Abstract
The so far largely uncharacterized central carbon metabolism of the yeast Pichia stipitis was explored in batch and glucose-limited chemostat cultures using metabolic-flux ratio analysis by nuclear magnetic resonance. The concomitantly characterized network of active metabolic pathways was compared to those identified in Saccharomyces cerevisiae, which led to the following conclusions. (i) There is a remarkably low use of the non-oxidative pentose phosphate (PP) pathway for glucose catabolism in S. cerevisiae when compared to P. stipitis batch cultures. (ii) Metabolism of P. stipitis batch cultures is fully respirative, which contrasts with the predominantly respiro-fermentative metabolic state of S. cerevisiae. (iii) Glucose catabolism in chemostat cultures of both yeasts is primarily oxidative. (iv) In both yeasts there is significant in vivo malic enzyme activity during growth on glucose. (v) The amino acid biosynthesis pathways are identical in both yeasts. The present investigation thus demonstrates the power of metabolic-flux ratio analysis for comparative profiling of central carbon metabolism in lower eukaryotes. Although not used for glucose catabolism in batch culture, we demonstrate that the PP pathway in S. cerevisiae has a generally high catabolic capacity by overexpressing the Escherichia coli transhydrogenase UdhA in phosphoglucose isomerase-deficient S. cerevisiae.
The two yeasts Saccharomyces cerevisiae and Pichia stipitis exhibit fundamentally different modes of metabolic regulation in glucose-containing media. At high extracellular concentrations of glucose, one observes simultaneous fermentation and respiration (respiro-fermentative metabolism) in S. cerevisiae at high growth rates even under fully aerobic conditions (11, 34, 53). In Crabtree-positive yeasts, such as S. cerevisiae, elevated glucose concentrations induce the carbon catabolite repression response (16), resulting in low levels of transcription of genes involved in respiration and in the tricarboxylic acid (TCA) cycle. In contrast, the Crabtree-negative P. stipitis exhibits predominantly respirative metabolism even at high glucose concentrations (33). These obvious differences in metabolic regulation between the two yeasts provide motivation for investigation of potential differences in the central carbon pathways. Stable isotope labeling experiments appear to be a promising approach, since it reveals in vivo activity of pathways and reactions (10, 43). Biosynthetically directed fractional (BDF) 13C labeling of amino acids can provide comprehensive insight into central carbon metabolism, yielding a network of active pathways and quantification of intracellular flux ratios (36, 37, 44, 46); its use in the present study promises to expand on the results of earlier labeling studies in yeast that focused on intermediates or products of individual pathways or reactions (14, 17, 20, 22, 41, 57).
BDF 13C labeling is achieved by growing cells on mixtures of unlabeled and uniformly 13C-labeled [U-13C6]glucose (40, 44). Using two-dimensional (2D) nuclear magnetic resonance (NMR) spectroscopy, the multiplet fine structures due to 13C-13C scalar coupling between adjacent carbons in proteinogenic amino acids are analyzed in hydrolysates from such fractionally labeled cells. Decomposition of the fine structures into their multiplet components allows calculation of the relative abundance of contiguous carbon fragments originating from a single source molecule of glucose (44). Such intact carbon fragments are then balanced within a metabolic network to identify the network of active biosynthetic pathways and to determine metabolic-flux ratios (44). Although it was initially conceived for studies of bacterial metabolism, metabolic-flux ratio (METAFoR) analysis has recently been applied also for the analysis of eukaryotic metabolism (27), where the spatial separation of metabolic subnetworks in cytosol and mitochondria as well as the intercompartmental transport fluxes of pyruvate (PYR), acetyl coenzyme A (ACoA), and oxaloacetate (OAA) were taken into consideration.
Here we use METAFoR analysis by NMR for metabolic profiling of S. cerevisiae (27) and the much less well characterized xylose-fermenting yeast P. stipitis in both glucose-limited chemostat and batch culture. The present investigation focuses on the metabolic impact of glucose repression (the Crabtree effect) and pentose phosphate (PP) pathway operation. To gain further insight into the PP pathway, we further investigated phosphoglucose isomerase (Pgi)-deficient S. cerevisiae, which catabolizes glucose exclusively via the PP pathway (3).
MATERIALS AND METHODS
Strains and growth conditions.
The laboratory yeast strains S. cerevisiae CEN.PK 113.7D (MATaMAL2-8c SUC2) and P. stipitis CBS6054 were used for all labeling experiments. The former was obtained from P. Kötter (Institute of Microbiology, Johann Wolfgang Goethe-University, Frankfurt, Germany) and the latter was from B. Hahn-Hägerdal (Department of Applied Microbiology, Lund University, Lund, Sweden). The strains ENY.WA-1A (MATα ura3-52 leu2-3 trp1-289 his3-Δ1 MAL2-8c MAL3 SUC3) and EBY44 (ENY.WA-1A pgi1-1Δ::URA3) (kindly provided by E. Boles, Institut für Mikrobiologie, Heinrich Heine Universität, Düsseldorf, Germany) were used for UdhA overexpression experiments. The soluble Escherichia coli transhydrogenase UdhA (4) was cloned as a BamHI-HindIII fragment from an E. coli overexpression vector (7) under the control of the constitutive, truncated HXT7 promoter of p425HXT7 (LEU2) (21) (provided by E. Boles). The resulting construct, p425-udhA, was transformed in S. cerevisiae ENY.WA-1A and in its Pgi-deficient derivative.
Batch cultures of 100 ml were grown in 1-liter baffled shake flasks on a rotary shaker at 30°C and 300 rpm in yeast minimal medium, which contained 0.5% (wt/vol) glucose and 6.7 g of yeast nitrogen base without amino acids (Difco) per liter. Uracil (50 mg/liter), tryptophan (50 mg/liter), histidine (50 mg/liter), and leucine (250 mg/liter; to avoid hidden Leu limitations [6]) were supplemented where necessary.
Glucose-limited chemostat cultures were grown in a 1.5-liter bioreactor (Bioengineering, Wald, Switzerland) at 30°C with a working volume of 1 liter. The filter-sterilized chemostat medium contained the following components (per liter of distilled water): glucose, 3.6 g; (NH4)2SO4, 5 g; MgSO4 · 7H2O, 0.5 g; KH2PO4, 3 g; 1 ml of vitamin solution (biotin, 0.05 mg/liter; calcium pantothenate, 1 mg/liter; nicotinic acid, 1 mg/liter; inositol, 25 mg/liter; thiamine-HCl, 1 mg/liter; pyridoxine-HCl, 1 mg/liter; para-aminobenzoic acid, 0.2 mg/liter); and 1 ml of trace element solution (EDTA, 15 mg/liter; ZnSO4 · 7H2O, 4.5 mg/liter; CoCl2 · 6H2O, 0.3 mg/liter; MnCl2 · 4H2O, 1 mg/liter; CuSO4 · 5H2O, 0.3 mg/liter; CaCl2 · 2H2O, 4.5 mg/liter; FeSO4 · 7H2O, 3 mg/liter; NaMoO4 · 2H2O; 0.4 mg/liter; H3BO3, 1 mg/liter; KI, 0.1 mg/liter) (51). To prevent foaming, 2 ml of polypropylene glycol 2000 was added (1:10 diluted in H2O) per liter of medium. Anaerobic cultures were supplemented with filter-sterilized Tween 80-ergosterol solution (1.25 ml/liter) that contained 8 g of ergosterol and 336 g of Tween 80 per liter of ethanol. The pH was maintained at 5.5 by the addition of 3 M KOH, and the fermentation volume was kept constant by using a weight-controlled pump. Aerobic conditions were achieved with a constant airflow of 0.5 liters/min and an agitation speed of 1,200 rpm. To establish anaerobic conditions, air was substituted with nitrogen at the same flow rate and the agitation speed was reduced to 600 rpm.
Analytical procedures.
Cell growth was monitored spectrophotometrically by determining the optical density of the cultures at 600 nm. Cellular dry weight (cdw) was determined from six parallel 10-ml culture aliquots that were centrifuged for 20 min in preweighed glass tubes at 3,000 × g, washed once with water, and dried at 90°C for 24 h to a constant weight. Concentrations of glucose and fermentation products in the culture broth were determined with commercial enzymatic kits (Beckman) or by high-performance liquid chromatography. Physiological parameters were calculated as described previously (37).
13C labeling experiments.
In batch culture, BDF 13C-labeling of cellular amino acids was achieved by growth on a 5-g/liter glucose mixture consisting of 90% (wt/wt) unlabeled and 10% (wt/wt) uniformly labeled [U-13C6]glucose (degree of 13C labeling > 99% [wt/wt]; Martek Biosciences Corp., Columbia, Md.). These cultures were inoculated with less than 1% (vol/vol) of a mid-exponential-phase culture in minimal medium so that the presence of unlabeled biomass could be neglected. Labeled biomass aliquots were taken from cultures in the mid-exponential phase at an optical density at 600 nm of 1 (37).
