeping genes such as those involved in cell metabolism (e.g., amino acid and carbohydrate metabolism [8%]), cell structure (e.g., cell wall and cytoskeleton [5%]) and protein metabolism (4%). The remaining clusters either show similarity to hypothetical proteins (817 clusters) or have no significant similarity to any protein sequence in the databases (no matches, 2,780 clusters). Among the 817 clusters that show similarity to hypothetical proteins, we identified 633 clusters that only show similarity to fungal ESTs. A high percentage of clusters showed no matches with known protein sequences (59%). This most probably reflects the absence in the databases of a completely sequenced genome related to filamentous fungi. Thus, we decided to compare our 2,780 no-match sequences with the predicted proteins of the last version (version 3) of the N. crassa genome project (http://www-genome.wi.mit.edu/annotation/fungi/neurospora) and S. pombe (ftp://ftp.sanger.ac.uk/pub/yeast/pombe/CDS_bases) by using BLASTX (2). Among the 2,780 no-match clusters, we were able to identify 2,001 P. brasiliensis sequences that display significant similarity to N.crassa hypothetical proteins (509 of them also display significant similarity to S. pombe hypothetical proteins). Accordingly, the P. brasiliensis no-match sequences decreased to 779 sequences (17%), and the hypothetical proteins increased to 2,818 sequences (60%) (see Fig. 1). Thus, we found 17% putative P. brasiliensis-specific sequences. The 633 clusters that show similarity only to fungal ESTs plus the 2,001 P. brasiliensis sequences that display significant similarity with N. crassa and S. pombe hypothetical proteins will result in 2,634 P. brasiliensis sequences that display similarity only with fungal proteins (which correspond to 56% of the P. brasiliensis sequences). The database sequence matches (putative homologs) were divided into highly significant (P ≤ 10−19 [1,242 ESTs]), moderately significant (1e-10 to 1e-19 [466 ESTs]), weakly significant (1e-03 to 1e-10 [2,195 ESTs]), and not significant (>1e-03 [789 ESTs]) classes. The complete list of clusters classified into functional groups is available online (http://143.107.203.68/est/default.html).
FIG. 1.
Classification of the 4,692 clusters of P. brasiliensis ESTs with BLASTX or TBLASTX E-values ≤10−3 are presented according to the classification developed at TIGR (66). Unclassified are ESTs that show similarity to a sequence with known function but do not fall into any of the classification schemes utilized; hypothetical proteins refer to sequences that show similarity to protein or DNA sequences to which no cellular role has yet been assigned; no match indicates new sequences with no significant similarity to protein or DNA sequences in the databases
Identification of putative virulence and pathogenicity genes.
In an attempt to identify additional putative virulence and pathogenicity factors in our P. brasiliensis database, we performed a direct BLAST comparison with a collection of about 60 genes that proved to be involved in virulence and pathogenicity in C. albicans, the best-studied human pathogenic fungus that displays a dimorphic behavior (7, 15, 23, 42, 44, 45). Sequences from two other dimorphic fungi (H. capsulatum and B. dermatitidis) have also been used in these comparisons (5, 33, 68). However, although the latter are phylogenetically closer to P. brasiliensis, very few genetic determinants of virulence and pathogenicity have been identified in these organisms (5, 33, 68). These analyses allowed us to find 26 P. brasiliensis clusters that show C. albicans putative homologues (Table 2). These genes were divided into three groups: (i) metabolic, (ii) cell wall and adhesion, and (iii) transduction signal.
TABLE 2.
P. brasiliensis clusters similar to C. albicans genes involved in virulence and pathogenicity
| Gene group and P. brasiliensis cluster IDa | C. albicans homologue (NCBI protein accession no.) | Main feature and/or role | BLAST E-value/% identity/% similarity |
|---|---|---|---|
| Metabolic genes | |||
| PbCC003-B07 | ADE2 (AAC49742) | Nucleotide synthesis | 4e-21/58/77 |
| PbCC006-G01 | HIS1 (P46586) | Amino acid synthesis | 9e-57/61/73 |
| PbCC002-G09 | MLS1 (AAF34695) | Malate synthase, glyoxylate cycle | 8e-30/56/69 |
| PbCC009-C01 | ICL1 (AAF34690) | Isocitrate lyase, glyoxylate cycle | 4e-66/74/85 |
| PbCC015-E02 | HEM3 (094048) | Hemosynthesis | 5e-16/39/60 |
| PbCC001-E02 | 1e-14/41/63 | ||
| PbCS033-B09 | PLB1 (AAC72296) | Phospholipase | 1e-09/43/55 |
| Cell wall genes | |||
| PbCC011-D01 | CHS2 (S20538) | Chitin synthase; cell wall synthesis | 4e-23/51/73 |
| PbCC013-B07 | CHS3 (P30573) | 2e-21/72/88 | |
| PbCS003-C01 | 3e-31/43/60 | ||
| PbCS008-H07 | 5e-30/43/51 | ||
| PbCC004-H02 | HWP1 (P46593) | Transglutaminase, adhesion | 2e-08/39/50 |
| PbCS001-B02 | INT1 (P53705) | Integrin, adhesion | 1e-49/33/56 |
| PbCS006-A05 | MNT1 (CAA67930) | Mannosyl transferase | 1e-31/55/80 |
| PbCC003-D08 | PHR1 (AAG16995) | Glycosidase GPI-anchored; cell wall structure | 2e-70/49/68 |
| PbCC013-H06 | PHR2 (AAG16996) | 2e-10/35/54 | |
| PbCS011-F12 | PMT1 (AAC31119) | Mannosyl transferase | 1e-07/31/48 |
| PbCC004-C02 | RBT5 (AAG09790) | Secreted cell wall protein, GPI anchor | 1e-07/30/47 |
| PbCC008-G03 | 4e-16/31/45 | ||
| PbCC002-C04 | BGL2 (AAA21151) | β-Glucosyltransferase; cell wall synthesis | 1e-60/41/57 |
| Signal transduction genes | |||
| PbCS022-F02 | CST20 (AAB38875) | MEKK kinase hyphal formation | 2e-77/77/88 |
| PbCC004-A02 | CPP1 (P43078) | Phosphatase, hyphal formation | 6e-04/44/64 |
| PbCC011-F09 | CEK1 (A47211) | MAPK, hyphal formation | 8e-04/29/48 |
| PbCS023-G05 | Protein kinase A (AAF64072) | Hyphal formation | 3e-15/43/56 |
| PbCC010-A10 | CDC42 (AAB69764) | Cell division control protein, hyphal formation | 1e-15/64/76 |
| PbCS021-A08 | GEF (Q9UQX7) | Ras-like protein, hyphal formation | 4e-21/33/54 |
ID, identification.
The metabolic genes identified were ADE2, HIS1, HEM3, MLS1, ICL1, and PLB1 (Table 2). These genes have already been linked to fungal virulence because the homozygous null strains are attenuated in animal models of candidiasis (42).
Genes involved in the synthesis of the cell wall and in adhesion, such as CHS2, CHS3, and BGL2, were also identified (Table 2). We also identified several homologues that are involved directly or indirectly in adhesion, including HWP1, INT1, MNT1, PHR1, PHR2, PMT1, and RBT5 (Table 2). Several genes encoding proteins involved in the signal transduction pathway and implicated in the dimorphic switch were also identified (Table 2). We could recognize several factors from the MAPK pathway in P. brasiliensis, such as CST20, CPP1, CEK1, and CDC42. One of the subunits of protein kinase A and GEF, involved in the cAMP pathway, were also found (Table 2).
