Abstract
Telomeres, the chromosome ends, are maintained by a balance of activities that erode and replace the terminal DNA sequences. Furthermore, telomere-proximal genes are often silenced in an epigenetic manner. In Saccharomyces cerevisiae, average telomere length and telomeric silencing are reduced by loss of function of UPF genes required in the nonsense-mediated mRNA decay (NMD) pathway. Because NMD controls the mRNA levels of several hundred wild-type genes, we tested the hypothesis that NMD affects the expression of genes important for telomere functions. In upf mutants, high-density oligonucleotide microarrays and Northern blots revealed that the levels of mRNAs were increased for genes encoding the telomerase catalytic subunit (Est2p), in vivo regulators of telomerase (Est1p, Est3p, Stn1p, and Ten1p), and proteins that affect telomeric chromatin structure (Sas2p and Orc5p). We investigated whether overexpressing these genes could mimic the telomere length and telomeric silencing phenotypes seen previously in upf mutant strains. Increased dosage of STN1, especially in combination with increased dosage of TEN1, resulted in reduced telomere length that was indistinguishable from that in upf mutants. Increased levels of STN1 together with EST2 resulted in reduced telomeric silencing like that of upf mutants. The half-life of STN1 mRNA was not altered in upf mutant strains, suggesting that an NMD-controlled transcription factor regulates the levels of STN1 mRNA. Together, these results suggest that NMD maintains the balance of gene products that control telomere length and telomeric silencing primarily by maintaining appropriate levels of STN1, TEN1, and EST2 mRNA.
Telomeres, the ends of linear chromosomes, are important for chromosome integrity and are maintained by telomerase, a reverse transcriptase-like enzyme that includes an integral RNA template. The catalytic components of Saccharomyces cerevisiae telomerase (TLC1 RNA and Est2p), as well as gene products required for telomerase activity in vivo (e.g., Est1p, Est3p, Cdc13/Est4p, Ku70/80, Mec1p, MRX, Rap1p, Stn1p, Tel1p, and Ten1p), have been identified (reviewed in reference 12). However, mechanisms that regulate the expression and activity of telomerase components and modulators have not been explored.
The nonsense-mediated mRNA decay (NMD) pathway accelerates the degradation of mRNAs that prematurely terminate translation due to nonsense mutations, frameshifts, or translation of alternate open reading frames (ORFs) within the mRNA (21, 37). In S. cerevisiae, the products of UPF1, UPF2, and UPF3 are required for NMD and provide a surveillance function to lower the abundance of potentially deleterious protein fragments by degrading mRNAs that cannot be translated full length (42). However, the only known growth phenotype of upf mutants is deficient respiration (1). Interestingly, NMD also controls the expression of some wild-type genes. By using high-density oligonucleotide arrays (HDOAs), several hundred wild-type S. cerevisiae mRNAs with either increased or decreased steady-state levels in upf mutants were identified (23). NMD directly regulates the level of wild-type mRNAs for some genes, such as SPT10 and CPA1, through accelerated degradation triggered by translation of alternate ORFs within the mRNA (46, 50). Given the large number of genes controlled by NMD, including transcription factors such as Ppr1p (29, 45) and Ino4p (23), many wild-type mRNAs are likely to change in abundance as an indirect consequence of changes in the abundance of transcriptional regulators (23).
Previously it has been found that mutations in UPF1, UPF2, or UPF3 reduced telomere length and silencing of a telomere-adjacent reporter gene (25). It was hypothesized that NMD regulates telomeres by altering the levels of specific wild-type mRNAs important for telomere functions. To identify genes important for the telomere-related phenotypes of upf mutants, we screened the HDOA data of Lelivelt et al. (23) for S. cerevisiae genes thought to be important for telomere functions. Here we report that mRNAs encoding the catalytic subunit of telomerase, regulators of telomerase activity, and proteins that affect telomeric silencing are all controlled by NMD. Furthermore, extra copies of EST2, STN1, and TEN1 were sufficient to mimic the telomeric silencing and telomere length phenotypes of upf mutants.
MATERIALS AND METHODS
Yeast strains, plasmids, and growth conditions.
The yeast strain used for HDOA studies was LRSy307 (MATa his3-11,15 trp1-Δ1 leu2-Δ1 ura3-52 upf1::ura3[5-fluoroorotic acid {FOA} resistant] upf2::HIS3 upf3::TRP1) transformed with UPF plasmids as described in Lelivelt et al. (23). Data from the HDOA experiments can be found at the following website: http://144.92.19.47/default.htm. Northern analysis was done using the same strains or with YJB276 (MATa leu2-3,112 ura3-52 trp1-289 his3Δ ade2Δ), YJB2763 (MATa leu2-3,112 ura3-52 trp1-289 his3Δ ade2Δ nmd2::HIS3 adh4::URA3-TEL), or YJB487 (MATa leu2-3,112 ura3-52 his3Δ ade2Δ adh4::URA3-TEL) transformed with either pRS315-NMD2 or pRS315 (25). Telomeric silencing assays were performed as described previously (16) using YJB487 transformed with the indicated plasmids and strain YJB539 (MATa leu2-3,112 ura3-52 his3Δ ade2Δ rlf4-1 adh4::URA3-TEL) carrying a upf2/nmd2 mutation as a control strain. Telomere length assays, telomerase assays, and TLC1 RNase protection experiments were performed using strain YJB209 (MATα ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1, ura3-1) transformed with appropriate plasmids and crossed with strain YJB195 (MATa ura3-1 ade2-1 his3-11,15 leu2-3,112 can1-100 trp1-1) transformed with YEplac181, p2μm-STN1a, and p2μm-EST2, respectively, as described below and in the figure legends. RNA half-life experiments were conducted using strain AAY333 (MATα ADE2 ura3 his3-11,15 trp1-1 leu2-3,112 rpb1-1 upf1::URA3) complemented with either pRS315 or pRS315-UPF1.
Plasmid p2μm-EST1 (pVL157; provided by V. Lundblad) contains a 2.58-kb BamHI/SphI fragment of EST1 inserted in the BamHI/SphI site of YEp24. p2μm-EST2 contains a 3.75-kb BamHI-SacI fragment containing the entire coding sequence of EST2 in YEplac181 (15). p2μm-EST3 contains a 1.15-kb fragment containing the wild-type EST3 gene in YEplac181. p2μm-STN1a contains full-length STN1 cloned as a PvuII-SacI fragment into the SmaI-SacI sites in pRS423 (7). p2μm-STN1b contains full-length STN1 cloned into YEplac195 (provided by M. Charbonneau). p2μm-SAS2 (pDR1058; provided by D. Rivier) contains full-length SAS2 cloned into pRS425. p2μm-ORC5 (pAD002; provided by A. Dillon and J. Rine) contains ORC5 cloned as a XhoI-NotI fragment into pFAT1. p2μm-TEN1 (provided by M. Charbonneau) contains YLR010c and ∼300 nucleotides (nt) of the promoter sequence. Strains containing two plasmids were made for all pairwise combinations of the seven relevant genes (STN1, EST1, EST2, EST3, ORC5, SAS2, and TEN1), except for EST3 with EST2, ORC5, or SAS2 and ORC5 with SAS2 or TEN1.
Strains expressing green fluorescent protein (GFP) reporter fusion mRNAs were constructed by PCR-mediated homologous recombination (49). Each ORF was replaced from the start codon to the stop codon with the complete GFP ORF. PEST1-GFP, PEST2-GFP, and PEST3-GFP were made by using pVL368, pVL296, and pVL298 (all provided by V. Lundblad), respectively, as templates. PSTN1-GFP was made by using pSE1393, which contains the STN1 gene in YEplac181.
Telomere length and yeast telomerase assays.
Average telomere length was determined by Southern blot analysis of PstI-digested genomic DNA as described previously (25). To assess telomerase activity, yeast whole-cell extracts and DEAE fractions were prepared as previously described (8, 32). DEAE fractions were tested for telomerase activity by using two different primers in standard primer extension assays, and the results were quantified by PhosphorImager analysis (32).