Chemostat cultures in physiological steady state were BDF 13C-labeled by substituting unlabeled glucose in the feed medium with the above-mentioned mixture containing 10% (wt/wt) [U-13C6]glucose. Labeled biomass aliquots were withdrawn after about one culture volume change so that about 60% of the biomass was fractionally 13C-labeled.
Biomass aliquots of about 100 mg (cdw) were harvested from all experiments by centrifugation of the culture broth at 3,000 × g and 4°C for 10 min, washed once with 20 mM Tris-HCl (pH 7.6), centrifuged again at the previous settings, and resuspended in 3 ml of 20 mM Tris-HCl (pH 7.6). After addition of 6 ml of 6 M HCl, hydrolysis was performed in sealed glass tubes at 110°C for 24 h, and the solutions were filtered through 0.2-μm-pore-size filters (Millex-GP; Millipore), lyophilized, and dissolved in 700 μl of 0.1 M 2HCl in 2H2O for the NMR measurements.
NMR spectroscopy and data analysis.
2D proton-detected heteronuclear single-quantum 13C-1H correlation NMR spectroscopy (COSY) was employed to detect aliphatic and aromatic resonances of amino acids at a 1H resonance frequency of 500 MHz on a Bruker DRX500 spectrometer as described previously (37, 44). The overall degree of 13C labeling was determined by integrating resolved 13C satellites in 1D 1H NMR spectra (27, 44). The thus-obtained degree of 13C labeling was in close agreement with the value calculated from the composition of the minimal medium, and was further confirmed by analysis of the 13C scalar coupling fine structure of Leu-β (44).
R. Glaser's program FCAL (version 2.3.0) (46) was used for integration of 13C-13C scalar coupling fine structures observed for the resonances of 48 carbon positions in the amino acids. The relative abundances, f, of intact carbon fragments originating from a single source molecule of glucose were calculated from the relative intensities, I, of the multiplets in the 13C-13C scalar coupling fine (27, 43, 44). These f values (see the appendix for supplementary material) provide information on the metabolic origin of the amino acid precursor molecules in the central metabolism. Tracing intact labeled carbon fragments in these metabolites leads to identification of the active metabolic pathways and to quantification of the ratios of fluxes converging to one particular substrate (45). In cases where only two reactions contribute to one metabolite pool and where one of the contributions is assessed from the NMR data, the remaining fraction of the total pool can be attributed to the competing reaction(s).
Biochemical reaction network for S. cerevisiae and P. stipitis.
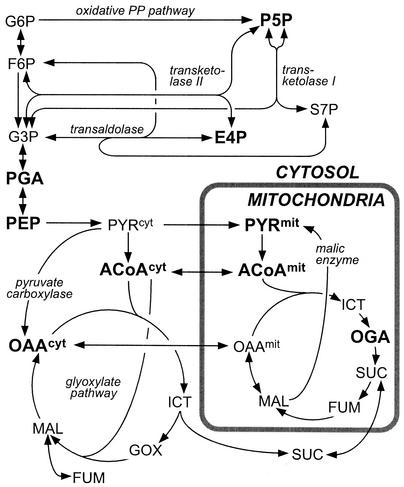
The biochemical reaction network considered for S. cerevisiae growing on glucose in batch culture was recently described (27). Eukaryotic compartmentation into mitochondrial and cytosolic subsystems was included in the model by considering distinct pools of PYR, OAA, and ACoA in both compartments (Fig. 1). Firstly, the transport of PYR into the mitochondria is driven by the proton motive force and was thus considered to be unidirectional (15). Secondly, although likewise proton driven, the transport of OAA into the mitochondria is reversible (27, 32), and intercompartmental exchange between the two OAA pools may additionally occur via shuttle transport mechanisms of TCA cycle intermediates (1, 26, 31). Hence, OAA transport was a priori considered to be bi-directional. Thirdly, mitochondrial transport of ACoA was considered reversible because it is mediated by facilitated diffusion via the so-called carnitine shuttle (48). Although much less is known about central carbon metabolism of P. stipitis compared to that of baker's yeast, we found no evidence in the literature that the metabolic network of S. cerevisiae needs to be modified for P. stipitis. In fact, the extensive body of 13C-labeling data obtained for P. stipitis in the present study was in agreement with the network proposed for S. cerevisiae.
FIG. 1.
Biochemical reaction network for yeast central carbon metabolism. The arrows indicate reaction directionality. Letters in boldface type indicate metabolites for which the 13C-labeling pattern can be accessed through METAFoR analysis. Abbreviations: G6P, glucose-6-phosphate; F6P, fructose-6-phosphate; P5P, pentose-5-phosphates; E4P, erythrose-4-phosphate; S7P, seduheptulose-7-phosphate; G3P, glyceraldehyde-3-phosphate; PGA, 3-phosphoglycerate; ICT, isocitrate; OGA, oxoglutarate; SUC, succinate; MAL, malate; GOX, glyoxylate.
RESULTS
Central metabolism during growth in batch culture.
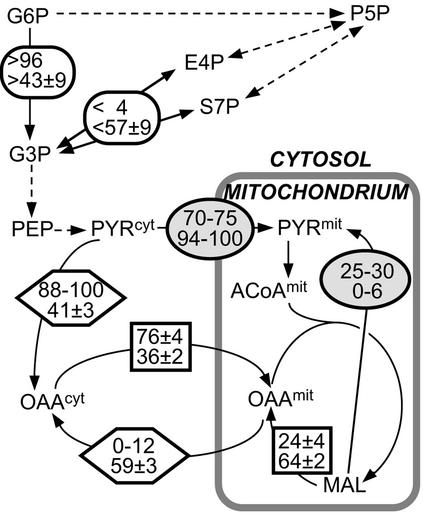
METAFoR analysis provides direct information on the biosynthetic pathways for amino acids (23), which are required to infer the 13C-labeling patterns of metabolic intermediates from those detected in the amino acids. For P. stipitis we obtain the result that amino acids are synthesized along the same pathways as in S. cerevisiae. As a continuation of our previous investigation of S. cerevisiae (27), we first compared the results of this earlier study with the flux ratios in central carbon metabolism of exponentially growing P. stipitis in aerobic batch culture. In these batch cultures with excess glucose concentrations, the maximum specific growth rates were 0.4 and 0.3 h−1 for S. cerevisiae and P. stipitis, respectively. Using BDF 13C-labeling, [13C,1H]-COSY, and METAFoR analysis, we quantified for several intracellular metabolites the fraction of the total pool that was derived from the specified substrates (Table 1). In turn, this quantifies the ratio of the fluxes that converge to a particular metabolite (Fig. 2). In cases where only two reactions contribute to one metabolite pool and where one of contributions is assessed from the NMR data, the remaining fraction of the total pool can be attributed to the competing reaction. Five key differences between the two yeast species in batch culture could be identified.
TABLE 1.
Origins of metabolic intermediates during aerobic exponential growth of P. stipitis and S. cerevisiae batch culturesf
| Metabolite | % Fraction of total pool (mean ± 2.5 SD)
|
|
|---|---|---|
| P. stipitis | S. cerevisiaea | |
| Cytosolic | ||
| PEP derived through at least one TKb (ub) | 57 ± 9 | 0-4 |
| P5P from glucose (lb) | 43 ± 2 | 32 ± 2 |
| P5P from G3P and S7P (TK reaction) | 57 ± 2 | 68 ± 2 |
| P5P from E4P (TK and TAc reactions) | 35 ± 2 | 10 ± 2 |
| E4P from F6P (lb) | 9 ± 6 | 15 ± 4 |
| PEP from OAAcyt (PEP carboxykinase reaction) | <5 | NDd |
| OAAcyt from PYRcyt | 41 ± 3e | 88-100e |
| OAAcyt reversibly converted to FUM at least once (cytosolic or intercompartmental exchange) | 24 ± 10 | 0-4 |
| Mitochondrial | ||
| PYRmit from MAL | <6 | 25-30 |
| OAAmit from PEP (anaplerosis) | 36 ± 2 | 76 ± 4 |
| OAAmit reversibly converted to FUM at least once (in the TCA cycle) | 58 ± 12 | 35 ± 4 |
Data taken from Maaheimo et al. (27).
TK, transketolase.
TA, transaldolase.
ND, not detectable because the fragment needed for tracing this activity is absent.
Values calculated assuming absence of cytosolic OAA-to-FUM conversion.
The experimental error was estimated from the analysis of redundant 13C-13C scalar coupling fine structures and the signal-to-noise ratio of the [13C,1H]-COSY spectra employing the Gaussian law of error propagation. In certain cases, the NMR data permit only the determination of upper (ub) or lower (lb) bounds on the origin of metabolites. Abbreviations are explained in the legend to Fig. 1.
FIG. 2.