In addition to the genes mentioned above, we were able to identify several genes that could play a role either in temperature-induced dimorphic transition and/or in the infection process (for the DNA sequences and annotation, see http://143.107.203.68/est/default.html). During the M-Y transition there is an increase in the chitin content in the cell wall (4). Accordingly, we identified a series of chitin synthases (Table 2), along with other genes involved in chitin synthesis, such as chitin synthase regulatory factor, chitin biosynthesis protein, and chitinase (Table 3). We could also recognize a gene that encodes a protein involved in cell wall biogenesis and architecture whose regulation could play a role during the M-Y transition (Table 3). Genes encoding proteins that are induced upon and/or are important for the heat shock response were also recognized. Besides several heat shock proteins (three of which are listed in Table 4), we identified the S. cerevisiae SIS1 homologue, the S. cerevisiae HFM1 homologue that encodes the heat shock-inducible inhibitor of cell growth, and the S. cerevisiae STI1 homologue (Table 3).
TABLE 3.
P. brasiliensis clusters that could play a role either in temperature-induced dimorphic transition or in the infection process
| Gene or protein type and P. brasiliensis cluster IDa | Homologue (NCBI protein accession no.) | Main feature and/or role | BLAST E-value/ % identity/ % similarity |
|---|---|---|---|
| Cell wall genes | |||
| PbCC011-D01 | C. albicans (BAA19847) | Chitin synthase regulatory factor | 4e-23/51/73 |
| PbCS022-H12 | S. cerevisiae CHS6 (NP_012436) | Chitin biosynthesis protein | 4e-04/30/49 |
| PbCC002-F03 | H. capsulatum (AAG41982) | Chitinase | 4e-20/39/50 |
| PbCC012-F03 | S. cerevisiae ECM27 (NP_012640) | Protein involved in the cell wall biogenesis and arquitecture | 6e-09/46/71 |
| Heat shock proteins | |||
| PbCC003-E08 | S. cerevisiae SIS1 (NP_014391) | Induced by heat shock | 5e-07/71/83 |
| PbCC004-A04 | S. cerevisiae HFM1 (NP_010978.1) | Heat shock - inducible inhibitor of cell growth | 7e-27/48/66 |
| PbCC003-G05 | S. cerevisiae STI1 (NP_014670) | Induced by heat shock | 2e-23/52/65 |
| Signal transduction genes | |||
| PbCC002-D10 | C. albicans PKC1 (P43057) | Protein kinase C | 3e-25/55/75 |
| PbCC011-H05 | H. capsulatum (AAC27509) | Calmodulin | 3e-27/39/62 |
| PbCS011-H09 | S. cerevisiae VTC1 (NP_010995) | Protein that acts as a negative regulator of the CDC42 protein | 2e-21/68/80 |
| Miscellaneous | |||
| PbCC006-H02 | M. grisea (AAK73019) | Ceramide glucosyl synthase | 8e-49/48/66 |
| PbCC028-E03 | S. cerevisiae YCF1 (NP_010419) | Glutathione S-conjugate vacuolar transporter | 2e-14/60/75 |
ID, identification.
TABLE 4.
P. brasiliensis contigs that were used for the design of TaqMan probes
| ID clustera | Contig (BLAST E-value/% identity/ % similarity) |
|---|---|
| PbCC003-E02 | UBI, polyubiquitin C. albicans (e-162/99/99) |
| PbCC006-F12 | OXI, alternative oxidase H. capsulatum (e-151/73/81) |
| PbCC004-F01 | Hydro, hydrophobin precursor A. nidulans (5e-10/40/70) |
| PbCC008-G03 | RBT, RBT5 C. albicans (4e-16/31/45) |
| PbCC006-G05 | HSP70, Hsp70 C. albicans (0.0/77/85) |
| PbCC005-F04 | HSP82, Hsp82 H. capsulatum (4e-87/68/72) |
| PbCC012-B06 | HSP104, Hsp104 C. albicans (0.0/55/73) |
| PbCC003-D08 | GPI, PHR1 glycosidase GPI-anchored C. albicans (2e-70/49/68) |
| PbCC005-H06 | ISSO, ICL1 C. albicans Isocltrate lyase (1e-33/52/68) |
| PbCC002-G09 | MAL, MLS1 C. albicans (1e-29/56/69) |
| PbCC011-G03 | OLE1, OLE1 S. cerevisiae delta-9-desaturase (0.0/87/91) |
| PbCC012-A11 | Pb1067, unknown |
| PbCC012-H05 | Pb1145, unknown |
ID, identification.
Another aspect of the dimorphic switch in P. brasiliensis is the change in the composition of the cell membrane lipids. There are documented differences in the composition of GLSs in the cell membrane (31, 61) in the dimorphic transition. GLSs are synthesized from glucosylceramides (GlcCer), and the biosynthesis of GlcCer is catalyzed by glucosylceramide synthase (29). We identified the P. brasiliensis ceramide glucosyl synthase homologue (Table 3).
Our knowledge about signal transduction pathways in P. brasiliensis is very limited compared to other fungal pathogens. In addition to the genes mentioned in Table 2, we were able to identify the protein kinase C homologue, a calmodulin gene (Table 3), and nine and four genes encoding putative serine-threonine kinases and phosphatases, respectively, with high identity to their respective homologues (data not shown). In addition, we have also seen the S. cerevisiae VTC1 homologue, which encodes a protein that acts as a negative regulator of the CDC42 protein (Table 3).
Multidrug resistance is becoming an important issue in fungal infections. Several genetic determinants of drug resistance were identified, and most of them encode transporters, mainly of the ATP-binding (ABC) cassette family (for a review, see reference 67). Based on sequence identity, we could identify 90 transporters in our project; 11 of these belong to the family of ABC transporters, whereas one of them is a major facilitator superfamily transporter. An interesting representative of the ABC family was the cluster PbCS028-E03, which encodes the homologue of the S. cerevisiae YCF1 glutathione S-conjugate vacuolar transporter (Table 3).
Expression analysis of some of the P. brasiliensis genes during dimorphic transition.
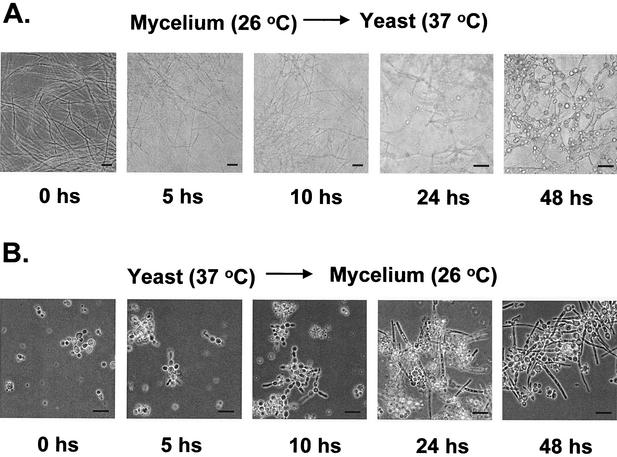
The morphological and functional transition in P. brasiliensis, in contrast to other dimorphic fungi such as C. albicans that depend on more complex stimuli (15), can be induced by simply shifting the incubation temperatures from 26 to 37°C. To verify the mRNA expression of some of the C. albicans homologues and some other selected P. brasiliensis genes (see Tables 2 and 4) during dimorphic transition, we isolated RNA from fungi undergoing M-Y and Y-M dimorphic transitions as described in Materials and Methods. As observed in Fig. 2A, there is a progressive transformation of mycelia to spherical or oval yeast cells after 5 to 10 h at 37°C, and this reaches >50% transformation after 48 and 120 h. Yeast cells start forming pseudohyphae after 5 h of incubation at 26°C; the mycelial transition progresses until reaching >50% transformation after 48 h of incubation at 26°C (Fig. 2B, lower panel).