RNA methods.
For half-life and steady-state experiments, total RNA was extracted as described by Leeds et al. (21). To measure mRNA half-lives, strain AAY333 (provided by A. Atkin, University of Nebraska, Lincoln), which carries the rbp1-1 temperature-sensitive allele encoding RNA polymerase II (36), was used. Transcription was terminated by shifting cells from 25 to 37°C. Cells were collected at intervals following temperature shift. After extraction, RNA was denatured by using glyoxal and dimethyl sulfoxide, separated on 1% agarose gels, and transferred to GeneScreen Plus (Dupont, NEN Research Products, Boston, Mass.) (4). RNA was detected by hybridization to radiolabeled DNA probes or riboprobes, which were prepared by in vitro transcription of template DNA in the presence of [α-32P]UTP (800 or 3,000 Ci/mmol; Amersham Life Science, Arlington Heights, Ill.) by using the Riboprobe System (Promega Corp., Madison, Wis.). DNA templates for riboprobe synthesis were prepared by PCR amplification of genomic DNA by using standard conditions. The ACT1 riboprobe is 224 nt in length and contains sequences complementary to nt 30 to 528 of the ORF. The CDC13 riboprobe is 270 nt in length and contains sequences complementary to nt 2 to 271 of the ORF. The CPA1 riboprobe is 697 nt in length and contains sequences complementary to nt 126 to 822 of the ORF. The EST1 riboprobe is 691 nt in length and contains sequences complementary to nt 706 to 139 of the ORF. The EST2 riboprobe is 645 nt in length and contains sequences complementary to nt 1011 to 1655 of the ORF. The EST3 riboprobe is 597 nt in length and contains sequences complementary to nt −29 through 568 relative to the first nucleotide of the ORF. The GFP riboprobe is 363 nt in length and contains sequences complementary to nt 222 to 584 of the ORF. The ORC5 riboprobe is 430 nt in length and contains sequences complementary to nt 506 to 935 of the ORF. The PGK1 riboprobe is 95 nt in length and contains sequences complementary to nt 1157 to 1251 of the ORF. The SAS2 riboprobe is 308 nt in length and contains sequences complementary to nt 353 to 660 of the ORF. The STN1 riboprobe is 377 nt in length and contains sequences complementary to nt 551 to 927 of the ORF. The TEN1 riboprobe is 366 nt in length and contains sequences complementary to nt 2 to 344 of the ORF. By using template DNA fragments obtained from restriction digestion or PCR, CYH2 or TRP1 probes were prepared and hybridized as described previously (2).
For analysis of TLC1 RNA levels in telomerase fractions, the TLC1 gene (nt 1 to 1301) was amplified by PCR and cloned between the BamHI and EcoRV sites of pBluescript II KS+. The resulting plasmid was linearized by digestion with HinfI, and antisense RNA encompassing residues 1097 to 1301 of the TLC1 gene was generated by T3 RNA polymerase in the presence of 12 μM [α-32P]GTP (31). Total RNAs from DEAE fractions were isolated and combined with the probe (100,000 cpm), precipitated with ethanol, hybridized, digested with RNase T1, RNase A, and proteinase K, and analyzed by gel electrophoresis (33).
RESULTS AND DISCUSSION
Steady-state levels of EST1, EST2, EST3, STN1, TEN1, SAS2, and ORC5 mRNA are elevated in upf mutants.
We hypothesized that NMD regulates the steady-state mRNA levels of specific wild-type genes that are important for telomere function (25). To identify wild-type mRNAs that accumulate in upf mutant strains and are responsible for the associated telomere-related phenotypes, we focused on HDOA data for a subset of ∼80 ORFs that encode proteins with known or suspected telomere function (Table 1) (23). Those with a combined knockout index (CKI) score of >0.5 or <−0.5 were selected for analysis. The CKI score is an indicator of how consistently the level of a specific mRNA is elevated or decreased in upf mutant strains relative to that in an isogenic UPF parental strain (23). By this criterion, the mRNA levels of the majority of the selected set of telomere-related mRNAs were not significantly affected in upf mutants. However, 7 of the ∼80 mRNAs (encoded by EST1, EST2, STN1, TEN1, SAS2, ORC5, and MSI1/CAC3) had CKI scores of >0.5, with the average change in mRNA levels in upf mutants being an increase of 1.75- to 3.3-fold relative to the levels in UPF strains (Table 1). None of the mRNAs had a score of <−0.5. To confirm the results of the HDOA studies and to extend them to relevant RNAs not present on the HDOA, we compared steady-state levels of mRNAs in total RNA prepared from wild-type and upf mutant strains on Northern blots. As predicted from the HDOA experiments, the levels of EST1, EST2, STN1, and TEN1 mRNA in the upf strains were increased by at least twofold relative to those in wild-type strains (Table 2).
TABLE 1.
Expression of selected genes with possible telomere-related functions in upf mutant strains relative to that in UPF strains
| ORF | Gene name | CKIa | AFCb |
|---|---|---|---|
| YLR010C | TEN1 | 1.00 | 3.3 |
| YMR127C | SAS2 | 0.88 | 3.20 |
| YNL261W | ORC5 | 0.67 | 1.75 |
| YDR082W | STN1 | 0.59 | 2.55 |
| YBR195C | MSI1/CAC3 | 0.56 | 2.29 |
| YLR318W | EST2 | 0.56 | 2.45 |
| YLR233C | EST1 | 0.53 | 2.25 |
| YCL011C | RLF6 | 0.44 | 1.30 |
| YMR106C | HDF2 | 0.25 | 1.40 |
| YML061C | PIF1 | 0.22 | 2.05 |
| YLR453C | RIF2 | 0.22 | 1.90 |
| YMR284W | HDF1 | 0.19 | 1.53 |
| YPR018W | CAC1/RLF2 | 0.13 | 0.25 |
| YPL001W | HAT1 | 0.13 | 0.98 |
| YLR223C | IFH1 | 0.13 | 0.98 |
| YNL250W | RAD50 | 0.13 | 7.03 |
| YPL128C | TBF1 | 0.13 | 3.70 |
| YGR099W | TEL2 | 0.13 | 1.02 |
| YML102W | CAC2 | 0.09 | 1.06 |
| YMR224C | MRE11 | 0.09 | 1.32 |
| YPR162C | ORC4 | 0.09 | 1.46 |
| YPL153C | RAD53 | 0.09 | 4.60 |
| YKR101W | SIR1 | 0.09 | 1.42 |
| YOR351C | MEK1 | 0.07 | 1.07 |
| YOL051W | GAL11 | 0.06 | 1.13 |
| YEL056W | HAT2 | 0.06 | 1.13 |
| YOR025W | HST3 | 0.06 | 1.33 |
| YDR191W | HST4 | 0.06 | 1.26 |
| YNL330C | RPD3 | 0.06 | 1.03 |
| YDR369C | XRS2 | 0.06 | 1.03 |
| YNL102W | CDC17 | 0.03 | 1.15 |
| YOR038C | HIR2 | 0.03 | 0.98 |
| YGL058W | RAD6 | 0.03 | 1.06 |
| YBL088C | TEL1 | 0.03 | 1.12 |
| YOR229W | WTM2 | 0.03 | 0.87 |
| YDL220C | CDC13 | 0.00 | 1.14 |
| YOR217W | CDC44 | 0.00 | 0.99 |
| YER088C | DOT6 | 0.00 | 1.08 |
| YDR225W | HTA1 | 0.00 | 0.90 |
| YDR224C | HTB1 | 0.00 | 0.86 |