Metabolic-flux ratios in S. cerevisiae (top values in the insets) and P. stipitis (bottom values) during batch growth with glucose as the sole carbon source. Connected ratios of fluxes that converge to a particular metabolite are presented in identical geometrical shapes. For abbreviations see the legend to Fig. 1.
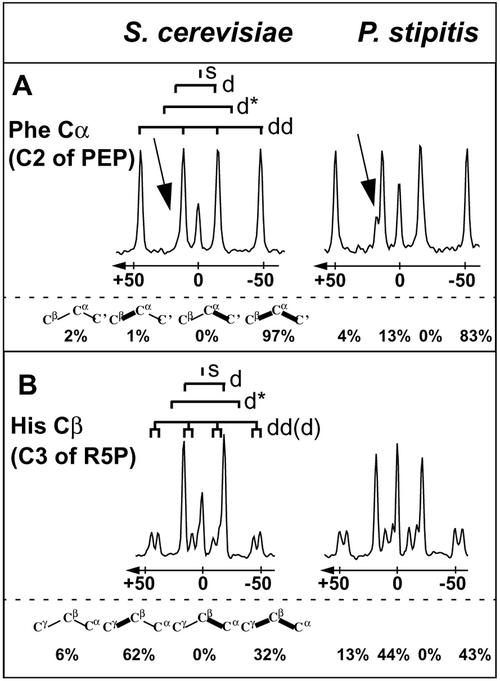
First, the upper bound on the fraction of phosphoenolpyruvate (PEP) molecules derived through at least one transketolase reaction was widely different (Table 1). This value provides information about the relative contributions of the PP pathway and the glucose catabolism via glycolysis to the generation of trioses. Only an upper bound for the relative catabolic activity of the PP pathway can be obtained because glycolytically derived G3P may be exchanged with intermediates of the nonoxidative PP pathway via the transketolase reaction. This exchange reaction generates the same intact fragments in PEP as glucose catabolism through the PP pathway. In P. stipitis, the upper bound on the relative PP pathway flux is less than 57% (Table 1), which is well in the range of values typically seen in prokaryotic cultures (15, 36, 37). In S. cerevisiae, in contrast, PEP synthesis from pentoses was not observed, which demonstrates that glucose catabolism proceeds almost exclusively through glycolysis (Fig. 2) (27). This key difference in glucose catabolism between the two yeast species becomes readily apparent by visual inspection of the 13C-13C scalar coupling fine structures of Phe Cα, which reflects the labeling pattern of PEP (Fig. 3A). The doublet component with the smaller coupling constant (Fig. 3A) corresponds to PEP molecules in which the C1 carbon and the C2-C3 fragment originate from different glucose source molecules, and its presence thus provides evidence for the contribution of transketolase to glucose catabolism. Despite the absence of glucose catabolism through the PP pathway in the batch culture of S. cerevisiae (27), evidence was obtained for significant anabolic flux via the oxidative PP pathway to the formation of the biomass precursor P5P: more than 32% of the P5P molecules originate directly from glucose (Table 1) (27). This anabolic activity of the PP pathway is directly apparent in the cross-sections of His Cβ, where an additional doublet splitting of the doublet of doublets (Fig. 3B) documents the presence of intact C5-fragments from glucose in the pool of pentoses (43, 44). Consistent with the putative increase of PP activity in P. stipitis, the minimal fraction of P5P derived from glucose is 43% (Table 1).
FIG. 3.
13C-13C scalar coupling fine structures along ω1 (13C) of a 2D [13C,1H]-COSY spectrum at the resonance frequency of Phe Cα (A) and His Cβ (B). The spectra were recorded with amino acids obtained from exponentially growing aerobic S. cerevisiae (left panels) and P. stipitis (right panels) batch cultures. The corresponding carbon atom in the precursor metabolites is given in parentheses. Carbon fragments that can be inferred primarily from a given multiplet components are depicted below the left cross-section. The arrow (A) indicates the multiplet component used to trace the generation of PEP from the PP pathway (see text). Preserved bonds from the source molecule are shown in bold. The relative abundances of the various carbon fragments calculated from integration of the fine structures (44, 46) are indicated below each spectrum. Abbreviations: s, singlet; d and d*, doublets with different coupling constants; dd, doublet of doublets.
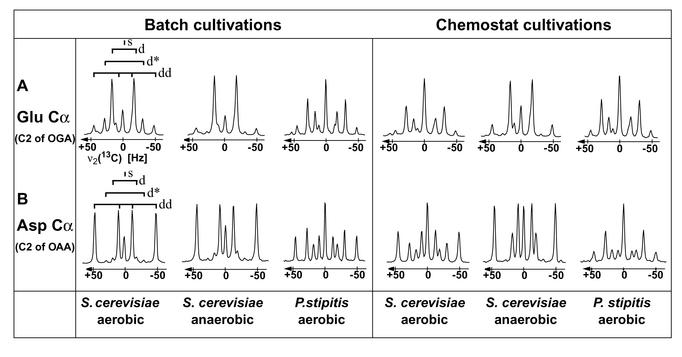
Second, the fraction of OAAmit (where the superscript indicates mitochondrial localization of the metabolite pool) that is derived from PEP via the anaplerotic PYR carboxylase reaction differs by a factor of 2 in the two yeasts: anaplerosis contributes 70% in S. cerevisiae but only 36% in P. stipitis to OAAmit synthesis (Table 1). This value specifies the relative flux to OAAmit required to satisfy the biosynthetic demands of the TCA cycle. The competing flux from ACoAmit to OAAmit is used for complete oxidation of carbon substrates to CO2 within the TCA cycle. In yeast, PYR carboxylase is localized exclusively in the cytosol (49). Following (27), anaplerosis is calculated from the fraction of OAAmit derived from PEP (Fig. 1), since the PYRcyt (where the superscript indicates cytosolic localization of the metabolite pool) pool is not directly assessable by BDF labeling because PYR-derived amino acids are synthesized from the PYRmit pool. We conclude that in P. stipitis the TCA cycle operates predominantly for respiration (Fig. 2). The 13C-13C scalar coupling fine structures of Glu Cα provide direct evidence for this salient difference (Fig. 4A, left panel). The higher contribution of the anaplerotic reaction to glutamate synthesis in S. cerevisiae is reflected by the higher intensity of the doublet with the smaller coupling constant and the doublet of doublets arising predominantly from carboxylation of triply 13C-labeled PYR (44).
FIG. 4.
13C-13C scalar coupling fine structures along ω1 (13C) of a 2D [13C,1H]-COSY spectrum at the resonance frequency of Glu Cα (A) and Asp Cα (B). The spectra were recorded with amino acids obtained from batch and chemostat cultures of S. cerevisiae and P. stipitis. The corresponding carbon atom in the precursor metabolites is given in parentheses. As indicated in the left-most cross-sections, all these multiplets consist of a singlet (s) representing the 12C′-13Cα-12Cβ isotopomer, a doublet (d) originating from 12C′-13Cα-13Cβ, a second doublet (d*) for 13C′-13Cα-12Cβ, and a doublet of doublets (dd) arising from 13C′-13Cα-13Cβ.
Third, while synthesis of OAAcyt, which is required for cytosolic biosynthesis of the Asp amino acid family and anaplerosis of the TCA cycle, originates nearly exclusively from PEP in S. cerevisiae (27), less than half of OAAcyt originates from PEP in P. stipitis (Table 1; Fig. 2). This becomes readily apparent when comparing the 13C-13C scalar coupling fine structures of Asp Cα (Fig. 4B, left panel). Specifically, the large doublet of doublets in the S. cerevisiae cross section demonstrates that a high proportion of intact C3-fragments arise from carboxylation of PYR. On the other hand, the doublet d in Fig. 4B documents that most of the C2-C3 bonds in OAAcyt of P. stipitis were newly formed. This is characteristic for OAAcyt synthesized either from oxoglutarate in the TCA cycle or through the glyoxylate pathway.
Fourth, the fraction of cytosolic or mitochondrial OAA that was once reversibly converted to fumarate (FUM) is much higher for P. stipitis than for S. cerevisiae (Table 1). For OAAmit, this conversion occurs via malate dehydrogenase and fumarase in the TCA cycle. The increased exchange flux might relate to a predominantly respirative metabolism in P. stipitis. The reversible conversion of OAAcyt, however, may occur either in the cytosol via the glyoxylate pathway enzyme MAL dehydrogenase in combination with the cytosolic isoform of fumarase (28) or by intercompartmental exchange of OAA molecules (Fig. 1). The small extent of reversible conversion of OAAcyt to FUM in S. cerevisiae thus demonstrates the absence of glyoxylate shunt activity and indicates that the flux of OAAcyt to OAAmit is unidirectional (27). In P. stipitis, however, about 20% of the OAAcyt molecules are converted to FUM at least once, and thus one or both of the two above-mentioned flux scenarios must be invoked.