FIG. 2.
Temperature-induced morphological switch in P. brasiliensis. Mycelial and yeast cells growing in the early exponential phase were induced to undergo morphological transformation by changing the temperature of incubation. (A) M-Y transition at 26 to 37°C for 0, 5, 10, 24, and 48 h. (B) Y-M transition at 37 to 26°C for 0, 5, 10, 24, and 48 h. Magnifications: the photomicrographs are reproduced at ×13 for 0, 5, and 10 h in the M-Y transition and at ×26 for 24 and 48 h in the M-Y transition, and are reproduced at ×26 for 0, 5, 10, 24, and 48 h in the Y-M transition. The bar at the bottom of each microphotograph represents 10 μm.
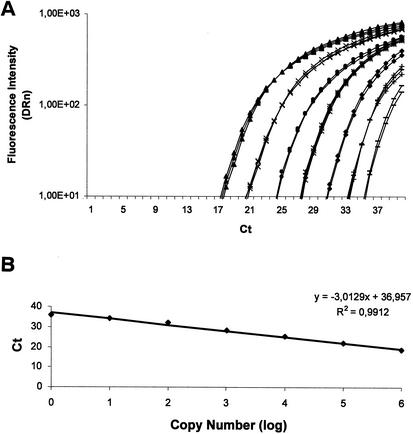
Real-time RT-PCR with fluorescent TaqMan probes was used to quantify the relative transcript levels of 13 P. brasiliensis genes (Table 4). The primers and fluorescent probes designed are shown in Table 1. All of the P. brasiliensis RT-PCR products yielded a single band with the expected size, as visualized on agarose gels (data not shown). Figure 3 shows the amplification plots and the respective standard curves for the α-tubulin gene (PbCC004-F01). By using these fluorescent probes, we were able to detect between 1 and 10 copies of each gene. No PCR products were detected in any negative control samples that contained all reagents except for the P. brasiliensis genomic DNA, even after 40 rounds of amplification (data not shown). Since there is no ideal control for gene expression, we first compared the α-tubulin and the 18S ribosomal probes as normalizers for the expression of these genes. We have seen no difference with either normalizer. However, we cannot completely exclude the possibility that the α-tubulin and 18S ribosomal gene expression are variable during the transition. Accordingly, the calibrator gene used in the expression experiments was the α-tubulin gene (data not shown).
FIG. 3.
Amplification plot (A) and the respective standard curve (B) for the α-tubulin gene from P. brasiliensis. The upper and lower panels in A and B correspond to the amplification plot and the standard curve for the α-tubulin gene from P. brasiliensis. The amplification plot corresponds to PCRs carried out with increasing amounts of genomic DNA, from left to right, 107 (σ), 106 (×), 105 (λ), 104 (θ), 103 (υ), 102 (+), and 101 (−) copies (based on an assumed genome size of 30 Mb). The DRn (ΔRn) is the normalized reporter signal minus the baseline established in the first few cycles of PCR. Ct is defined as the first amplification cycle at which fluorescence indicating PCR products becomes detectable. The data from the amplification plot were used for the construction of the standard curves (i.e., Ct values plotted against the logarithm of the DNA copy number). A linear relationship was obtained by plotting the resulting threshold cycle values (Ct) against the logarithm of the initial template amount. The best-fit line was determined via linear regression analysis. The results are the averages of four determinations. For each gene used for quantitative RT-PCR analyses, a similar amplification plot and standard curve was established.
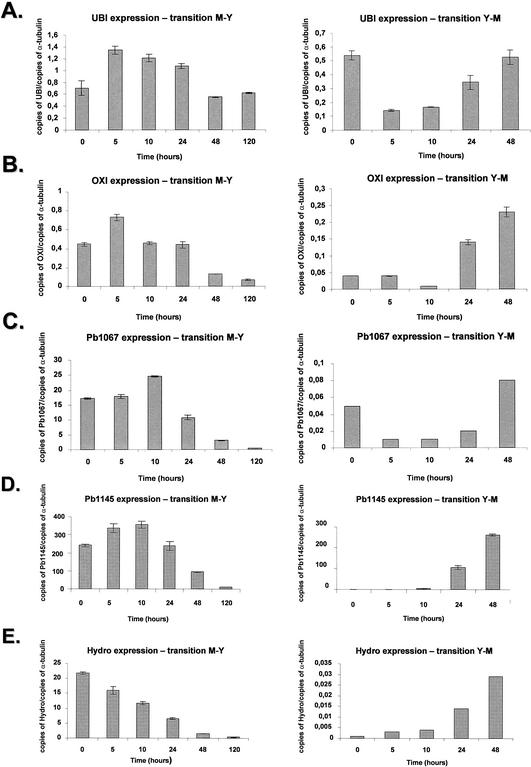
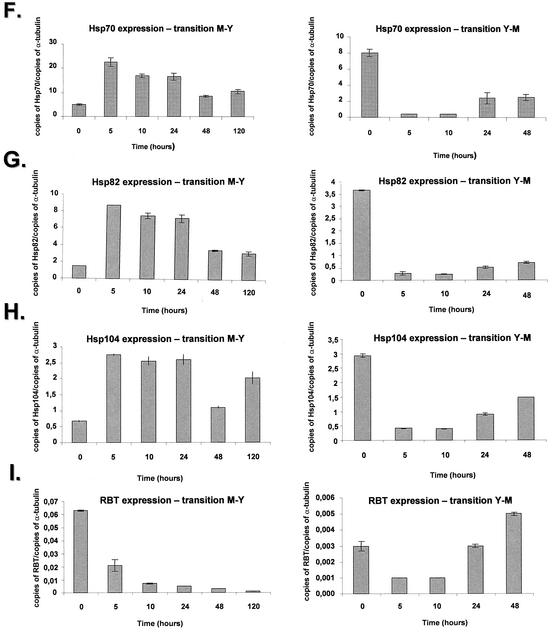
The polyubiquitin gene is induced at transcriptional level in several organisms upon heat shock stress (47). The P. brasiliensis UBI gene (contig PbCC003-E02) showed an increase of approximately twofold after 5 h, followed by constant levels that decreased after 48 h to the initial levels (Fig. 4A, right panel). In contrast, during the Y-M transition there was a fivefold decrease of mRNA at 5 and 10 h, followed by a steady increase until the initial mRNA levels of the yeast phase at time zero were reached (Fig. 4A, left panel).
FIG. 4.