| YJL076W | NET1 | 0.00 | 1.05 |
| YML065W | ORC1 | 0.00 | 1.06 |
| YLL004W | ORC3 | 0.00 | 1.15 |
| YKL113C | RAD27 | 0.00 | 0.94 |
| YML032C | RAD52 | 0.00 | 0.88 |
| YDR217C | RAD9 | 0.00 | 0.20 |
| YOR217W | RFC1 | 0.00 | 0.99 |
| YBR275C | RIF1 | 0.00 | 0.97 |
| YBL092W | RPL32 | 0.00 | 0.93 |
| YDR227W | SIR4 | 0.00 | 1.33 |
| YLR234W | TOP3 | 0.00 | 0.00 |
| YER151C | UBP3 | 0.00 | 1.41 |
| YDR440W | DOT1 | −0.03 | 0.87 |
| YBL008W | HIR1 | −0.03 | 1.37 |
| YHR013C | ARD1 | −0.06 | 1.13 |
| YJL115W | ASF1 | −0.06 | 1.15 |
| YDL160C | DHH1 | −0.06 | 1.22 |
| YNL021W | HDA1 | −0.06 | 0.00 |
| YBR009C | HHF1 | −0.06 | 0.93 |
| YBR060C | ORC2 | −0.06 | 1.51 |
| YBL052C | SAS3 | −0.06 | 1.03 |
| YOR230W | WTM1 | −0.06 | 0.82 |
| YNL031C | HHT2 | −0.07 | 0.90 |
| YJR138W | HIR3 | −0.09 | 1.66 |
| YIL010W | DOT5 | −0.13 | 1.00 |
| YNL216W | RAP1 | −0.13 | 0.77 |
| YLR442C | SIR3 | −0.13 | 0.96 |
| YPL139C | WTM3 | −0.13 | 0.91 |
| YDL042C | SIR2 | −0.16 | 0.85 |
| YNL030W | HHF2 | −0.19 | 0.71 |
| YBR010W | HHT1 | −0.19 | 0.88 |
| YBL002W | HTB2 | −0.19 | 0.80 |
| YDL040C | NAT1 | −0.19 | 1.08 |
| YHR119W | SET1 | −0.19 | 1.01 |
| YGL173C | KEM1 | −0.22 | 0.84 |
| YBL003C | HTA2 | −0.25 | 0.81 |
CKI (combined knockout index) was defined by Lelivelt and Culbertson (23). For each mRNA, numerical weights were assigned to GeneChip difference calls for four trials for each of the four upf mutant strains compared to four trials for an isogenic UPF strain as follows: increased signal (+2), statistically marginal increase (+1), no change (0), statistically marginal decrease (−1), and decrease (−2). The numerical values of the difference calls were summed across all trials and then divided by the maximum potential score to yield the CKI score. Scores of ≥0.5 indicate that the given mRNA was increased in abundance with relative consistency from trial to trial, whereas scores of ≤−0.5 indicate that a given mRNA was decreased in abundance with similar relative consistency.
AFC (average fold change) was measured independently of the CKI score. The AFC is the average increase or decrease in mRNA abundance from 16 trials with upf mutants compared with four trials with UPF strains. AFCs for some mRNAs with CKI scores between 0.5 and −0.5 are not reliable because the CKI score indicates poor consistency of calls across trials.
TABLE 2.
Average increases (n-fold) in mRNA levels measured by HDOA and Northern blot analysis
| Gene name | HDOA dataa | Northern blotting (upf1 upf2 upf3/ UPF1 UPF2 UPF3)b | Northern blotting (upf1/UPF1)c | GFP reporter Northern blotting (upf3/UPF3)d |
|---|---|---|---|---|
| EST1 | 2.1 ± 0.4 | 2.8 ± 0.4 | 3.6 | 2.1 ± 0.61 |
| EST2 | 2.3 ± 1.1 | 4.3 ± 0.4 | 7.7 | 1.8 ± 0.01 |
| EST3 | NT | 3.1 ± 0.3 | 4.7 | 2.0 ± 0.01 |
| ORC5 | 1.7 ± 0.3 | 2.0 ± 0.3 | 2.6 | ND |
| SAS2 | 3.2 ± 0.9 | 2.9 ± 0.4 | 2.8 | ND |
| STN1 | 2.6 ± 0.5 | 5.2 ± 0.5 | 6.1 | 3.1 ± 0.1 |
| TEN1 | 3.3 ± 0.7 | 4.8 ± 0.7e | NDf | ND |
| CDC13 | 1.1 ± 0.7 | 1.1 ± 0.2 | 1.1 | ND |
| MSI1 | 2.1 ± 1.0 | 1.3 ± 0.3 | 1.4 | ND |
| TLC1 | NT | 0.8 ± 0.3e | 0.9 | ND |
Values represent average fold change and sample standard deviation (Table 1). NT, EST3 and TLC1 were not represented on the microarrays.
mRNA levels were determined by using quantitative RNA blots like those shown in Fig. 1 for isogenic upf mutant strain LRSy307 (pRS316) compared with wild-type strain LRSy307 (pML1). Values represent average of data ± standard deviation from the four upf mutant strains relative to the wild type (n = 4). In all cases, mRNA levels were normalized to actin mRNA levels.
mRNA levels were determined by using quantitative RNA blots of strains ML51 (upf1 mutant) and ML34 (UPF1+).
GFP reporter mRNA levels were determined by quantitative RNA blots of strains YJB3758 and YJB4468.
TEN1 and TLC1 RNA levels were determined in strain YJB1471 (upf2 mutant) and were normalized to levels of PGK1 mRNA.
ND, not done.
EST1 and EST2 (24) are members of the telomerase epistasis group. EST2 encodes the catalytic subunit of telomerase. Est1p mediates the access of telomerase to the telomere through interactions with Cdc13/Est4p (11, 17, 43) and is required for in vivo, but not in vitro, telomerase activity (26). CDC13/EST4 mRNA levels in upf mutant strains were unchanged (Table 2). EST3 and TLC1, also members of the telomerase/EST epistasis group, were not present on the HDOA.
TLC1 encodes the RNA component of telomerase and was not included on the HDOA because it does not have an obvious ORF (47). High levels of TLC1 RNA, like upf mutations, cause telomere shortening and reduced telomeric silencing (47). Northern analysis of total RNA indicated that TLC1 RNA levels were not affected by upf mutations (Table 2), which is consistent with the idea that the NMD pathway acts on mRNAs during translation (2, 3) and not on untranslated RNAs like the product of TLC1. Thus, despite the similarity in the phenotypes of upf mutants and strains expressing high levels of TLC1 RNA, NMD does not affect telomere function by altering the steady-state levels of TLC1 RNA.
The EST3 ORF includes a programmed +1 frameshift (34) but was not included on the HDOA. Because EST3 includes a frameshift within the 5′ 50% of the mRNA that bypasses a premature stop codon, it was a good candidate for an mRNA that is degraded by the NMD pathway (21, 22). The result of Northern blot analysis was consistent with this prediction: EST3 mRNA levels in upf mutants increased approximately three- to fivefold relative to those in the isogenic UPF strains (Table 2).
STN1 and TEN1 are essential genes and have a role in chromosome capping and the prevention of deleterious degradation of chromosome ends (18, 19). STN1 encodes a high-copy suppressor of cdc13-1 (19), and TEN1 encodes a gene product that interacts physically with both Cdc13p and Stn1p to enhance the ability of Stn1p to negatively regulate telomerase activity at telomeres (18). Northern blot analysis confirmed that the mRNA levels of STN1 and TEN1 were elevated in upf mutant strains (Table 2). Thus, the mRNA levels of several telomerase subunits and regulators (Est1p, Est2p, Est3p, Stn1p, and Ten1p) are influenced by the NMD pathway, while the RNAs for others involved in the same telomere-related processes (TLC1 RNA and Cdc13p) are not.