Fifth, although malic enzyme contributes significantly to mitochondrial PYR biosynthesis in batch cultures of S. cerevisiae (27), malic enzyme activity is low in P. stipitis (Table 1). The manifestation of this reaction in the labeling pattern of PYRmit would be consistent with mitochondrial localization of the malic enzyme not only in S. cerevisiae (2, 27) but in both yeasts.
Central metabolism during growth in glucose-limited chemostat cultures.
While catabolic fluxes are maximal during exponential growth in batch cultures, growth and metabolic fluxes are reduced in chemostat cultures by the limited availability of glucose. Flux patterns obtained from these different types of growth conditions have been shown to differ significantly in most microbes (15, 18, 37). To investigate flux responses to glucose limitation under conditions when no Crabtree effect is active, we cultivated both yeasts in aerobic chemostat cultures at a dilution rate of 0.1 h−1. P. stipitis grew somewhat more efficiently, as evidenced by biomass yields per g of glucose consumed (means ± standard deviations) of 0.45 ± 0.03 g and 0.38 ± 0.02 g (cdw) for P. stipitis and S. cerevisiae, respectively. For both yeasts the growth was predominantly respirative, with little ethanol formation (data not shown). When the S. cerevisiae culture was switched to anaerobic conditions (note that P. stipitis does not grow under such conditions), metabolism became entirely fermentative, with a strongly reduced biomass yield (mean ± standard deviation) of 0.09 ± 0.00 g (cdw) per g of glucose and ethanol as the primary metabolic product. Glycerol, acetate, succinate, and PYR were also found in appreciable amounts in the anaerobic culture (data not shown). METAFoR analyses were performed with hydrolyzed biomass samples that were harvested from these chemostat cultures in physiological steady-state.
In aerobic chemostats, the upper bound of PEP molecules that were derived through at least one transketolase reaction was higher in P. stipitis than in S. cerevisiae (Table 2), but for S. cerevisiae in both aerobic and anaerobic growth it was much higher than previously described for batch cultures of (27) (compare Tables 1 and 2). Thus, the difference in catabolic PP pathway usage is much less pronounced under glucose-limited growth conditions than in batch cultures because the chemostat cultures grew at only about one third of the maximum specific growth rate. Under anaerobic conditions, the fraction of PEP derived through at least one transketolase reaction is significantly reduced (Table 2), which indicates reduced catabolic use of the PP pathway, as was shown previously also for anaerobic batch cultures (27).
TABLE 2.
Origins of metabolic intermediates during growth of P. stipitis and S. cerevisiae in glucose-limited chemostat cultures at a dilution rate of 0.1 h−1c
| Metabolite | % Fraction of total pool (mean ± SD)
|
||
|---|---|---|---|
| P. stipitis (aerobic) |
S. cerevisiae
|
||
| Aerobic | Anaerobic | ||
| Cytosolic | |||
| PEP derived through at least one TK (ub) | 61 ± 11 | 40 ± 8 | 8 ± 5 |
| P5P from glucose (lb) | 28 ± 2 | 41 ± 2 | 11 ± 2 |
| P5P from G3P and S7P (TK reaction) | 72 ± 2 | 59 ± 2 | 89 ± 2 |
| P5P from E4P (TK and TA reactions) | 43 ± 2 | 33 ± 2 | 15 ± 2 |
| E4P from F6P (lb) | 27 ± 5 | 6 ± 6 | 44 ± 4 |
| PEP from OAAcyt (PEP carboxykinase reaction) | 0-3 | 2 ± 8 | NDa |
| OAAcyt from PYRcyt | 24 ± 3b | 62 ± 4b | 40 ± 10b |
| OAAcyt reversibly converted to FUM at least once (cytosolic or intercompartmental exchange) | 47 ± 16 | 0-8 | 37 ± 7 |
| Mitochondrial | |||
| PYRmit from MAL | <7 | <13 | >10 |
| OAAmit from PEP (anaplerosis) | 32 ± 2 | 31 ± 2 | 98 ± 2 |
| OAAmit reversibly converted to FUM at least once (in the TCA cycle) | 58 ± 14 | 56 ± 14 | 43 ± 4 |
During aerobic chemostat growth, both yeasts exhibit a comparable contribution of about 30% from the anaplerotic reaction to the synthesis of OAAmit (OAAmit from PEP in Table 2), while the remaining OAAmit originates from the TCA cycle. This flux partitioning is very similar to the value obtained for the P. stipitis batch culture, but it is in contrast to the values obtained for the S. cerevisiae batch culture (Table 1). Thus, P. stipitis appears to exhibit predominantly respirative metabolism in both glucose-limited chemostat and batch culture, whereas S. cerevisiae metabolism is respirative in the chemostat but respiro-fermentative in the batch culture, which is consistent with the physiological data. In anaerobic chemostat culture, the TCA cycle operates through two branches, which both serve exclusively anabolic functions (Table 2). As described above for anaplerosis, these flux responses are readily apparent by inspection of the Glu Cα cross-sections (Fig. 4A, right panel).
In contrast to the S. cerevisiae batch culture (Table 1), OAAcyt is not derived exclusively from PYRcyt in the chemostat cultures (Table 2). As discussed above for the batch cultures, this observation could be explained either by significant exchange between the OAAcyt and OAAmit pools, by high activity of the glyoxylate pathway, or by a combination of these two factors. Consistently, a significant fraction of OAAcyt was reversibly converted at least once to FUM in aerobic P. stipitis and anaerobic S. cerevisiae cultures (Table 2). Furthermore, the data indicate that a small fraction of PYRmit originates from MAL (Tables 1 and 2), thus demonstrating activity of the malic enzyme in S. cerevisiae and P. stipitis chemostat cultures. The relative contribution of malic enzyme to the synthesis of PYRmit in S. cerevisiae is, however, significantly reduced in the chemostat culture when compared with the batch culture (27) (Tables 1 and 2). The malic enzyme activity in aerobic and anaerobic S. cerevisiae cultures is comparable (Table 2).
Transhydrogenase expression compensates for Pgi knockout in S. cerevisiae.
The flux ratios in batch culture might suggest that S. cerevisiae has relatively low catabolic capacity of the PP pathway, which would be consistent with the apparent difference in pentose utilization by the two yeasts considered here (20, 25, 39, 52). To address this hypothesis, we used a Pgi-deficient S. cerevisiae strain which catabolizes glucose exclusively via the PP pathway. Since S. cerevisiae does not contain a transhydrogenase that could transfer electrons from NADPH to NAD+ (47), this mutant cannot reoxidize PP pathway-produced NADPH and, consequently, it cannot grow on glucose as the sole carbon source (3). To enable recycling of NADPH, we overexpressed the soluble transhydrogenase UdhA from E. coli (4, 7) in this Pgi mutant. As expected, the resulting Pgi− UdhA+ strain grows aerobically on glucose with a specific growth rate of 0.15 h−1, which is half the rate observed for the control strain with a functional Pgi. Hence, S. cerevisiae is fully capable of exclusive glucose catabolism via the PP pathway at a relatively high metabolic rate, provided that concomitantly produced NADPH is recycled.
DISCUSSION
Here, we report a metabolic network analysis of P. stipitis central carbon metabolism, which shows that S. cerevisiae network topology (27) allows consistent data interpretation also for P. stipitis. The following key factors were identified for the flux distributions: (i) There is significant in vivo activity of the nonoxidative PP pathway. (ii) There is evidence for PYR carboxylase activity in the anaplerotic reaction. (iii) There must be intercompartmental exchange of OAA if one assumes that the glyoxylate pathway is inactive. (iv) There is evidence for malic enzyme activity in the mitochondria, as was also reported for S. cerevisiae (2, 27). Furthermore, there was no evidence in the aerobic regime for activity of the gluconeogenic PEP carboxykinase in either of the two yeasts, which is consistent with the reported glucose repression of the PEP carboxykinase-encoding gene in S. cerevisiae (13, 35, 42). The present data thus suggest that a similar regulation operates in P. stipitis, and that the pathways for amino acid biosynthesis in P. stipitis are identical to those used in S. cerevisiae.