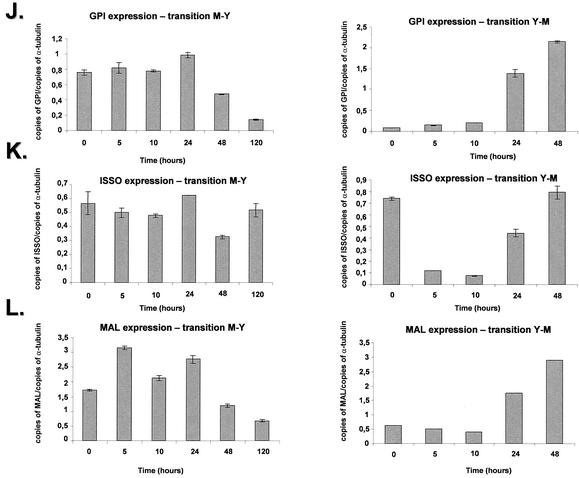
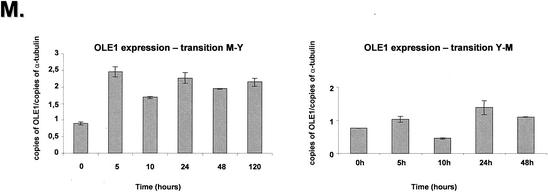
Quantitation of the mRNA expression of selected genes of P. brasiliensis during the M-Y-M transition. The measured quantity of each P. brasiliensis gene mRNA in each of the treated samples was normalized by using the CT values obtained for the α-tubulin RNA amplifications run in the same plate. The relative quantitation of each P. brasiliensis gene and α-tubulin gene expression was determined by a standard curve (i.e., Ct values plotted against the logarithm of the DNA copy number). The values represent the number of copies of the cDNAs of each P. brasiliensis gene divided by the number of copies of the cDNAs of the α-tubulin gene. The left and right panels represent the M-Y and the Y-M transitions, respectively. (A) UBI gene, ubiquitin; (B) OXI gene, alternative oxidase; (C) contig Pb1067; (D) contig Pb1145; (E) Hydro gene, hydrophobin gene; (F) HSP70 gene; (G) HSP82 gene; (H) HSP104 gene; (I) RBT gene; (J) GPI gene; (K) ISSO gene, isocitrate lyase; (L) MAL gene, malate dehydrogenase; (M) OLE1 gene, delta-9-desaturase. The results are the averages of three repetitions.
To probe mitochondrial gene expression during the transition, we decided to verify the quantitative expression of the P. brasiliensis OXI gene (contig PbCC006-F12), the homologue of the alternative oxidase. In the M-Y transition, there was a twofold increase of expression during the first 5 h that decreased after 10 to 24 h, followed by a fourfold decrease after 120 h (Fig. 4B, right panel); in the Y-M transition there was an increase of about fivefold after 48 h (Fig. 4B, left panel).
We have chosen two genes, Pb1067 (contig PbCC012-A11) and Pb1145 (contig PbCC012-H05) that encode unique proteins and were classified in the no-match category. In the M-Y transition, the Pb1067 gene showed a constant mRNA expression after 5 h, increasing ca. 50% after 10 h and decreasing dramatically (25-fold) after 120 h (Fig. 4C, left panel); in the Y-M transition, there was an initial fivefold decrease after 5 h, followed by an eightfold increase (Fig. 4C, right panel). The Pb1145 gene had an initial increase in the mRNA expression at the first 10 h, followed by a 30-fold decrease after 120 h in the M-Y transition (Fig. 4D, right panel); in the Y-M transition, there was an almost undetectable mRNA expression in the yeast phase that reached a 250-fold increase after 48 h (Fig. 4D, left panel). These results indicate that these two genes are predominantly expressed in the mycelial state.
The P. brasiliensis Hydro gene (contig PbCC004-F01) showed high similarity with the hydrophobin precursor of A. nidulans. The P. brasiliensis hydrophobin homologue displayed a decrease of ca. 25- to 30-fold in the mRNA expression after 48 to 120 h in the M-Y transition, whereas there was an increase of ca. 35-fold in the Y-M transition (Fig. 4E, right and left panels). These results indicate the Hydro gene is preferentially expressed in the mycelial state.
We verified the quantitative mRNA expression of the P. brasiliensis HSP70, HSP82, and HSP104 genes (Fig. 4F to H). These three genes showed a similar mRNA expression pattern in both transitions. In the M-Y transition, there was an increase of about four to fivefold at 5 h, followed by constant levels of expression and a decrease after 48 and 120 h (Fig. 4F to H, right panels). In the three genes, there was a three- to fourfold decrease in the mRNA expression during the Y-M transition (Fig. 4F to H, left panels). Our results confirm those of Da Silva et al. (13), who had shown that the HSP70 gene is differentially expressed in the transition, with a progressive increase in HSP70 expression during the period of conversion from mycelium to yeast.
We have also chosen two genes that have C. albicans homologues involved in adhesion, RBT and GPI (contigs PbCC008-G03 and PbCC003-D08, respectively), a GPI-anchored secreted cell wall protein, and a GPI-anchored glycosidase, linked to the cell wall structure, respectively. Two genes, ISSO and MAL, that encode putative C. albicans homologues from genes of the glyoxylate cycle, isocitrate lyase, ISSO, and malate synthase, MAL (contigs PbCC005-H06 and PbCC002-G09, respectively), and the S. cerevisiae OLE1 gene were also selected for further studies.
The P. brasiliensis RBT gene showed a 10-fold decrease during the M-Y transition (three times during the first five hours). As for the Y-M transition, we observed a fivefold increase in the mRNA expression during the 5- to 10-h period (Fig. 4I, right and left panels). The GPI gene showed a constant mRNA expression during the first 24 h of M-Y transition (Fig. 4J, left panel), decreasing two- and fourfold after 48 and 120 h, respectively. In the Y-M transition, the GPI mRNA expression increased ca. 22-fold after 48 h (Fig. 4J, right panel). The ISSO gene showed constant mRNA expression levels in the M-Y transition (Fig. 4K, right panel), while its expression dropped ∼7-fold at 5 to 10 h, followed by an increase of 5- and 7-fold at 24 and 48 h, respectively (Fig. 4K, left panel). The MAL gene showed a twofold increase after 5 h, which was kept constant after 24 h and decreased about threefold after 120 h of the M-Y transition; in the Y-M transition, the mRNA levels gradually increased until they reached an approximately sixfold induction after 48 h (Fig. 4K, right panel).
The P. brasiliensis OLE1 gene showed increased mRNA expression in the M-Y transition (about threefold from 5 to 120 h) and constant mRNA expression during the Y-M transition (Fig. 4 M, right and left panels).
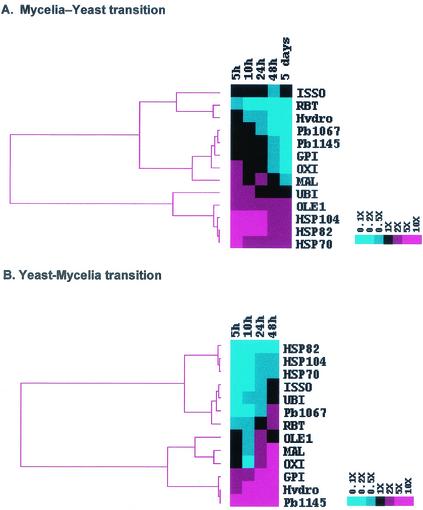
Hierarchical clustering is a method of analysis that brings out patterns of gene expression, by grouping genes with similar expression profiles. We decided to cluster the mRNA expression values of P. brasiliensis genes during the dimorphic transition Y-M-Y. Thus, mRNA expression values during the M-Y transition (Fig. 5A) clustered three different groups according to their mRNA expression pattern: (i) RBT, Hydro, and ISSO; (ii) MAL, Pb1067, Pb1145, GPI, and OXI; and (iii) UBI, OLE1, HSP70, HSP82, and HSP104. The first two groups displayed high mRNA expression in the mycelial phase, and a reduced expression during the M-Y transition, whereas the third group had lower mRNA expression in the mycelial phase, with increased expression during the M-Y transition (Fig. 4 and 5A).