In addition to ORFs with known effects on telomerase function, data from the HDOA analysis revealed three genes (SAS2, ORC5, and MSI1/CAC3) with known effects on telomeric silencing and chromatin structure that had elevated mRNA levels in upf mutant strains. The results from Northern blot experiments were consistent with an increase in SAS2 and ORC5 mRNA levels of at least twofold (Table 2). Sas2p is a putative histone acetyltransferase that is a positive regulator of silencing at telomeres (44). ORC5 encodes a component of the origin recognition complex (ORC) (10, 28) and influences silencing at telomeres and the HM loci (14). MSI1/CAC3 mRNA, which encodes a subunit of chromatin assembly factor I, exhibited only a 1.3-fold increase on the Northern blots compared with the 2.1 ± 1.0-fold increase measured with HDOAs (Table 2). Therefore, we did not study MSI1/CAC3 further. Also consistent with the HDOA data, the levels of RAP1, SIR3, SIR4, histone H4 (HHF1 and HHF2), and CAC1/RLF2 mRNAs in upf mutant strains did not change (data not shown). Thus, the NMD pathway affects the accumulation of mRNAs for at least seven genes (EST1, EST2, EST3, STN1, TEN1, SAS2, and ORC5) that are important for telomerase activity, telomere length control, and/or telomeric silencing.
Elevated levels of Est2p and Stn1p together phenocopy the telomeric silencing defect of upf mutant strains.
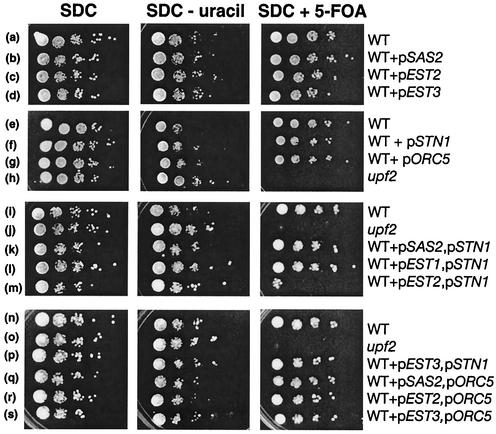
To test the hypothesis that the telomere-related phenotypes of upf mutants are caused by increases in the level(s) of one or more of the seven mRNAs (EST1, EST2, EST3, STN1, TEN1, SAS2, and ORC5), we investigated whether increasing the copy number of any one of the genes could mimic (phenocopy) the telomere-related phenotypes of upf mutants. Yeast 2μm vectors were used to provide multiple copies of each of the individual genes with their native promoters. Northern analysis confirmed that the levels of the individual mRNAs increased twofold or more in strains carrying these 2μm plasmids (data not shown). Thus, the increased steady-state level of mRNA in these experiments generally equaled or exceeded the magnitude of the increase of specific mRNAs seen in upf mutants. Using strains that overexpress the gene products, we first examined whether increased copies of any individual gene could reduce the normal silencing of a telomere-adjacent gene. In wild-type strains, a URA3 gene inserted near the left end of chromosome VII is subject to epigenetic telomeric silencing such that a proportion of the cells are able to grow on 5-FOA (synthetic dextrose complete [SDC] plus 5-FOA), which is toxic to Ura+ strains. In upf mutant strains, the telomere-adjacent URA3 is no longer silent; thus, upf mutant cells do not grow on SDC plus 5-FOA (Fig. 1). An elevated level of EST2, EST3, STN1, SAS2, or ORC5 expression had no obvious effect on silencing of the telomeric URA3 gene in these otherwise wild-type strains (Fig. 1, rows a to h). Similar results were observed when extra copies of TEN1 or EST1 were provided and telomeric silencing was monitored by use of a strain in which ADE2 was inserted near the right end of chromosome V (data not shown). This is consistent with the observation that the overproduction of wild-type Est1p or Est2p does not affect telomeric silencing (13). Thus, an elevated level of any one of these mRNAs was not sufficient to account for the telomeric silencing phenotype of upf mutant strains.
FIG. 1.
High-level expression of EST2 and STN1 results in reduced silencing of telomere-adjacent genes. The expression of a telomere-adjacent URA3 gene present in otherwise wild-type or upf mutant strains was determined by growth of the strain on complete medium (SDC), on medium lacking uracil (SDC − uracil), and on complete medium containing 5-FOA (SDC + 5-FOA) as indicated. Wild-type strain YJB487 was transformed with vector YEplac181 (row a), p2μm-SAS2 (row b), p2μm-EST2 (row c), p2μm-EST3 (row d), vector pRS423 (row e), p2μm-STN1 (row f), or p2μm-ORC5 (row g). The same wild-type strain was cotransformed with both YEPlac181 and pRS423 (rows i and n) or p2μm-SAS2 and p2μm-STN1 (row k), p2μm-EST1 and p2μm-STN1 (row l), p2μm-EST2 and p2μm-STN1 (row m), p2μm-EST3 and p2μm-STN1 (row p), p2μm-SAS2 and p2μm-ORC5 (row q), p2μm-EST2 and p2μm-ORC5 (row r), or p2μm-EST3 and p2μm-ORC5 (row s). upf2 mutant strain YJB539 was also transformed with YEplac181 (row h) or with both YEplac181 and pRS423 (rows j and o).
We next tested whether overexpression of pairs of genes could confer a reduced silencing phenotype (similar to the telomeric silencing phenotype of upf mutants). In each case, there was a greater-than-twofold average increase in the level of each RNA expressed (data not shown). Interestingly, only one combination of two plasmids, p2μm-STN1 with p2μm-EST2, resulted in reduced telomeric silencing (Fig. 1, rows i to s). In several independent transformants, telomeric silencing in strains containing both p2μm-STN1 and p2μm-EST2 (Fig. 1, row m) was similar to that seen in upf mutants, as indicated by an inability to grow in the presence of 5-FOA. Because the NMD pathway down-regulates PPR1 (21, 38), which is a positive regulator of URA3, the slight difference in growth observed for these two strains is most likely due to increased levels of PPR1 and URA3 expression in the upf mutant strains relative to their levels in the UPF strain containing both p2μm-STN1 and p2μm-EST2. However, the effect of extra copies of p2μm-STN1 and p2μm-EST2 was seen when telomeric silencing was detected using ADE2 to mark the right end of chromosome V. Thus, the effect is not dependent on URA3 or PPR1. We did not observe additional silencing when p2μm-TEN1 was expressed together with p2μm-EST2 or p2μm-STN1 by using the telomeric ADE2 marker on chromosome V (data not shown). Thus, extra copies of EST2 and STN1 together, which encode the catalytic subunit of telomerase and a regulator of telomerase function, respectively, were sufficient to recapitulate the telomeric silencing defect observed in upf mutant strains.
Our results support a connection between telomerase regulation and telomeric silencing and are consistent with the idea that titration of telomere-associated proteins influences telomeric silencing. TLC1 was cloned as a high-copy disruptor of telomeric silencing (47). This occurs through a 48-nt stem-loop structure in TLC1 RNA that most likely interacts with the Ku proteins (40), which are important for telomere organization within the nucleus as well as for telomeric silencing (20). Similarly, in upf mutant strains, high levels of Stn1p and Est2p may perturb telomeric silencing by altering the stoichiometry or function of telomere-associated proteins such as Cdc13p and/or Ku (17).
Increased levels of Stn1p phenocopy the telomeric length control defect of upf mutant strains.