Overall, comparative flux profiling by METAFoR analysis revealed distinctly different metabolic regimes for cultures growing either in batch or in glucose-limited chemostat mode. Metabolic differences between the two yeast species were minor in glucose-limited chemostats, where cultures exhibited a predominantly respirative mode of metabolism, but were significant in batch culture. First, in batch culture, S. cerevisiae exhibited only 24% cyclic (respiratory) TCA cycle flux (from oxoglutarate) compared to 76% anaplerotic flux (from OAAcyt) into the OAAmit pool of the TCA cycle, but the inverse ratio for these two fluxes was found for P. stipitis (Fig. 2), as was also reported for other yeasts (29). This flux ratio is consistent with the generally held view that sugar catabolism in aerobic batch cultures is respiro-fermentative in S. cerevisiae (53), and predominantly respirative in Crabtree-negative yeasts such as P. stipitis (33). Second, during respiro-fermentative metabolism of S. cerevisiae in batch culture, cytosolic OAA originates exclusively from PYR and is transported unidirectionally into the mitochondria (27). The predominantly respirative metabolism in all other aerobic cultures, however, generates the OAAcyt pool containing significant fractions of molecules that do not originate from PYRcyt but come either from intercompartmental exchange of TCA cycle intermediates or from the cytosolic glyoxylate pathway. Third, although some in vivo activity of the mitochondrial malic enzyme is detected in all cultures, the highest relative contribution to PYRmit synthesis was observed for respiro-fermentative S. cerevisiae in the batch culture. Fourth, our results reveal a major difference in the contribution of the PP pathway to glucose catabolism in batch cultures of P. stipitis and S. cerevisiae. The fraction of PEP molecules derived through at least one transketolase provides the sum of the net flux through the PP pathway and the reversible exchange of trioses with intermediates of the nonoxidative PP pathway. This value is close to zero in the S. cerevisiae batch culture (27), indicating absence of significant in vivo activity of the nonoxidative PP pathway branch in S. cerevisiae batch culture, which is thus different from P. stipitis batch culture (Table 1; Fig. 2).
From this exceptionally low catabolic PP pathway flux in S. cerevisiae one might conclude that the in vivo activity of this pathway limits catabolism of pentoses in recombinant S. cerevisiae (20, 25, 52), but this appears to be incompatible with the comparatively rapid growth of Pgi-deficient, transhydrogenase-overexpressing S. cerevisiae. Since it depends on exclusive glucose catabolism via the PP pathway, this strain grows at a specific rate that is half of that observed with the control. As a reference, a corresponding E. coli strain grew at about one third of the wild type rate (7). Thus, sufficient PP pathway capacity is potentially available in S. cerevisiae, although it might not be fully activated during pentose catabolism. The conclusion that the nonoxidative pathway does not principally limited pentose catabolism in recombinant S. cerevisiae agrees well with the results of a recent study to simultaneous overexpress all four pathway enzymes (24). Our results, however, do not exclude the possibility that the rate of pentose catabolism is affected by the activity of one or more PP pathway enzymes. Like in E. coli, our results reveal that UdhA catalyzes the electron transfer from NADPH to NAD+, as was observed previously during overexpression of another soluble transhydrogenase in S. cerevisiae (30).
Using a different methodology, Gombert et al. recently reported on network identification and flux quantification in the same S. cerevisiae strain that was used here (18), leading to a mutual identity of the metabolic network models in both cases. To facilitate direct comparison between the results of both studies, we calculated several metabolic-flux ratios from the net flux data of Gombert et al. (18) (Table 3). The flux ratios obtained with the two different approaches are in good agreement; i.e., both reveal increased glucose catabolism via the PP pathway and reduced anaplerosis in glucose-limited chemostat culture when compared to batch culture. Both cultures show some malic enzyme activity and very little if any glycine cleavage to CO2 (data not shown). In particular, the consistent flux distributions obtained for glucose-limited S. cerevisiae by these two and other methods support that both techniques provide reliable results (8, 9, 18).
TABLE 3.
Comparison of the flux ratios in S. cerevisiae batch and chemostat cultures determined by metabolic flux balancing (18) and by METAFoR analysis (Tables 1 and 2)
| Pathway and metabolite | % Fraction of total pool (mean ± SD)
|
|||
|---|---|---|---|---|
| Batch culture
|
Chemostat culture
|
|||
| Flux balancing | METAFoR analysis | Flux balancing | METAFoR analysis | |
| Glucose catabolism through the PP pathway (PEP derived through at least one TK reaction) | 14a | 0-4 | 52a | <40 ± 8 |
| Anaplerosis | ||||
| OAAmit from PEP | 100 | 76 ± 4 | 42 | 31 ± 2 |
| OAAcyt from PYRcyt | 82 | 88-100 | 60 | 62 ± 4 |
| Malic enzyme (PYRmit from MAL) | 2 | 28-42 | 5 | 2-13 |
Note that Gombert et al. estimated the split ratio of glycolysis to PP pathway. To compare this value to the value of PEP via TK, one needs to subtract the biosynthetic withdrawal of PP pathway intermediates.
Three apparent differences between the results obtained with the method used by Gombert et al. and with our approach concern primarily the batch growth of S. cerevisiae. First, Gombert et al. report exclusive operation of the TCA cycle as a two-branched pathway in batch culture that does not contribute to glucose catabolism, while we found previously that cyclic TCA cycle fluxes contribute about 24% of the total flux to OAAmit (27). Since the absolute flux to OAAmit is relatively low in batch culture, this minor net TCA cycle flux may have been missed by the flux estimation procedure due to its small contribution to the global target function of the isotopomer balancing. Second, our results suggest a significant contribution of malic enzyme to PYRmit synthesis in batch culture (27), where this activity is not reported by Gombert et al. (18). Similar to the above case, absolute fluxes to PYRmit are low under these respiro-fermentative conditions, so that the higher relative contribution of malic enzyme to PYRmit synthesis not necessarily reflect a higher absolute flux. Third, our data show that the catabolic flux through the PP pathway in batch cultures of S. cerevisiae contributes less than 4% to PEP biosynthesis (27), compared to an estimate of 14% calculated from the data of Gombert et al. (Table 3). Both estimates are consistent with the calculated minimal flux into the PP pathway of 2% of the total glucose consumption for NADPH formation with ammonia as the sole nitrogen source (5, 47).
Global interpretation of 13C-labeling and physiological data on the basis of isotopomer balancing (12, 38, 50, 56) as used by Gombert et al. extracts the maximum information from labeling experiments, but local features of the flux distribution may be inaccurately reflected as a result of the global error minimization function used. METAFoR analysis differs from this approach by providing a strictly local analysis of 13C-labeling data, which is independent of absolute fluxes and physiological data (15). Additionally, variations in the growth conditions used (i.e., 0.5 units pH difference, different composition of the minimal medium, and bioreactor versus shake flask cultivation) may have led to slightly different flux distributions. A possible dependence of catabolic PP pathway fluxes on environmental conditions may also explain, at least in part, previously reported differences in PP pathway flux estimates for S. cerevisiae on the basis of isotope labeling studies (17, 18, 27, 54; P. M. Bruinenberg, G. W. Waslander, J. P. van Dijken, and W. A. Scheffers, unpublished data [presented at the Physiological and genetic modulation of product formation, Como, 1986]). Overall, the combined use of global isotopomer balancing and local METAFoR analysis promises to further enhance the reliability of future flux determinations (10, 15, 55).
Acknowledgments
We thank Jay Bailey for his continuous support and Eckhard Boles for sharing the Pgi mutant and the HXT promoter with us.
Financial support for this work was partly obtained from the Swiss Priority Program in Biotechnology.
APPENDIX
The supplementary material mentioned in Materials and Methods is presented in Tables A1 and A2.
TABLE A1.