FIG. 5.
Hierarchical clustering showing the gene expression profiles of 13 different P. brasiliensis genes during the M-Y (A) and Y-M (B) transitions (Fig. 4). In both experiments, relative levels of each transcript were monitored by real-time RT-PCR with mRNA extracted at the beginning of the differentiation process (T = 0) and also at different time points after the temperature-induced switch. These values were used to calculate the relative expression ratios against time T = 0. These ratios were log transformed and submitted to a hierarchical clustering algorithm by using the default parameters of the software CLUSTER, kindly provided by M. Eisen and available at http://www.microarrays.org/software.html. In both cases, as shown by the scales in the right, a color scheme was used to designate genes that were downregulated (blue) or upregulated (red). The clustering, along with the resulting dendrogram, was displayed by using TREEVIEW, also available at the same URL mentioned above (14).
Gene clustering of the mRNA expression for the Y-M transition yielded two large groups: (i) HSP82, HSP104, HSP70, ISSO, UBI, Pb1067, and RBT and (ii) OLE1, MAL, OXI, GPI, Hydro, and Pb1145. The first group displayed high mRNA expression in the yeast state, with lowered expression during the Y-M transition, whereas the second group had lower mRNA expression in the yeast state, with increased expression during the Y-M transition (Fig. 4 and 5B).
DISCUSSION
We have identified in this study the partial sequences of 4,692 nonredundant expressed genes from P. brasiliensis yeast phase. By using pulsed-field gel electrophoresis, Cano et al. (8) and Montoya et al. (41) showed that P. brasiliensis has between four or five chromosomes, varying in size from 2 to 10 Mb, and the genome size was estimated to be 23 to 20 Mb, suggesting the presence of 10,000 to 15,000 genes. Thus, our work has provided information of ca. 30 to 50% of P. brasiliensis genes. Considering that the NCBI nonredundant database contains only over a hundred expressed sequences from P. brasiliensis, our data represent a significant increment in the number of known P. brasiliensis genes.
Gene discovery and categorization.
Gene discovery in eukaryotes has been usually achieved by sequencing cDNA libraries due to the large amounts of noncoding DNA. The computational approach of identifying genes relies on similarity searches, and the limitation of this approach is apparent when phylum- or species-specific new genes are encountered. In our survey ca. 60% of putative new genes code for hypothetical proteins with no function known but with similarity to described proteins of other organisms. The vast majority of these putative new genes has matches to fungal proteins and is thus phylum specific “maverick” proteins. In the no match category the P. brasiliensis specific new sequences classified as unknown represent 17% of the total. The other putative proteins with functions suggested by similarity represent 23% assigned to seven broad functional categories (Fig. 1). The genes found represent a very substantial increment in the described P. brasiliensis genes.
Virulence and pathogenicity genes.
Using C. albicans genes already assigned a role in virulence and pathogenicity, we detected a number of P. brasiliensis candidates (Table 2). The metabolic genes listed have to be viewed with caution since there is disagreement about including factors associated with metabolic pathways as virulence factors (7, 23). We decided, however, to include metabolic genes as virulence determinants since it was reasoned that there are several other metabolic genes that, when deleted, do not cause reduced virulence in the pathogen. The genes indicated are significant in animal models of fungal disease (43). In C. albicans, phagocytosis upregulates the principal enzymes of the glyoxylate cycle, isocitrate lyase (ILC1) and malate synthase (MLS1). C. albicans mutants lacking ICL1 are markedly less virulent in mice than the wild type (32). Deletion of the PLB1 gene also results in reduced virulence (28).
Some cell wall biosynthesis and adhesion genes were also found. There is no universal consensus about the causal connection between reduced virulence upon deletion of genes, such as CHS2, CHS3, and BGL2, that encode biosynthetic cell wall enzymes. However, Navarro-Garcia et al. (42) point out that subtle changes in the cell wall could modify adhesion and recognition by the immunological system of the host, thus leading to altered virulence. The putative adhesion genes identified, when deleted in C. albicans, resulted in reduced virulence as determined in standard survival curves (42). This finding opens the possibility that conserved mechanisms of adhesion during host infection could be present in P. brasiliensis and C. albicans.
Signal transduction pathway genes have been implicated in the dimorphic switch and some were found (Table 2). There is a great deal of information about signaling pathways involved in controlling morphological switching, mostly provided by studies about pseudohyphal growth in S. cerevisiae (19). These studies have revealed that the signaling pathways are controlled by both cAMP and MAPK pathways (3). The identification of these genes in P. brasiliensis suggests that these pathways could be operating in this fungus, probably controlling the morphological switch.
Nonetheless, in spite of all of the information so far obtained, efficient transformation and inactivation systems are yet to be developed for P. brasiliensis and, therefore, the unequivocal involvement of these genes in dimorphic transition and/or virulence will require the appropriate experimental demonstration.
Expression analysis of selected P. brasiliensis genes during the dimorphic transition.
To obtain more accurate data on gene expression in P. brasiliensis, we used real-time RT-PCR assays. This approach is preferable over conventional Northern blot analysis because of the narrower linear range associated with radioactively labeled probes. This approach has been successfully used to characterize gene expression of sigma factor genes in Mycobacterium tuberculosis (34) and, more recently, in the quantitative expression of ABC transporter encoding genes in A. nidulans (48).
As noted by Lambowitz et al. (27), the morphological transition may be viewed as a heat shock response, followed by cellular adaptation to higher temperatures. Thus, the transition from 25 to 37°C in H. capsulatum is accompanied by marked changes in metabolic processes, such as respiration, cysteine metabolism, and rapid decline in intracellular ATP levels, due to uncoupling of oxidative phosphorylation (35). This is possible, because the mitochondria of fungi possess an alternative form of respiration in addition to the cytochrome pathway, which is resistant to cyanide and is mediated by alternative oxidase, an enzyme that transfers electrons from reduced ubiquinone to molecular oxygen (for a review, see reference 63). When electrons produced from the oxidation of NADH flow through the alternative pathway, two of the three sites of energy conservation are bypassed and the level of ATP is decreased. The alternative oxidase expression in P. brasiliensis was shown to parallel the behavior of the enzyme in H. capsulatum, where the cytochrome system and the alternative oxidase decrease in parallel over the first 24 to 40 h during the M-Y transition (35). The heat shock proteins investigated (HSP70, HSP82 and HSP04) have similar patterns of expression in the dimorphic transition with increased expression during the M-Y transition. The pattern of expression of the polyubiquitin gene suggests that the UBI gene product be directly involved in the increased rate of protein turnover that possibly exists particularly at the M-Y transition. They are certainly important factors in the stabilization of proteins necessary for the morphological transition at a higher temperature.