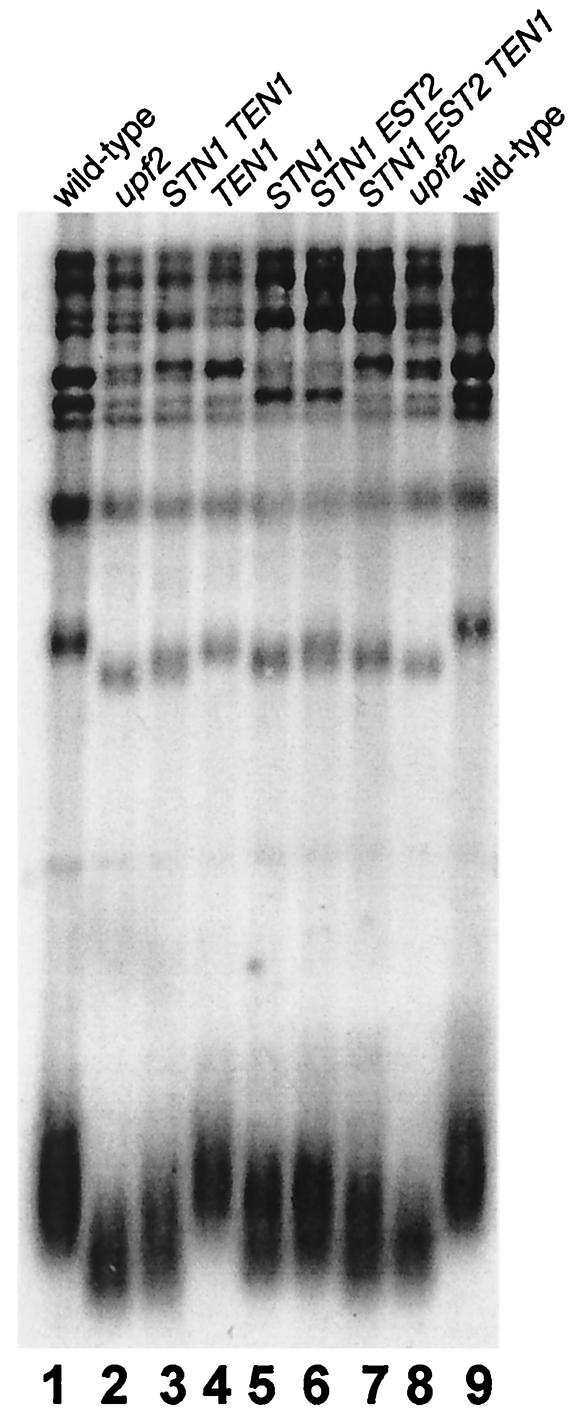
We next investigated whether additional copies of EST1, EST2, EST3, STN1, TEN1, SAS2, or ORC5 affected telomere length control to the degree seen in upf mutants. Telomere length was measured by digestion of genomic DNA with PstI, which releases a terminal ∼0.8-kb telomere fragment from the majority of the telomeres (those that contain a Y′ telomere-associated sequence [30]). Larger fragments that hybridize to the TG1-3/C1-3A probe correspond to non-Y′ telomeres and to subtelomeric fragments that include internal TG1-3/C1-3A repeats. The upf mutant strains carrying one or two control vectors exhibited an average telomere length that was ∼57 bp shorter than that in an isogenic wild-type strain (Fig. 2) (Table 3). Transformation with p2μm-STN1 alone resulted in a consistent decrease in the lengths of the shortest terminal telomere fragments (∼43 ± 19 bp shorter than those in the wild type [Fig. 2] [Table 3]), which were significantly different from the lengths of telomeres in the wild-type strain but not significantly different from the lengths of telomeres in upf mutant strains. In contrast, increasing the level of EST1, EST2, EST3, SAS2, ORC5, or TEN1 had no significant effect on telomere length in an otherwise wild-type strain (Fig. 2 and data not shown). Importantly, telomeres in strains that carried p2μm-STN1 together with either p2μm-TEN1 or p2μm-EST2 were 54 ± 26 bp and 52 ± 20 bp shorter, respectively, than wild-type telomeres (Fig. 2) (Table 3). When p2μm-STN1, p2μm-EST2, and p2μm-TEN1 were all present in the same strain, telomeres were 67 ± 28 bp shorter than in the wild-type and 10 ± 28 bp shorter than in the upf mutant strains. Since the levels of all three RNAs in this strain are higher than their levels in the upf2 mutant strain, this result suggests that increased levels of STN1 are required for the short-telomere phenotype of upf mutant strains and that increased levels of TEN1 and EST2 mRNAs contribute to the phenotype. Consistent with this notion, both Stn2p and Ten1p are negative regulators of telomerase (17, 18).
FIG. 2.
High-level expression of STN1 and TEN1 results in reduced telomere length. Genomic DNA was digested with PstI and analyzed on a 1% agarose gel, and telomere sequences were detected with a telomere repeat sequence probe on pCA75 (51). Lanes 1 and 9, wild-type strain (YJB209) transformed with control vector (YEp195) and crossed to YJB3011; lanes 2 and 8, upf2 mutant strain (YJB7178); lanes 3, 4, and 7, YJB209 transformed with p2μm-TEN1 (YLR010) and crossed with strains YJB3234, YJB3011, and YJB3345, respectively; lanes 5 and 6, YJB209 transformed with YEplac195 and crossed with YJB3234 and YJB3345, respectively.
TABLE 3.
Average telomere length in strains carrying extra copies of STN1, TEN1, and EST2
| Relevant genotypea | Avg lengthb | Difference (bp) from:
|
|
|---|---|---|---|
| Wild type | upf2 mutant | ||
| WT | 703 ± 21 (14) | 0 | 57 |
| upf2 | 646 ± 9 (6) | −57 | 0 |
| STN1 | 660 ± 19 (6)c | −43 | 14 |
| TEN1 | 723 ± 23 (6)d | 20 | 77 |
| STN1 TEN1 | 649 ± 26 (12)c | −54 | 3 |
| STN1 EST2 | 651 ± 20 (6)c | −52 | 5 |
| STN1 EST2 TEN1 | 636 ± 28 (6)c | −67 | −10 |
Genotypes as described in the legend to Fig. 2. Genes shown in capital letters were provided on 2μm plasmids.
Average length in base pairs of PstI fragment ± standard deviation. The numbers in parentheses indicate the numbers of independent experiments done.
Not significantly different from the upf2 mutant according to the rank sum test (47).
Not significantly different from wild type according to the rank sum test (47).
Three NMD-sensitive genes, STN1, TEN1, and EST2, appear to account for the telomeric phenotypes of upf mutant strains. The effect of NMD on telomere length requires STN1, and the effect of NMD on telomeric silencing involves STN1 and EST2. Thus, NMD confers the two phenotypes through increased levels of STN1 RNA in combination with other gene products. The involvement of Stn1p in both telomeric silencing and telomeric length control is especially interesting in light of the proposed role for Stn1p as the primary effector of chromosome end protection (39). Through interactions with Cdc13p, Stn1p also has a role in coupling lagging-strand synthesis (of the telomeric C strand) to telomerase extension of the 3′ end (telomeric G strand) of the chromosome (6). Thus, NMD-mediated control of the STN1 mRNA level appears to be critical for both the telomere length and telomeric silencing phenotypes of upf mutant strains.
Sequences necessary for NMD-mediated control of telomere-related mRNA abundance.
The NMD pathway controls the levels of specific mRNAs either directly or indirectly. Direct effects, in which the decay rate of an mRNA is affected by NMD, result when an mRNA contains a built-in premature stop codon. In wild-type mRNAs that are normally subject to NMD, this may occur when a translatable upstream ORF is present in the 5′ leader (46) or when leaky scanning leads to translation initiation at an out-of-frame AUG codon which brings a premature stop codon into register (50). Indirect effects on mRNA accumulation can result when the mRNA is transcriptionally regulated by the product of another mRNA that is affected by the NMD pathway (9, 23). Thus, mRNAs involved in telomere function could be either direct targets whose mRNA decay rates depend on NMD or indirect targets whose transcription rates depend on NMD.