Relative abundances of intact C2 and C3 fragments in the biosynthesis of fractionally 13C-labeled amino acids for chemostat cultivations of S. cerevisiae and P. stipitisa
| Carbon position | Chemostat | Relative abundance
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
|
S. cerevisiae
|
P. stipitis
|
||||||||
| f(1) | f(2) | f(2∗) | f(3) | f(1) | f(2) | f(2∗) | f(3) | ||
| Ala Cα | Aerobic | 0.04 | 0.11 | 0.04 | 0.81 | 0.01 | 0.15 | 0.03 | 0.81 |
| Anaerobic | 0.00 | 0.07 | 0.00 | 0.94 | |||||
| Ala Cβ | Aerobic | 0.08 | 0.92 | 0.06 | 0.94 | ||||
| Anaerobic | 0.01 | 0.99 | |||||||
| Arg Cβ | Aerobic | 0.66 | 0.32 | 0.00 | 0.02 | 0.63 | 0.00 | 0.33 | 0.04 |
| Anaerobic | 0.07 | 0.93 | 0.00 | 0.00 | |||||
| Arg Cδ | Aerobic | 0.14 | 0.86 | 0.09 | 0.91 | ||||
| Anaerobic | 0.26 | 0.74 | |||||||
| Asp Cα | Aerobic | 0.14 | 0.10 | 0.19 | 0.57 | 0.24 | 0.14 | 0.42 | 0.20 |
| Anaerobic | 0.01 | 0.22 | 0.01 | 0.76 | |||||
| Asp Cβ | Aerobic | 0.17 | 0.65 | 0.16 | 0.02 | 0.26 | 0.25 | 0.41 | 0.08 |
| Anaerobic | 0.01 | 0.79 | 0.00 | 0.20 | |||||
| Glu Cα | Aerobic | 0.28 | 0.22 | 0.42 | 0.08 | 0.24 | 0.22 | 0.45 | 0.09 |
| Anaerobic | 0.02 | 0.75 | 0.03 | 0.20 | |||||
| Glu Cβ | Aerobic | 0.69 | 0.31 | 0.00 | 0.68 | 0.32 | 0.00 | ||
| Anaerobic | 0.00 | 1.00 | 0.00 | ||||||
| Glu Cγ | Aerobic | 0.08 | 0.00 | 0.92 | 0.00 | 0.02 | 0.00 | 0.98 | 0.00 |
| Anaerobic | 0.14 | 0.00 | 0.86 | 0.00 | |||||
| Gly Cα | Aerobic | 0.19 | 0.81 | 0.18 | 0.82 | ||||
| Anaerobic | 0.10 | 0.90 | |||||||
| His Cα | Aerobic | 0.04 | 0.02 | 0.04 | 0.90 | 0.03 | 0.00 | 0.02 | 0.95 |
| Anaerobic | 0.02 | 0.01 | 0.00 | 0.97 | |||||
| His Cβ | Aerobic | 0.10 | 0.49 | 0.00 | 0.41 | 0.12 | 0.60 | 0.00 | 0.28 |
| Anaerobic | 0.07 | 0.82 | 0.00 | 0.11 | |||||
| His Cδ2 | Aerobic | 0.32 | 0.68 | 0.43 | 0.57 | ||||
| Anaerobic | 0.15 | 0.85 | |||||||
| Ile Cα | Aerobic | 0.25 | 0.00 | 0.75 | 0.00 | 0.37 | 0.01 | 0.62 | 0.00 |
| Anaerobic | 0.23 | 0.00 | 0.77 | 0.00 | |||||
| Ile Cγ2 | Aerobic | 0.12 | 0.88 | 0.06 | 0.94 | ||||
| Anaerobic | 0.03 | 0.97 | |||||||
| Ile Cγ1 | Aerobic | 0.82 | 0.18 | 0.00 | 0.52 | 0.48 | 0.00 | ||
| Anaerobic | 0.82 | 0.18 | 0.00 | ||||||
| Ile Cδ1 | Aerobic | 0.84 | 0.16 | 0.53 | 0.47 | ||||
| Anaerobic | 0.91 | 0.09 | |||||||
| Leu Cα | Aerobic | 0.10 | 0.00 | 0.90 | 0.00 | 0.05 | 0.00 | 0.95 | 0.00 |
| Anaerobic | 0.11 | 0.00 | 0.89 | 0.00 | |||||
| Leu Cβ | Aerobic | 0.95 | 0.01 | 0.04 | 1.00 | 0.00 | 0.00 | ||
| Anaerobic | 1.00 | 0.00 | 0.00 | ||||||
| Leu Cδ1 | Aerobic | 0.24 | 0.76 | 0.17 | 0.83 | ||||
| Anaerobic | 0.13 | 0.87 | |||||||
| Leu Cδ2 | Aerobic | 1.00 | 0.00 | 1.00 | 0.00 | ||||
| Anaerobic | 1.00 | 0.00 | |||||||
| Lys Cα | Aerobic | 0.07 | 0.05 | 0.85 | 0.03 | 0.06 | 0.03 | 0.91 | 0.00 |
| Anaerobic | 0.18 | 0.00 | 0.82 | 0.00 | |||||
| Lys Cβ | Aerobic | 0.70 | 0.30 | 0.00 | 0.70 | 0.29 | 0.01 | ||
| Anaerobic | 0.03 | 0.97 | 0.00 | ||||||
| Lys Cγ | Aerobic | 0.66 | 0.34 | 0.00 | 0.68 | 0.32 | 0.00 | ||
| Anaerobic | 0.04 | 0.96 | 0.00 | ||||||
| Lys Cδ | Aerobic | 0.11 | 0.79 | 0.10 | 0.05 | 0.95 | 0.00 | ||
| Anaerobic | 0.22 | 0.77 | 0.01 | ||||||
| Lys Cɛ | Aerobic | 0.14 | 0.86 | 0.07 | 0.93 | ||||
| Anaerobic | 0.24 | 0.76 | |||||||
| Met Cα | Aerobic | 0.14 | 0.12 | 0.19 | 0.55 | 0.20 | 0.22 | 0.32 | 0.26 |
| Anaerobic | 0.03 | 0.20 | 0.02 | 0.75 | |||||
| Phe Cα | Aerobic | 0.04 | 0.09 | 0.01 | 0.86 | 0.04 | 0.10 | 0.00 | 0.86 |
| Anaerobic | 0.03 | 0.02 | 0.00 | 0.95 | |||||
| Phe Cβ | Aerobic | 0.03 | 0.97 | 0.00 | 0.00 | 0.02 | 0.98 | 0.00 | 0.00 |
| Anaerobic | 0.02 | 0.98 | 0.00 | 0.00 | |||||
| Pro Cδ | Aerobic | 0.12 | 0.88 | 0.09 | 0.91 | ||||
| Anaerobic | 0.22 | 0.78 | |||||||
| Pro Cα | Aerobic | 0.30 | 0.22 | 0.43 | 0.05 | 0.27 | 0.28 | 0.33 | 0.12 |
| Anaerobic | 0.07 | 0.75 | 0.00 | 0.18 | |||||
| Pro Cβ | Aerobic | 0.66 | 0.34 | 0.00 | 0.67 | 0.33 | 0.00 | ||
| Anaerobic | 0.02 | 0.18 | 0.80 | ||||||
| Pro Cγ | Aerobic | 0.09 | 0.88 | 0.03 | 0.06 | 0.91 | 0.03 | ||
| Anaerobic | 0.20 | 0.76 | 0.04 | ||||||
| Ser Cα | Aerobic | 0.06 | 0.07 | 0.30 | 0.57 | 0.05 | 0.08 | 0.23 | 0.64 |
| Anaerobic | 0.02 | 0.02 | 0.34 | 0.62 | |||||
| Ser Cβ | Aerobic | 0.39 | 0.61 | 0.30 | 0.70 | ||||
| Anaerobic | 0.39 | 0.61 | |||||||
| Thr Cα | Aerobic | 0.15 | 0.11 | 0.18 | 0.56 | 0.25 | 0.14 | 0.41 | 0.20 |
| Anaerobic | 0.02 | 0.23 | 0.02 | 0.73 | |||||
| Thr Cβ | Aerobic | 0.17 | 0.41 | 0.42 | 0.27 | 0.68 | 0.05 | ||
| Anaerobic | 0.04 | 0.82 | 0.14 | ||||||
| Thr Cγ2 | Aerobic | 0.83 | 0.17 | 0.50 | 0.50 | ||||
| Anaerobic | 0.83 | 0.17 | |||||||
| Tyr Cα | Aerobic | 0.05 | 0.10 | 0.00 | 0.85 | 0.04 | 0.12 | 0.00 | 0.84 |
| Anaerobic | 0.03 | 0.03 | 0.00 | 0.94 | |||||
| Tyr Cβ | Aerobic | 0.034 | 0.96 | 0.00 | 0.00 | 0.05 | 0.95 | 0.00 | 0.00 |
| Anaerobic | 0.03 | 0.97 | 0.00 | 0.00 | |||||
| Tyr Cδ1 | Aerobic | 0.05 | 0.95 | 0.00 | 0.04 | 0.96 | 0.00 | ||
| Anaerobic | 0.04 | 0.96 | 0.00 | ||||||
| Tyr Cɛ1 | Aerobic | 0.31 | 0.08 | 0.18 | 0.43 | 0.24 | 0.05 | 0.16 | 0.55 |
| Anaerobic | 0.29 | 0.00 | 0.27 | 0.44 | |||||
| Val Cα | Aerobic | 0.14 | 0.00 | 0.83 | 0.03 | 0.12 | 0.01 | 0.83 | 0.04 |
| Anaerobic | 0.10 | 0.00 | 0.90 | 0.00 | |||||
| Val Cγ1 | Aerobic | 0.11 | 0.89 | 0.06 | 0.94 | ||||
| Anaerobic | 0.02 | 0.98 | |||||||
| Val Cγ2 | Aerobic | 0.97 | 0.03 | 0.96 | 0.04 | ||||
| Anaerobic | 0.96 | 0.04 | |||||||
The fractions f(i), named according to reference 44 and corresponding to the relative amount of Ci intact fragments, were calculated from the relative intensities of the 13C multiplet components using the probabilistic equations described by Szyperski (44). Note that in reference 44 f(2a) and f(2b) correspond to f(2) and f(2∗), respectively.
TABLE A2.