The RBT-, GPI-, ISSO-, MAL-, and OLE1-like genes were analyzed in their expression since the first four genes have already been shown to be required for fungal virulence (32, 42), whereas the OLE1 encodes the delta-9-desaturase gene, which is a key enzyme involved in regulating membrane fluidity in animal cells and microorganisms (20). The behavior of the OLE1 gene is marked by increased expression at the onset of the mycelium-to-yeast phase and a more constant expression in the Y-M transition. The fine regulation of this enzyme is believed to be responsible for major adjustments in the membrane composition in response to temperature or nutritional changes (35, 65). The H. capsulatum OLE1 is expressed in the yeast morphotype of all H. capsulatum strains examined but is rendered transcriptionally silent in the mycelia of some but not all strains (20). Evidence suggests that cells exposure to an abrupt increase in temperature causes their membranes to undergo a rapid decrease in molecular order (i.e., fluidity increases) (65). Chatterjee et al. (11) showed that the percentage of cellular unsaturated fatty acids increased when S. cerevisiae cells grown at 25°C were shifted to 37°C. Carratù et al. (10) complemented an S. cerevisiae mutant that has a disruption in its OLE1 gene with constructs that expressed the H. capsulatum OLE1 coding region under the control of different promoters. The manipulation of the expression of the H. capsulatum OLE1 gene altered the fatty acid composition of the cell membrane, and the different saturated and unsaturated fatty acid ratios had an important effect on HSP70 and HSP82 transcription. The authors of that study proposed that changes in the membrane order are an important factor in determining heat shock gene expression.
The behavior of two hypothetical and apparently species-specific genes, Pb1067 and Pb1145, revealed a predominant expression in the mycelial state, a behavior analogous to that exhibited by the hydrophobin gene. Fungal hydrophobins are secreted proteins that respond to the external environment. They are normally abundant proteins that are found in aerial hyphae, on the surface of fungal spores and infection structures, or on the surface of fruiting bodies (for a review, see reference 25). Hydrophobins have already been identified in plant pathogenic fungi (26) and in the human pathogen A. fumigatus (43).
The hierarchical clustering during the M-Y transition revealed three groups: (i) RBT, Hydro, and ISSO; (ii) MAL, Pb1067, Pb1145, GPI, and OXI; and (iii) UBI, OLE1, HSP70, HSP82, and HSP104. Although groups i and ii suggest that these genes are mainly expressed in the mycelial phase, group iii supports the hypothesis that changes in membrane order and ability to monitor protein folding and degradation are important factors in determining the dynamics of entry into the yeast phase, since P. brasiliensis OLE1, HSP70, HSP82, HSP104, and UBI genes display the same expression profile. Expression clustering at the Y-M transition resulted in two groups: (i) HSP82, HSP104, HSP70, ISSO, UBI, Pb1067, and RBT and (ii) OLE1, MAL, OXI, GPI, Hydro, and Pb1145. Most of the genes showed an inverse pattern of clustering in the two transitions (Fig. 5).
The correlation between the expression of specific genes in the yeast form and the morphological phase is complicated by the fact that a temperature shift is used to promote the dimorphic transition. Dissociation of temperature shift responses from morphogenesis in dimorphic fungi was achieved in Sporothrix schenckii in a study seeking to correlate certain polysaccharide structures with the Y-M transition (38). Yeast cells were independently obtained either by temperature shift to 37°C or by growing the cells in a synthetic medium with ammonium carbonate in tightened closed flasks with shaking at room temperature. Both cultures yielded yeasts with identical morphology, which synthesized identical polysaccharide structures that differed from those synthesized by the mycelium phase also grown at room temperature. In P. brasiliensis no attempts have yet been made to obtain the yeast phase at room temperature and therefore follow the expression of specific genes in cultures that have not been submitted to a temperature shift.
Overall, the results presented herein show, to our knowledge, the first case in which a high-throughput EST analysis was used to examine gene expression in the pathogenic state of P. brasiliensis. Classical genetic tools, such as DNA-mediated transformation and modulation of gene expression either by deletion or replacement or inactivation by RNA interference or antisense RNA have not been developed yet for P. brasiliensis. Consequently, this type of genomic approach, coupled with transcriptional profiling by microarrays and proteomics may represent an efficient alternative to identify genes involved in virulence or pathogenicity and provide a wider platform for a better understanding of P. brasiliensis biology and the role of several genes during the dimorphic transition and the onset of virulence and pathogenicity in PCM.
.
Acknowledgments
We thank the Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP), the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), and MCT-PRONEX (Brazil) for financial support.
REFERENCES
- 1.Almeida, I. C., D. C. A. Neville, A. Mehlert, A. Treumann, M. A. J. Ferguson, J. O. Previato, and L. R. Travassos. 1996. Structure of the N-linked oligosaccharide of the main diagnostic antigen of the pathogenic fungus Paracoccidioides brasiliensis. Glycobiology 6:507-515. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and Psi-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borges-Walsmley, M. I., and A. R. Walmsley. 2000. cAMP signalling in pathogenic fungi: control of dimorphic switching and pathogenicity. Trends Microbiol. 8:133-141. [DOI] [PubMed] [Google Scholar]
- 4.Borges-Walmsley, M. I., D. Chen, X. Shu, and A. R. Walmsley. 2002. The pathobiology of Paracoccidioides brasiliensis. Trends Microbiol. 10:80-87. [DOI] [PubMed] [Google Scholar]
- 5.Brandhorst, T. T., P. J. Rooney, T. D. Sullivan, and B. S. Klein. 2001. Using new genetic tools to study the pathogenesis of Blastomyces dermatitidis. Trends Microbiol. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 6.Brown, A. J. P., and N. A. R. Gow. 1999. Regulatory networks controlling Candida albicans morphogenesis. Trends Microbiol. 7:333-338. [DOI] [PubMed] [Google Scholar]
- 7.Calderone, R. A., and W. A. Fonzi. 2001. Virulence factors of Candida albicans. Trends Microbiol. 9:327-335. [DOI] [PubMed] [Google Scholar]
- 8.Cano, M. I. N., P. S. Cisalpino, I. Galindo, J. L. Ramirez, R. A. Mortara, and J. F. da Silveira. 1998. Electrophoretic karyotypes and genome sizing of the pathogenic fungus Paracoccidioides brasiliensis. J. Clin. Microbiol. 36:742-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carmona, A. K., R. Puccia, M. C. F. Oliveira, E. Rodrigues, L. Juliano, and L. R. Travassos. 1995. Characterization of an exocellular serine-thiol proteinase activity in Paracoccidioides brasiliensis. Biochem. J. 309:209-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carratù, L., S. Franceschelli, C. L. Pardini, G. S. Kobayashi, I. Horvarth, L. Vigh, and B. Maresca. 1996. Membrane lipid perturbation modifies the set point of the temperature of heat shock response in yeast. Proc. Natl. Acad. Sci. USA 93:3870-3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chatterjee, M. T., S. A. Khalawan, and B. P. G. Curran. 1997. Alterations in cellular lipids may be responsible for the transient nature of the yeast heat shock response. Microbiology 143:3063-3068. [DOI] [PubMed] [Google Scholar]