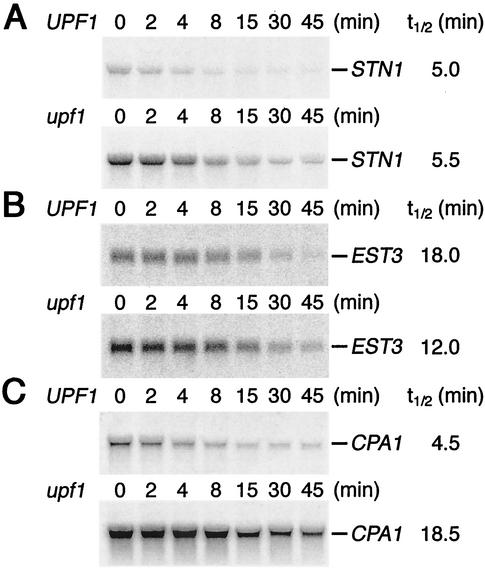
We determined that NMD affects the expression of seven genes important for telomere functions and that a subset of these genes (EST2, STN1, and TEN1) can phenocopy a upf mutant strain. Because STN1 contributed significantly to both of the telomere-related phenotypes of upf mutant strains, experiments were done to address the effect of NMD on the STN1 mRNA levels. To determine whether STN1 mRNA is specifically targeted for degradation by NMD, we compared the half-lives of STN1 mRNA in wild-type and upf1 mutant strains. The half-lives of STN1 mRNA were not significantly different when measured in UPF1 and upf1 strains (Fig. 3A), whereas the control CPA1 mRNA (50) exhibited an approximately fourfold difference (Fig. 3C), indicating that STN1 mRNA does not appear to be a direct substrate of NMD. If STN1 mRNA is indirectly regulated by one or more NMD-sensitive transcription factors, then the STN1 promoter sequence is expected to confer NMD control to a reporter gene inserted in place of the STN1 coding sequence. To determine whether the STN1 promoter is sufficient for NMD control of the mRNA, we constructed a plasmid that fused 300 nt of DNA upstream to and including the STN1 start codon to the ORF for GFP (PSTN1-GFP) (27). Epifluorescence microscopy indicated that wild-type cells carrying PSTN1-GFP appeared slightly green due to expression of GFP from the STN1 sequence (data not shown). These cells were crossed to a upf3::HIS3 strain and sporulated. Epifluorescence analysis of asci containing four spores revealed an apparent segregation of two bright green and two dim green spores (data not shown). Northern blot analysis of RNA levels in sister spores from this cross (using a GFP riboprobe) indicated that levels of reporter mRNA were 3.1-fold higher in upf3 mutant strains than in UPF3 sister spores (Table 2). Analysis of isogenic upf3::HIS3 and UPF3 progeny by fluorimetry confirmed that upf3::HIS3 spores emitted significantly more GFP fluorescence than the UPF3 spores (data not shown). These results indicate that the STN1 promoter sequences are sufficient to confer NMD-dependent control on STN1 mRNA levels. Taken together, our results suggest that the promoter of STN1 is subject to control by NMD through an indirect mechanism involving the modulation of transcription levels. We propose that NMD controls the stability of an mRNA corresponding to an upstream regulator of STN1 expression.
FIG. 3.
STN1 and EST3 mRNA half-lives in wild-type and upf1 mutant strains are not different. mRNA half-lives (t1/2) were determined by Northern blot analysis of total RNA (15 μg) extracted from UPF1 (AAY333 plus pRS315UPF1) and upf1 mutant (AAY333 plus vector) strain cultures collected at the indicated times after termination of transcription (see Materials and Methods). Blots were hybridized with a riboprobe complementary to the STN1 mRNA (A), EST3 mRNA (B), or CPA1 mRNA (C). In two independent experiments, the STN1 mRNA at time zero was 3.4- and 4.0-fold more abundant in the upf1 mutant strain. Similarly, the EST3 mRNA was 2.0- and 2.1-fold more abundant, and the CPA1 mRNA was 4.1- and 3.8-fold more abundant in the upf1 mutant strain. Half-life values determined from the experiment shown are indicated at right. In independent experiments, the STN1 half-lives were 5.0 min in the UPF1 strain and 4.5 min in the upf1 mutant strain, the EST3 half-lives were 12.0 min in the UPF1 strain and 15.0 min in the upf1 mutant strain, and the CPA1 half-lives were 4 min in the UPF1 strain and 16 min in the upf1 mutant strain.
Using GFP-reporter fusions, we compared EST1 and EST2 expression in wild-type and upf mutant strains. We constructed and analyzed the expression of a fusion construct containing EST1 sequence 5′ to the start codon fused to the GFP ORF. GFP fluorescence levels were too low for quantitation by epifluorescence microscopy, fluorimetry, or flow cytometry. Northern analysis revealed a twofold increase in the levels of reporter mRNA in upf3 spores relative to the levels in UPF3 sister spores (Table 2), suggesting that, like STN1, promoter sequences 5′ to the EST1 ORF contribute to the increased level of EST1 mRNA in upf mutant strains. Similar results were obtained when the EST2 ORF was replaced with the GFP ORF: the reporter mRNA levels were 1.8-fold higher in upf3 mutants than in UPF3 strains (Table 2). This indicates that sequences 5′ of the EST1 and EST2 ORFs are sufficient to account for all of the EST1 and most of the EST2 mRNA accumulation, respectively, in upf mutant strains relative to the levels in wild-type strains. However, we could not discern whether EST1 and EST2 mRNAs are regulated directly or indirectly, because the mRNA half-lives could not be measured.
The EST3 ORF was also replaced with that of GFP, and the accumulation of reporter mRNA levels was twofold greater in upf3 mutants than in UPF3 strains (Table 2), indicating that the increased level of EST3 mRNA in upf mutant strains was not due to sequences within the EST3 ORF. This was surprising because the EST3 ORF has an internal stop codon and a +1 programmed frameshift (34), suggesting that it might be degraded by NMD. Furthermore, the EST3 mRNA decay rates in upf1 mutant and UPF1 strains were similar (Fig. 3B), indicating that EST3 is not regulated at the level of mRNA decay. This implies that the internal stop codon in the ORF is not responsible for the effects of NMD on this mRNA. This in-frame stop codon could fail to trigger NMD for several reasons, including its position within the ORF relative to downstream sequences required for NMD or an interplay between the internal stop codon and the programmed frameshift site that allows translation to bypass the stop codon frequently. In either case, EST3 appears to be affected by NMD because of indirect effects on EST3 transcription initiation.
Role of NMD in controlling the level of telomerase activity.
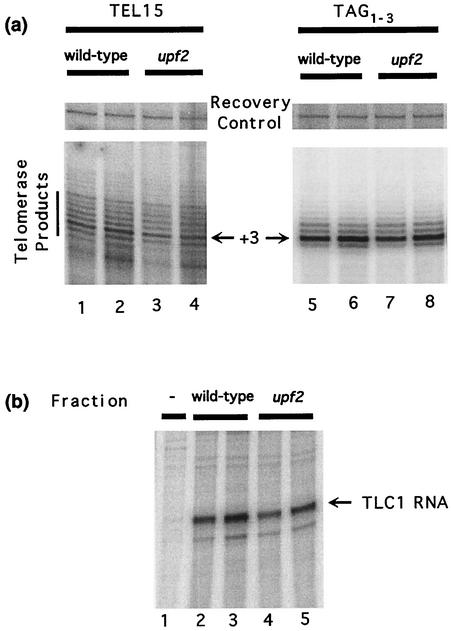
To determine whether telomere length in upf mutant strains was due to altered telomerase activity, we compared the ability of partially purified telomerase extracts prepared from wild-type and upf2 mutant strains to extend telomere sequence primers. Extracts from the two strains exhibited nearly identical activity in the primer extension assays with two different oligonucleotides as substrates (Fig. 4a). Consistent with the results of our Northern blot analysis of total RNA (Table 2), the amounts of TLC1 RNA in partially purified telomerase extracts were comparable in the two strains (Fig. 4B). Thus, loss of NMD (and the resulting increased levels of EST2 mRNA) does not affect the amount of extractable telomerase activity or the amount of TLC1 RNA in the telomerase fraction. In S. cerevisiae, in vitro telomerase activity requires Est2p and TLC1 but does not require Est1p or Est3p (26). Since neither in vitro telomerase activity nor the amount of TLC1 RNA is altered in upf mutant strains, it is possible that extractable telomerase activity in upf mutant cells may be limited by the levels of TLC1 RNA.
FIG. 4.