Relative abundances of intact C2 and C3 fragments in the biosynthesis of fractionally 13C-labeled amino acids for batch cultivations of S. cerevisiae and P. stipitisa
| Carbon positionb | Relative abundance
|
|||||||
|---|---|---|---|---|---|---|---|---|
|
S. cerevisiae
|
P. stipitis
|
|||||||
| f(1) | f(2) | f(2∗) | f(3) | f(1) | f(2) | f(2∗) | f(3) | |
| Ala Cα | 0.03 | 0.05 | 0.05 | 0.87 | 0.05 | 0.10 | 0.02 | 0.83 |
| Ala Cβ | 0.09 | 0.91 | 0.08 | 0.92 | ||||
| Arg Cβ | 0.26 | 0.00 | 0.70 | 0.04 | 0.59 | 0.00 | 0.36 | 0.05 |
| Arg Cδ | 0.05 | 0.95 | 0.08 | 0.92 | ||||
| Asp Cα | 0.02 | 0.02 | 0.02 | 0.94 | 0.19 | 0.15 | 0.27 | 0.39 |
| Asp Cβ | 0.03 | 0.96 | 0.00 | 0.01 | 0.20 | 0.46 | 0.27 | 0.07 |
| Glu Cα | 0.05 | 0.61 | 0.21 | 0.13 | 0.27 | 0.24 | 0.39 | 0.10 |
| Glu Cβ | 0.25 | 0.75 | 0.00 | 0.66 | 0.34 | 0.00 | ||
| Glu Cγ | 0.03 | 0.00 | 0.97 | 0.00 | 0.07 | 0.00 | 0.93 | 0.00 |
| Gly Cα | 0.07 | 0.93 | 0.23 | 0.77 | ||||
| His Cα | 0.02 | 0.00 | 0.00 | 0.98 | 0.07 | 0.00 | 0.04 | 0.89 |
| His Cβ | 0.06 | 0.62 | 0.00 | 0.32 | 0.13 | 0.44 | 0.00 | 0.43 |
| His Cδ2 | 0.11 | 0.89 | 0.35 | 0.65 | ||||
| Ile Cα | 0.05 | 0.00 | 0.94 | 0.01 | 0.32 | 0.00 | 0.68 | 0.00 |
| Ile Cγ2 | 0.12 | 0.88 | 0.08 | 0.92 | ||||
| Ile Cγ1 | 0.98 | 0.00 | 0.02 | 0.00 | 0.69 | 0.00 | 0.31 | 0.00 |
| Ile Cδ1 | 1.00 | 0.00 | 0.68 | 0.32 | ||||
| Leu Cα | 0.03 | 0.00 | 0.96 | 0.01 | 0.07 | 0.00 | 0.93 | 0.00 |
| Leu Cβ | 0.99 | 0.00 | 0.01 | 1.00 | 0.00 | 0.00 | ||
| Leu Cδ1 | 0.18 | 0.82 | 0.18 | 0.82 | ||||
| Leu Cδ2 | 1.00 | 0.00 | 1.00 | 0.00 | ||||
| Lys Cα | 0.04 | 0.00 | 0.96 | 0.00 | 0.05 | 0.02 | 0.92 | 0.01 |
| Lys Cβ | 0.26 | 0.74 | 0.00 | 0.67 | 0.33 | 0.00 | ||
| Lys Cγ | 0.18 | 0.78 | 0.04 | 0.60 | 0.38 | 0.02 | ||
| Lys Cδ | 0.04 | 0.95 | 0.01 | 0.06 | 0.90 | 0.04 | ||
| Lys Cɛ | 0.05 | 0.95 | 0.10 | 0.90 | ||||
| Met Cα | 0.03 | 0.05 | 0.05 | 0.87 | 0.20 | 0.20 | 0.19 | 0.41 |
| Phe Cα | 0.02 | 0.01 | 0.00 | 0.97 | 0.04 | 0.13 | 0.00 | 0.83 |
| Phe Cβ | 0.02 | 0.98 | 0.00 | 0.00 | 0.04 | 0.96 | 0.00 | 0.00 |
| Pro Cδ | 0.04 | 0.96 | 0.07 | 0.93 | ||||
| Pro Cα | 0.07 | 0.63 | 0.22 | 0.08 | 0.25 | 0.24 | 0.41 | 0.10 |
| Pro Cβ | 0.26 | 0.74 | 0.00 | 0.67 | 0.33 | 0.00 | ||
| Pro Cγ | 0.02 | 0.93 | 0.05 | 0.04 | 0.96 | 0.00 | ||
| Ser Cα | 0.01 | 0.01 | 0.19 | 0.79 | 0.05 | 0.11 | 0.12 | 0.72 |
| Ser Cβ | 0.21 | 0.79 | 0.21 | 0.79 | ||||
| Thr Cα | 0.02 | 0.02 | 0.03 | 0.93 | 0.21 | 0.13 | 0.28 | 0.38 |
| Thr Cβ | 0.01 | 0.99 | 0.00 | 0.22 | 0.74 | 0.04 | ||
| Thr Cγ2 | 1.00 | 0.00 | 0.65 | 0.35 | ||||
| Tyr Cα | 0.03 | 0.01 | 0.00 | 0.96 | 0.04 | 0.12 | 0.00 | 0.84 |
| Tyr Cβ | 0.03 | 0.97 | 0.00 | 0.00 | 0.04 | 0.96 | 0.00 | 0.00 |
| Tyr Cδ1 | 0.04 | 0.96 | 0.00 | 0.04 | 0.96 | 0.00 | ||
| Tyr Cɛ1 | 0.31 | 0.02 | 0.25 | 0.42 | 0.33 | 0.06 | 0.23 | 0.38 |
| Val Cα | 0.09 | 0.01 | 0.89 | 0.01 | 0.17 | 0.01 | 0.81 | 0.01 |
| Val Cγ1 | 0.10 | 0.90 | 0.08 | 0.92 | 0.01 | |||
| Val Cγ2 | 0.93 | 0.07 | 0.98 | 0.02 | 0.07 | |||
The data for S. cerevisiae are those of reference 27 for comparison. The fractions f(i), named according to reference 46 and corresponding to the relative amount of Ci intact fragments, were calculated from the relative intensities of the 13C multiplet components using the probabilistic equations described by Szyperski (44). Note that in reference 46 f(2a) and f(2b) correspond to f(2) and f(2∗), respectively.
All cultures aerobic.
REFERENCES
- 1.Bakker, B. M., K. M. Overkamp, A. J. A. van Maris, P. Kötter, M. A. H. Luttik, J. P. van Dijken, and J. T. Pronk. 2001. Stoichiometry and compartmentation of NADH metabolism in Saccharomyces cerevisiae. FEMS Microbiol. Rev. 25:15-37. [DOI] [PubMed] [Google Scholar]
- 2.Boles, E., P. de Jong-Gubbels, and J. T. Pronk. 1998. Identification and characterization of MAE1, the Saccharomyces cerevisiae structural gene encoding mitochondrial malic enzyme. J. Bacteriol. 180:2875-2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boles, E., W. Lehnert, and F. K. Zimmermann. 1993. The role of the NAD-dependent glutamate dehydrogenase in restoring growth on glucose of a Saccharomyces cerevisiae phosphoglucose isomerase mutant. Eur. J. Biochem. 217:469-477. [DOI] [PubMed] [Google Scholar]
- 4.Boonstra, B., C. E. French, I. Wainwright, and N. C. Bruce. 1999. The udhA gene of Escherichia coli encodes a soluble pyridine nucleotide transhydrogenase. J. Bacteriol. 181:1030-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bruinenberg, P. M., J. P. van Dijken, and W. A. Scheffers. 1983. A theoretical analysis of NADPH production and consumption in yeasts. J. Gen. Microbiol. 129:953-964. [Google Scholar]
- 6.Çakar, Z. P., U. Sauer, and J. E. Bailey. 1999. Metabolic engineering of yeast: the perils of auxotrophic hosts. Biotechnol. Lett. 21:611-616. [Google Scholar]
- 7.Canonaco, F., T. A. Hess, S. Heri, T. Wang, T. Szyperski, and U. Sauer. 2001. Metabolic flux response to phosphoglucose isomerase knock-out in Escherichia coli and impact of overexpression of the soluble transhydrogenase UdhA. FEMS Microbiol. Lett. 204:247-252. [DOI] [PubMed] [Google Scholar]
- 8.Christensen, B., T. Christiansen, A. K. Gombert, and J. Nielsen. 2001. Simple and robust method for estimation of the split ratio between the oxidative pentose phosphate pathways and the Embden-Meyerhof-Arnas pathway in microorganisms. Biotechnol. Bioeng. 74:517-523. [DOI] [PubMed] [Google Scholar]
- 9.Christensen, B., A. K. Gombert, and J. Nielsen. 2002. Analysis of flux estimates based on 13C-labeling experiments. Eur. J. Biochem. 269:2795-2800. [DOI] [PubMed] [Google Scholar]
- 10.Christensen, B., and J. Nielsen. 1999. Metabolic network analysis. Adv. Biochem. Eng. Biotechnol. 66:209-231. [PubMed] [Google Scholar]
- 11.Cortassa, S., and M. A. Aon. 1998. The onset of fermentative metabolism in continuous culture depends on the catabolite repression properties of Saccharomyces cerevisiae. Enz. Microb. Technol. 22:705-712. [Google Scholar]
- 12.Dauner, M., J. E. Bailey, and U. Sauer. 2001. Metabolic flux analysis with a comprehensive isotopomer model in Bacillus subtilis. Biotechnol. Bioeng. 76:144-156. [DOI] [PubMed] [Google Scholar]
- 13.de Jong-Gubbels, P., P. A. Vanrolleghem, J. J. Heijnen, J. P. van Dijken, and J. T. Pronk. 1995. Regulation of carbon metabolism in chemostat cultures of Saccharomyces cerevisiae grown on mixtures of glucose and ethanol. Yeast 11:407-418. [DOI] [PubMed] [Google Scholar]
- 14.Dickinson, J. R., S. J. Harrison, and M. J. Hewlins. 1998. An investigation of the metabolism of valine to isobutyl alcohol in Saccharomyces cerevisiae. J. Biol. Chem. 273:25751-25756. [DOI] [PubMed] [Google Scholar]