- 12.Cisalpino, P. S., R. Puccia, L. M. Yamauchi, M. I. N. Cano, J. F. da Silveira, and L. R. Travassos. 1996. Cloning, characterization, and epitope expression of the major diagnostic antigen of Paracoccidioides brasiliensis. J. Biol. Chem. 271:4553-4560. [DOI] [PubMed] [Google Scholar]
- 13.Da Silva, S. P., M. I. Borges-Walmsley, I. S. Pereira, C. M. A. Soares, A. R. Walmsley, and M. S. S. Felipe. 1999. Differential expression of na hsp70 gene during transition from the mycelial to the infective yeast form of the human pathogenic fungus Paracoccidioides brasiliensis. Mol. Microbiol. 31:1039-1050. [DOI] [PubMed] [Google Scholar]
- 14.Eisen, M. B., P. T. Spellman, P. O. Brown, and D. Botstein. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95:14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ernst, J. F. 2000. Regulation of dimorphism in Candida albicans, p. 98-111. In J. F. Ernst and A. Schimdyt (ed.), Dimorphism in human pathogenic and apathogenic yeasts. Karger, Basel, Switzerland. [DOI] [PubMed]
- 16.Ewing, B., L. Hillier, M. C. Wendl, and P. Green. 1998. Base-calling of automated sequencer traces using Phred. I. Accuracy assessment. Genome Res. 8:175-185. [DOI] [PubMed] [Google Scholar]
- 17.Ewing, B., and P. Green. 1998. Base calling of automated sequencer traces using Phred. II. Error probabilities. Genome Res. 8:194-196. [PubMed] [Google Scholar]
- 18.Freeman, W. M., S. J. Walker, and K. E. Vrana. 1999. Quantitative RT-PCR: pitfalls and potential. BioTechniques 26:112-125. [DOI] [PubMed] [Google Scholar]
- 19.Gancedo, J. 2001. Control of pseudohyphae formation in Saccharomyces cerevisiae. FEMS Microbiol. Rev. 25:107-123. [DOI] [PubMed] [Google Scholar]
- 20.Gargano, S., G. Di Lallo, G. S. Kobayashi, and B. Maresca. 1995. A temperature-sensitive strain of Histoplasma capsulatum has an altered delta 9-fatty acid desaturase gene. Lipids 30:899-906. [DOI] [PubMed] [Google Scholar]
- 21.Gordon, D., C. Abajian, and P. Green. 1998. Consed: a graphical tool for sequence finishing. Genome Res. 8:195-202. [DOI] [PubMed] [Google Scholar]
- 22.Hanna, S. A., J. L. M. da Silva, and M. J. S. M. Giannini. 2000. Adherence and intracellular parasitism of Paracoccidioides brasiliensis in Vero cells. Microbes Infect. 2:877-884. [DOI] [PubMed] [Google Scholar]
- 23.Haynes, K. 2001. Virulence in Candida species. Trends Microbiol. 9:591-596. [DOI] [PubMed] [Google Scholar]
- 24.Huang, X., and A. Madan. 1999. CAP3: a DNA sequence assembly program. Genome Res. 9:868-877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kershaw, M. J., and N. J. Talbot. 1998. Hydrophobins and repellents: proteins with fundamental roles in fungal morphogenesis. Fungal Genet. Biol. 23:18-33. [DOI] [PubMed] [Google Scholar]
- 26.Knogge, W. 1998. Fungal pathogenicity. Curr. Opin. Plant Biol. 1:324-328. [DOI] [PubMed] [Google Scholar]
- 27.Lambowitz, A. M., G. S. Kobayashi, A. Painter, and G. Medoff. 1983. Possible relationship of morphogenesis in pathogenic fungus, Histoplasma capsulatum, to heat shock response. Nature 303:806-808. [DOI] [PubMed] [Google Scholar]
- 28.Leidich, S. D., A. S. Ibrahim, Y. Fu, A. Koul, C. Jessup, J. Vitullo, W. Fonzi, F. Mirbod, S. Nakashima, Y. Nozawa, and M. A. Ghannoum. 1998. Cloning and disruption of caPLB1, a phospholipase B gene involved in the pathogenicity of Candida albicans. J. Biol. Chem. 273:26078-26086. [DOI] [PubMed] [Google Scholar]
- 29.Leipelt, M., D. Warnecke, U. Zahringer, C. Ott, F. Muller, B. Hube, and E. Geinz. 2001. Glucosylceramide synthases, a gene family responsible for the biosynthesis for the biosynthesis of glucosphingolipids in animals, plants, and fungi. J. Biol. Chem. 36:33621-33629. [DOI] [PubMed] [Google Scholar]
- 30.Lengeler, K. B., R. C. Davidson, C. D'Souza, T. Harshima, W.-C. Shen, P. Wang, X. Pan, M. Waugh, and J. Heitman. 2000. Signal transduction cascades regulating fungal development and virulence. Microbiol. Mol. Biol. Rev. 64:746-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Levery, S. B., M. S. Toledo, A. H. Straus, and H. K. Takahashi. 1998. Structure elucidation of sphingolipids from the mycopathogen Paracoccidioides brasiliensis: an immunodominant β-galacto-furanose residue is carried by a novel glycosylinositol phosphorylceramide antigen. Biochemistry 37:8764-8775. [DOI] [PubMed] [Google Scholar]
- 32.Lorenz, M. C., and G. R. Fink. 2001. The glyoxylate cycle is required for fungal virulence. Nature 412:83-86. [DOI] [PubMed] [Google Scholar]
- 33.Magrini, V., and W. E. Goldman. 2001. Molecular mycology: a genetic toolbox for Histoplasma capsulatum. Trends Microbiol. 9:541-546. [DOI] [PubMed] [Google Scholar]
- 34.Manganelli, R., E. Dubnau, S. Tyagi, F. R. Kramer, and I. Smith. 1999. Differential expression of 10 sigma factor genes in Mycobacterium tuberculosis. Mol. Microbiol. 31:715-724. [DOI] [PubMed] [Google Scholar]
- 35.Maresca, B., and G. S. Kobayashi. 2000. Dimorphism in Histoplasma capsulatum and Blastomyces dermatitidis, p. 201-216. In J. F. Ernst and A. Schimdyt (ed.), Dimorphism in human pathogenic and apathogenic yeasts. Karger, Basel, Switzerland. [DOI] [PubMed]
- 36.Marra, M. A., T. A. Kucaba, L. W. Hillier, and R. H. Waterston. 1999. High throughput plasmid DNA purification for 3 cents per sample. Nucleic Acids Res. 27:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Medoff, G., G. S. Kobayashi, A. Painter, and S. Travis. 1987. Morphogenesis and pathogenicity of Histoplasma capsulatum. Infect. Immun. 55:1355-1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mendonça, L., P. A. J. Gorin, K. O. Lloyd, and L. R. Travassos. 1976. Polymorphism of Sporothrix schenckii surface polysaccharides as a function of morphological differentiation. Biochemistry 15:2423-2431. [DOI] [PubMed] [Google Scholar]
- 39.Mendes-Gianinni, M. J. S., J. P. Bueno, M. A. Shikanai-Yasuda, W. Ferreira, and A. Masuda. 1989. Detection of the 43,000 molecular weight glycoprotein in sera of patients with paracoccidioidomycosis. J. Clin. Microbiol. 27:2842-2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mendes-Gianinni, M. J. S., M. L. Taylor, J. B. Bouchara, E. Burger, V. L. Calich, E. D. Escalantee, S. A. Hanna, H. L. Leenzi, M. P. Machado, M. Miyaii, J. L. Monteiro da Silva, E. M. Mota, A. Restrepo, S. Restrepo, G. Tronchin, L. R. Vincenzi, C. F. Xidieh, and E. Zenteno. 2000. Pathogenesis. II. Fungal responses to host responses: interaction of host cells with fungi. Med. Mycol. 1:113-123. [PubMed] [Google Scholar]
- 41.Montoya, A. E., M. N. Moreno, A. Restrepo, and J. G. McEwen. 1999. Electrophoretic karyotype of clinical isolates of Paracoccidioides brasiliensis. Fungal Genet. Biol. 21:223-227. [DOI] [PubMed] [Google Scholar]
- 42.Navarro-Garcia, F., M. Sánchez, C. Nombela, and J. Pla. 2001. Virulence genes in the pathogenic yeast Candida albicans. Microbiol. Rev. 25:245-268. [DOI] [PubMed] [Google Scholar]
- 43.Parta, M., Y. Chang, S. Rulong, P. Pinto da Silva, and K. J. Kwon-Chung. 1994. Hyp1, a hydrophobin gene from Aspergillus fumigatus complements the rodletless phenotype in Aspergillus nidulans. Infect. Immun. 62:4389-4395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Perfect, J. R., and G. M. Cox. 2000. Virulence mechanisms for fungi. I. Clin. Microbiol. Newsl. 22:113-119. [Google Scholar]
- 45.Perfect, J. R., and G. M. Cox. 2000. Virulence mechanisms for fungi. II. Clin. Microbiol. Newsl. 22:121-125. [Google Scholar]
- 46.Pinto, A. R., R. Puccia, S. N. Diniz, M. F. Franco, L. R. Travassos. 2000. DNA-based vaccination against murine paracoccidioidomycosis using the gp43 gene from Paracoccidioides brasiliensis. Vaccine 26:3050-3058. [DOI] [PubMed] [Google Scholar]
- 47.Piper, P. 1997. The yeast heat shock response, p. 75-100. In S. Hohmann and W. H. Mager (ed.), Yeast stress responses. Springer-Verlag, Berlin, Germany.