In vitro telomerase activity and TLC1 RNA levels are not dependent on UPF2. (a) Primer extension assays for yeast telomerase were carried out using either TEL15 primer (lanes 1 to 4) or TAG1-3 primer (lanes 5 to 8) and either l μl (lanes 1, 3, 5, and 7) or 10 μl (lanes 2, 4, 6, and 8) of DEAE fractions derived from wild-type (YJB209; lanes 1, 2, 5, and 6) or upf2 mutant (YJB1274; lanes 3, 4, 7, 8) strains. Each reaction also contained a prelabeled 46-mer oligonucleotide that served as a recovery control. Under the standard reaction conditions, telomerase consistently gave rise to the product at the primer +3 position (arrows). (b) RNase protection assays using either 100 μl (lanes 2 and 4) or 200 μl (lanes 3 and 5) of DEAE fractions derived from either wild-type (YJB209; lanes 2 and 3) or upf2 mutant (YJB1274; lanes 4 and 5) strains. Lane 1 contains 20 μg of yeast tRNA as a negative control (48).
In vivo, EST1, EST3, STN1, and TEN1 contribute to telomerase-dependent telomere length control, presumably by regulating the access of the chromosomal terminus to telomerase (6, 17, 18, 39; reviewed in reference 12). Despite the fact that the NMD pathway controls levels of EST2 mRNA, which encodes the catalytic subunit of telomerase, telomere length and levels of extractable telomerase activity are not affected by increased levels of EST2 mRNA (Fig. 2 and 3). Thus, the shorter telomeres in upf mutant strains are not due to increased levels of EST2 or to increased levels of telomerase activity. Rather, we propose that upf mutant strains have short telomeres because Stn1p (together with Ten1p) limits the accessibility of the telomeres to telomerase. Our results suggest that NMD affects telomeric silencing by increasing the levels of Stn1p and Est2p, which may titrate other factors that interact with the chromosome end complex and/or with telomerase itself. One candidate for the titrated factor is Cdc13p, which interacts with both telomerase and Stn1p (17, 39). Another candidate is the Ku70/Ku80 complex, which is also required for telomere length control and telomeric silencing (5, 20, 35, 41).
In summary, the level of expression of several telomerase components and regulators, including EST1, EST2, EST3, STN1, and TEN1, but not CDC13/EST4, depend on NMD. Increasing the levels of EST2, STN1, and TEN1, which encode the catalytic subunit of telomerase and two negative regulators of telomerase recruitment to the telomere, is sufficient to account for the telomeric silencing and telomere shortening phenotypes of upf mutants. While levels of mRNAs that regulate telomerase are altered in upf mutants, the levels of TLC1 RNA and of in vitro telomerase activity are not changed. This implies that the telomere length phenotype of upf mutants is due to changes in the access of telomerase to the telomere rather than to changes in the amount of telomerase activity. For EST1, EST2, EST3, and STN1, the sequences upstream of the ORF are sufficient to confer an NMD-mediated effect upon reporter mRNA levels. The effect of NMD on STN1 and EST3 mRNA levels most likely occurs via transcription initiation, since mRNAs were not stabilized in upf mutant strains. For EST3, this implies that the +1 programmed frameshift in the Est3p coding sequence (34) does not trigger UPF-mediated mRNA decay. We propose that NMD controls the mRNA stability for one or more upstream regulators of STN1 and possibly EST3. Thus, the NMD pathway affects telomere length and telomeric silencing by regulating the levels of EST2, STN1, and TEN1 mRNAs, primarily through an indirect effect on transcription levels.
Acknowledgments
J.N.D. and J.L.-S. contributed equally to this publication.
We thank Sara Johnson and Angela Williams for technical assistance; Audrey Atkin for strain AAY333; M. Charbonneau, A. Dillon, V. Lundblad, and J. Rine for providing plasmids, and Kirk Anders for helpful discussions regarding Ppr1p.
This work was supported by grants from the National Institutes of Health (GM38636 to J.B. and GM65172 to M.R.C.), the National Science Foundation (MCB-9870313 to M.R.C. and MCB-0091300 to J.N.D.), and the American Cancer Society (RPG-99-048-01-GMC to N.L.). A.F. was supported by the McIntyre-Stennis HATCH grant WIS04308 awarded to M.R.C.
REFERENCES
- 1.Altamura, N., O. Groudinsky, G. Dujardin, and P. P. Slonimski. 1992. NAM7 nuclear gene encodes a novel member of a family of helicases with a Zn-ligand motif and is involved in mitochondrial functions in Saccharomyces cerevisiae. J. Mol. Biol. 224:575-587. [DOI] [PubMed] [Google Scholar]
- 2.Atkin, A. L., N. Altamura, P. Leeds, and M. R. Culbertson. 1995. The majority of yeast UPF1 co-localizes with polyribosomes in the cytoplasm. Mol. Biol. Cell 6:611-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atkin, A. L., L. R. Schenkman, M. Eastham, J. N. Dahlseid, M. J. Lelivelt, and M. R. Culbertson. 1997. Relationship between yeast polyribosomes and Upf proteins required for nonsense mRNA decay. J. Biol. Chem. 272:22163-22172. [DOI] [PubMed] [Google Scholar]
- 4.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1993. Current protocols in molecular biology. Greene Publishing Associates and Wiley Interscience, New York, N.Y.
- 5.Boulton, S. J., and S. P. Jackson. 1998. Components of the Ku-dependent non-homologous end-joining pathway are involved in telomeric length maintenance and telomeric silencing. EMBO J. 17:1819-1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chandra, A., T. R. Hughes, C. I. Nugent, and V. Lundblad. 2001. Cdc13 both positively and negatively regulates telomere replication. Genes Dev. 15:404-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Christianson, T. W., R. S. Sikorski, M. Dante, J. H. Shero, and P. Hieter. 1992. Multifunctional yeast high-copy-number shuttle vectors. Gene 110:119-122. [DOI] [PubMed] [Google Scholar]
- 8.Cohn, M., and E. H. Blackburn. 1995. Telomerase in yeast. Science 269:396-400. [DOI] [PubMed] [Google Scholar]
- 9.Culbertson, M. R. 1999. RNA surveillance. Unforeseen consequences for gene expression, inherited genetic disorders and cancer. Trends Genet. 15:74-80. [DOI] [PubMed] [Google Scholar]
- 10.Dillin, A., and J. Rine. 1998. Roles for ORC in M phase and S phase. Science 279:1733-1737. [DOI] [PubMed] [Google Scholar]
- 11.Evans, S. K., and V. Lundblad. 1999. Est1 and Cdc13 as comediators of telomerase access. Science 286:117-120. [DOI] [PubMed] [Google Scholar]
- 12.Evans, S. K., and V. Lundblad. 2000. Positive and negative regulation of telomerase access to the telomere. J. Cell Sci. 113:3357-3364. [DOI] [PubMed] [Google Scholar]
- 13.Evans, S. K., M. L. Sistrunk, C. I. Nugent, and V. Lundblad. 1998. Telomerase, Ku, and telomeric silencing in Saccharomyces cerevisiae. Chromosoma 107:352-358. [DOI] [PubMed] [Google Scholar]
- 14.Fox, C. A., A. E. Ehrenhofer-Murray, S. Loo, and J. Rine. 1997. The origin recognition complex, SIR1, and the S phase requirement for silencing. Science 276:1547-1551. [DOI] [PubMed] [Google Scholar]
- 15.Gietz, R. D., and A. Sugino. 1988. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74:527-534. [DOI] [PubMed] [Google Scholar]
- 16.Gottschling, D. E., O. M. Aparicio, B. L. Billington, and V. A. Zakian. 1990. Position effect at S. cerevisiae telomeres: reversible repression of pol II transcription. Cell 63:751-762. [DOI] [PubMed] [Google Scholar]