- 48.Pizeta-Semighini, C., M. Marins, M. H. S. Goldman, and G. H. Goldman. 2002. Quantitative analysis of the relative transcript levels of ABC-transporter atr genes in Aspergillus nidulans by real-time RT-PCR assay. Appl. Environ. Microbiol. 68:1351-1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Puccia, R., S. Schenckman, P. A. J. Gorin, and I. R. Travassos. 1986. Exocellular components of Paracoccidioides brasiliensis: identification of a specific antigen. Infect. Immun. 53:199-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Puccia, R., A. K. Carmona, J. L. Gesztesi, L. Juliano, and L. R. Travassos. 1998. Exocellular proteolytic activity of Paracoccidioides brasiliensis: cleavage of components associated with the basement membrane. Med. Mycol. 36:345-348. [PubMed] [Google Scholar]
- 51.Puccia, R., M. A. Juliano, L. Juliano, L. R. Travassos, and A. K. Carmona. 1999. Detection of the basement membrane-degrading proteolytic activity of Paracoccidioides brasiliensis after SDS-PAGE using agarose overlays containing Abz-MKALTLQ-EDDnp. Braz. J. Med. Biol. 32:645-649. [DOI] [PubMed] [Google Scholar]
- 52.Restrepo, A., J. G. McEwen, and E. Castañeda. 2001. The habitat of Paracoccidioides brasiliensis: how far from solving the riddle? Med. Mycol. 39:233-241. [DOI] [PubMed] [Google Scholar]
- 53.Rocha, C. R. C., K. Schroppel, D. Harcus, A. Macil, D. Dignard, B. N. Taylor, D. Y. Thomas, M. Whiteway, and E. Leberer. 2001. Signalling through adenyl cyclase is essential for hyphal growth and virulence in the pathogenic fungus Candida albicans. Mol. Biol. Cell 12:3631-3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rooney, P. J., and B. S. Klein. 2002. Linking fungal morphogenesis with virulence. Cell. Microbiol. 4:127-137. [DOI] [PubMed] [Google Scholar]
- 55.San-Blas, G. 1982. The cell wall of fungal human pathogens: its possible role in host-parasite relationship—a review. Mycopathologia 79:159-184 [DOI] [PubMed] [Google Scholar]
- 56.San-Blas, G., and G. Niño-Veja. 2001. Paracoccidioides brasiliensis: virulence and host response, p. 205-226. In R. L. Cihlar and R. A. Calderone (ed.), Fungal pathogenesis: principles and clinical applications. Marcel Dekker, Inc., New York, N.Y.
- 57.Sandoval, M. P., G. B. Del Negro, M. J. S. Mendes-Gianinni, and T. Brito. 1996. Distribution of exoantigens and 43 kDa glycoprotein (gp43) in yeast and mycelial forms of Paracoccidioides brasiliensis. J. Mycol. Med. 6:1-6. [Google Scholar]
- 58.Soares, R. M. A., F. Costa e Silva-Filho, S. Rozental, J. Angluster, W. de Souza, C. S. Alviano, and L. R. Travassos. 1998. Anionogenic groups and surfaces sialoglyconjugate structures of yeast forms of the human pathogen Paracoccidioides brasiliensis. Microbiology 144:309-314. [DOI] [PubMed] [Google Scholar]
- 59.Straus, A. H., E. Freymuller, L. R. Travassos, and H. K. Takahashi. 1996. Immunochemical and subcellular localization of the 43 kDa glycoprotein antigen of Paracoccidioides brasiliensis with monoclonal antibodies. J. Med. Vet. Mycol. 34:181-186. [DOI] [PubMed] [Google Scholar]
- 60.Tekaia, F., G. Blandin, A. Malpertuy, B. Llorente, P. Durrens, C. Tofffano-Nioche, O. Ozier-Kalogeropoulos, E. Bom, C. Gaillardin, M. Aigle, M. Bolotin-Fukuhara, S. Casaregola, J. de Montigny, A. Lepingle, C. Neuveglise, S. Potier, J. Souciet, M. Wesolowski-Louvel, and B. Dujon. 2000. Genomic exploration of the hemiascomycetous yeasts. III. Methods and strategies used for sequence analysis and annotation. FEBS Lett. 487:17-30. [DOI] [PubMed] [Google Scholar]
- 61.Toledo, M. S., S. B. Levery, A. H. Straus, E. Suzuki, M. Momany, J. Glushka, J. M. Moulton, and H. K. Takahashi. 1999. Characterization of sphingolipids from mycopathogens: factors correlating with expression of 2-hydroxy fatty acyl (E)-Δ[3]-unsaturation in cerebrosides of Paracoccidioides brasiliensis and Aspergillus fumigatus. Biochemistry 38:7294-7306. [DOI] [PubMed] [Google Scholar]
- 62.Tyagi, S., D. P. Bratu, and F. R. Kramer. 1998. Multicolor molecular beacons for allele discrimination. Nat. Biotechnol. 16:49-53. [DOI] [PubMed] [Google Scholar]
- 63.Vanlerbeghe, G. C., and L. McIntosh. 1997. Alternative oxidase: from gene to function. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48:703-734. [DOI] [PubMed] [Google Scholar]
- 64.Vicentini, A. P., J. L. Gesztesi, M. Franco, W. Souza, J. Moraes, L. R. Travassos, and J. D. Lopes. 1994. Binding of Paracoccidioides brasiliensis to laminin through surface glycoprotein gp43 leads to enhancement of fungal pathogenesis. Infect. Immun. 62:4-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vigh, L., B. Maresca, and J. L. Horwood. 1998. Does the membrane physical state control the expression of heat shock and other genes? Trends Biochem. Sci. 23:369-374. [DOI] [PubMed] [Google Scholar]
- 66.White, O., and A. R. Kerlavage. 1996. TDB: new databases for biological discovery. Methods Enzymol. 266:27-40. [DOI] [PubMed] [Google Scholar]
- 67.White, T. C., K. A. Marr, and R. A. Bowden. 1998. Clinical, cellular, and molecular factors that contribute to antifungal drug resistance. Clin. Microbiol. Rev. 11:382-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wood, J. P. 2002. Histoplasma capsulatum molecular genetics, pathogenesis, and responsiveness to its environment. Fungal Genet. Biol. 35:81-97. [DOI] [PubMed] [Google Scholar]