- 17.Grandin, N., C. Damon, and M. Charbonneau. 2000. Cdc13 cooperates with the yeast Ku proteins and Stn1 to regulate telomerase recruitment. Mol. Cell. Biol. 20:8397-8408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grandin, N., C. Damon, and M. Charbonneau. 2001. Ten1 functions in telomere end protection and length regulation in association with Stn1 and Cdc13. EMBO J. 20:1173-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grandin, N., S. I. Reed, and M. Charbonneau. 1997. Stn1, a new Saccharomyces cerevisiae protein, is implicated in telomere size regulation in association with Cdc13. Genes Dev. 11:512-527. [DOI] [PubMed] [Google Scholar]
- 20.Laroche, T., S. G. Martin, M. Gotta, H. C. Gorham, F. E. Pryde, E. J. Louis, and S. M. Gasser. 1998. Mutation of yeast Ku genes disrupts the subnuclear organization of telomeres. Curr. Biol. 8:653-656. [DOI] [PubMed] [Google Scholar]
- 21.Leeds, P., S. W. Peltz, A. Jacobson, and M. R. Culbertson. 1991. The product of the yeast UPF1 gene is required for rapid turnover of mRNAs containing a premature translational termination codon. Genes Dev. 5:2303-2314. [DOI] [PubMed] [Google Scholar]
- 22.Leeds, P., J. M. Wood, B. S. Lee, and M. R. Culbertson. 1992. Gene products that promote mRNA turnover in Saccharomyces cerevisiae. Mol. Cell. Biol. 12:2165-2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lelivelt, M. J., and M. R. Culbertson. 1999. Yeast Upf proteins required for RNA surveillance affect global expression of the yeast transcriptome. Mol. Cell. Biol. 19:6710-6719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lendvay, T. S., D. K. Morris, J. Sah, B. Balasubramanian, and V. Lundblad. 1996. Senescence mutants of Saccharomyces cerevisiae with a defect in telomere replication identify three additional EST genes. Genetics 144:1399-1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lew, J. E., S. Enomoto, and J. Berman. 1998. Telomere length regulation and telomeric chromatin require the nonsense-mediated mRNA decay pathway. Mol. Cell. Biol. 18:6121-6130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lingner, J., T. R. Cech, T. R. Hughes, and V. Lundblad. 1997. Three Ever Shorter Telomere (EST) genes are dispensable for in vitro yeast telomerase activity. Proc. Natl. Acad. Sci. USA 94:11190-11195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Longtine, M. S., A. McKenzie III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 28.Loo, S., C. A. Fox, J. Rine, R. Kobayashi, B. Stillman, and S. Bell. 1995. The origin recognition complex in silencing, cell-cycle progression, and DNA replication. Mol. Biol. Cell 6:741-756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Losson, R., R. P. Fuchs, and F. Lacroute. 1985. Yeast promoters URA1 and URA3. Examples of positive control. J. Mol. Biol. 185:65-81. [DOI] [PubMed] [Google Scholar]
- 30.Louis, E. J., and J. E. Haber 1992. The structure and evolution of subtelomeric Y′ repeats in Saccharomyces cerevisiae. Genetics 131:559-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lue, N. F., and R. D. Kornberg. 1987. Accurate initiation at RNA polymerase II promoters in extracts from Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 84:8839-8843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lue, N. F., and Y. Peng. 1998. Negative regulation of yeast telomerase activity through an interaction with an upstream region of the DNA primer. Nucleic Acids Res. 26:1487-1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Melton, D. A., P. A. Krieg, M. R. Rebagliati, T. Maniatis, K. Zinn, and M. R. Green. 1984. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 12:7035-7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morris, D. K., and V. Lundblad. 1997. Programmed translational frameshifting in a gene required for yeast telomere replication. Curr. Biol. 7:969-976. [DOI] [PubMed] [Google Scholar]
- 35.Nugent, C. I., G. Bosco, L. O. Ross, S. K. Evans, A. P. Salinger, J. K. Moore, J. E. Haber, and V. Lundblad. 1998. Telomere maintenance is dependent on activities required for end repair of double-strand breaks. Curr. Biol. 8:657-660. [DOI] [PubMed] [Google Scholar]
- 36.Parker, R., D. Herrick, S. Peltz, and A. Jacobson. 1991. Measurement of mRNA decay rates in Saccharomyces cerevisiae. Methods Enzymol. 194:415-423. [DOI] [PubMed] [Google Scholar]
- 37.Peltz, S. W., A. H. Brown, and A. Jacobson. 1993. mRNA destabilization triggered by premature translational termination depends on at least three cis-acting sequence elements and one trans-acting factor. Genes Dev. 7:1737-1754. [DOI] [PubMed] [Google Scholar]
- 38.Peltz, S. W., and A. Jacobson. 1993. mRNA turnover in Saccharomyces cerevisiae, p. 291-327. In G. Brawerman and J. Belasco (ed.), Control of messenger RNA stability. Academic Press, San Diego, Calif.
- 39.Pennock, E., K. Buckley, and V. Lundblad. 2001. Cdc13 delivers separate complexes to the telomere for end protection and replication. Cell 104:387-396. [DOI] [PubMed] [Google Scholar]
- 40.Peterson, S. E., A. E. Stellwagen, S. J. Diede, M. S. Singer, Z. W. Haimberger, C. O. Johnson, M. Tzoneva, and D. E. Gottschling. 2001. The function of a stem-loop in telomerase RNA is linked to the DNA repair protein Ku. Nat. Genet. 27:64-67. [DOI] [PubMed] [Google Scholar]
- 41.Porter, S. E., P. W. Greenwell, K. B. Ritchie, and T. D. Petes. 1996. The DNA-binding protein Hdf1p (a putative Ku homolog) is required for maintaining normal telomere length in Saccharomyces cerevisiae. Nucleic Acids Res. 24:582-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pulak, R., and P. Anderson. 1993. mRNA surveillance by the Caenorhabditis elegans smg genes. Genes Dev. 7:1885-1897. [DOI] [PubMed] [Google Scholar]
- 43.Qi, H., and V. A. Zakian. 2000. The Saccharomyces telomere-binding protein Cdc13p interacts with both the catalytic subunit of DNA polymerase alpha and the telomerase-associated Est1 protein. Genes Dev. 14:1777-1788. [PMC free article] [PubMed] [Google Scholar]
- 44.Reifsnyder, C., J. Lowell, A. Clarke, and L. Pillus. 1996. Yeast SAS silencing genes and human genes associated with AML and HIV-1 Tat interactions are homologous with acetyltransferases. Nat. Genet. 14:42-49. [DOI] [PubMed] [Google Scholar]
- 45.Roy, A., and R. Losson. 1990. cis- and trans-acting regulatory elements of the yeast URA3 promoter. Mol. Cell. Biol. 10:5257-5270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ruiz-Echevarria, M. J., and S. W. Peltz. 2000. The RNA binding protein Pub1 modulates the stability of transcripts containing upstream open reading frames. Cell 101:741-751. [DOI] [PubMed] [Google Scholar]
- 47.Singer, M. S., and D. E. Gottschling. 1994. TLC1: template RNA component of Saccharomyces cerevisiae telomerase. Science 266:404-409. [DOI] [PubMed] [Google Scholar]
- 48.Snedecor, G. W., and W. G. Cochran. 1980. Statistical methods, 7th ed. Iowa State University Press, Ames.
- 49.Wach, A. 1996. PCR-synthesis of marker cassettes with long flanking homology regions for gene disruptions in S. cerevisiae. Yeast 12:259-265. [DOI] [PubMed] [Google Scholar]
- 50.Welch, E. M., and A. Jacobson. 1999. An internal open reading frame triggers nonsense-mediated decay of the yeast SPT10 mRNA. EMBO J. 18:6134-6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wellinger, R. J., A. J. Wolf, and V. A. Zakian. 1993. Origin activation and formation of single-strand TG1-3 tails occur sequentially in late S phase on a yeast linear plasmid. Mol. Cell. Biol. 13:4057-4065. [DOI] [PMC free article] [PubMed] [Google Scholar]