- 15.Emmerling, M., M. Dauner, A. Ponti, J. Fiaux, M. Hochuli, T. Szyperski, K. Wüthrich, J. E. Bailey, and U. Sauer. 2002. Metabolic flux responses to pyruvate kinase knockout in Escherichia coli. J. Bacteriol. 184:152-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gancedo, J. M. 1998. Yeast carbon catabolite repression. Microbiol. Mol. Biol. Rev. 62:334-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gancedo, J. M., and R. Lagunas. 1973. Contribution of the pentose-phosphate pathway to glucose metabolism in Saccharomyces cerevisiae: a critical analysis on the use of labelled glucose. Plant Sci. Lett. 1:193-200. [Google Scholar]
- 18.Gombert, A. K., M. M. dos Santos, B. Christensen, and J. Nielsen. 2001. Network identification and flux quantification in the central metabolism of Saccharomyces cerevisiae under different conditions of glucose repression. J. Bacteriol. 183:1441-1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gupta, R. N., T. Hemscheidt, B. G. Sayer, and I. D. Spenser. 2001. Biosynthesis of vitamin B6 in yeast: incorporation pattern of glucose. J. Am. Chem. Soc. 123:11353-11359. [DOI] [PubMed] [Google Scholar]
- 20.Hahn-Hägerdahl, B., C. F. Wahlbom, M. Gardonyi, W. H. van Zyl, R. R. Cordero Otero, and L. J. Jönsson. 2001. Metabolic engineering of Saccharomyces cerevisiae for xylose utilization. Adv. Biochem. Eng. Biotechnol. 73:53-84. [DOI] [PubMed] [Google Scholar]
- 21.Hauf, J., F. K. Zimmermann, and S. Müller. 2000. Simultaneous genomic overexpression of seven glycolytic enzymes in the yeast Saccharomyces cerevisiae. Enz. Microb. Technol. 26:688-698. [DOI] [PubMed] [Google Scholar]
- 22.Herve, M., B. Buffin-Meyer, F. Bouet, and S. Tran-Dinh. 2000. Detection of modifications in the glucose metabolism induced by genetic mutations in Saccharomyces cerevisiae by 13C- and 1H-NMR spectroscopy. Eur. J. Biochem. 267:3337-3344. [DOI] [PubMed] [Google Scholar]
- 23.Hochuli, M., H. Patzelt, D. Oesterhelt, K. Wüthrich, and T. Szyperski. 1999. Amino acid biosynthesis in the halophilic archaeon Haloarcula hispanica. J. Bacteriol. 181:3226-3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johansson, B., and B. Hahn-Hägerdahl. 2002. The non-oxidative pentose phosphate pathway controls the fermentation rate of xylulose but not of xylose in Saccharomyces cerevisiae TMB3001. FEMS Yeast Res. 2:277-282. [DOI] [PubMed] [Google Scholar]
- 25.Kötter, P., and M. Ciriacy. 1993. Xylose fermentation by Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 38:776-783. [Google Scholar]
- 26.Lancar-Benda, J., B. Foucher, and M. Saint-Macary. 1996. Characterization, purification and properties of the yeast mitochondrial dicarboxylate carrier. Biochimie 78:195-200. [DOI] [PubMed] [Google Scholar]
- 27.Maaheimo, H., J. Fiaux, Z. P. Çakar, J. E. Bailey, U. Sauer, and T. Szyperski. 2001. Central carbon metabolism of Saccharomyces cerevisiae explored by biosynthetic fractional 13C labeling of common amino acids. Eur. J. Biochem. 268:2464-2479. [DOI] [PubMed] [Google Scholar]
- 28.McAlister-Henn, L., and W. C. Small. 1997. Molecular genetics of yeast TCA cycle isoenzmyes. Prog. Nucleic Acid Res. 57:317-339. [DOI] [PubMed] [Google Scholar]
- 29.Møller, K., B. Christensen, J. Förster, J. Piskur, J. Nielsen, and L. Olsson. 2002. Aerobic glucose metabolism of Saccharomyces kluyveri: growth, metabolite production, and quantification of metabolic fluxes. Biotechnol. Bioeng. 77:186-193. [DOI] [PubMed] [Google Scholar]
- 30.Nissen, T. L., M. Anderlund, J. Nielsen, J. Villadsen, and M. C. Kielland-Brandt. 2001. Expression of a cytoplasmic transhydrogenase in Saccharomyces cerevisiae results in formation of 2-oxoglutarate due to depletion of the NADPH pool. Yeast 18:19-32. [DOI] [PubMed] [Google Scholar]
- 31.Palmeri, L., F. M. Lasorsa, A. De Palma, F. Palmeri, M. J. Runswick, and J. E. Walker. 1997. Identification of the yeast ACR1 gene product as a succinate-fumarate transporter essential for growth on ethanol or acetate. FEBS Lett. 417:114-118. [DOI] [PubMed] [Google Scholar]
- 32.Palmeri, L., A. Vozza, G. Agrimi, V. De Marco, M. J. Runswick, F. Palmieri, and J. E. Walkers. 1999. Identification of the yeast mitochondrial transporter for oxaloacetate and sulfate. J. Biol. Chem. 274:22184-22190. [DOI] [PubMed] [Google Scholar]
- 33.Passoth, V., M. Zimmermann, and U. Klinner. 1996. Peculiarities of the regulation of fermentation and respiration in the crabtree-negative xylose-fermenting yeast Pichia stipitis. Appl. Biochem. Biotechnol. 57-58:201-212. [DOI] [PubMed] [Google Scholar]
- 34.Postma, E., C. Verduyn, W. A. Scheffers, and J. P. van Dijken. 1989. Enzymic analysis of the crabtree effect in glucose-limited chemostat cultures of Saccharomyces cerevisiae. Appl. Environ. Microbiol. 55:468-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Proft, M., D. Grzesitza, and K.-D. Entian. 1995. Identification and characterization of regulatory elements in the phosphoenolpyruvate carboxykinase gene PCK1 of Saccharomyces cerevisiae. Mol. Gen. Genet. 246:367-373. [DOI] [PubMed] [Google Scholar]
- 36.Sauer, U., V. Hatzimanikatis, J. E. Bailey, M. Hochuli, T. Szyperski, and K. Wüthrich. 1997. Metabolic fluxes in riboflavin-producing Bacillus subtilis. Nat. Biotechnol. 15:448-452. [DOI] [PubMed] [Google Scholar]
- 37.Sauer, U., D. R. Lasko, J. Fiaux, H. M., R. Glaser, T. Szyperski, K. Wüthrich, and J. E. Bailey. 1999. Metabolic flux ratio analysis of genetic and environmental modulations of Escherichia coli central carbon metabolism. J. Bacteriol. 181:6679-6688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmidt, K., M. Carlsen, J. Nielsen, and J. Villadsen. 1997. Modeling isotopomer distributions in biochemical networks using isotopomer mapping matrices. Biotechnol. Bioeng. 55:831-840. [DOI] [PubMed] [Google Scholar]
- 39.Senac, T., and B. Hahn-Hägerdal. 1990. Intermediary metabolite concentrations in xylulose- and glucose-fermenting Saccharomyces cerevisiae cells. Appl. Environ. Microbiol. 56:120-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Senn, H., B. Werner, B. A. Messerle, C. Weber, R. Traber, and K. Wüthrich. 1989. Stereospecific assignment of the methyl 1H-NMR lines of valine and isoleucine in polypeptides by nonrandom 13C labeling. FEBS Lett. 249:113-118. [Google Scholar]
- 41.Skoog, K., B. Hahn-Hägerdal, H. Degn, J. P. Jacobsen, and H. S. Jacobsen. 1992. Ethanol reassimilation and ethanol tolerance in Pichia stipitis CBS6054 as studied by 13C nuclear magnetic resonance spectroscopy. Appl. Environ. Microbiol. 58:2552-2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stückrath, I., H. C. Lange, P. Kötter, W. M. van Gulik, K.-D. Entian, and J. J. Heijnen. 2002. Characterization of null mutants of the glyoxylate cycle and gluconeogenic enzymes in S. cerevisiae through metabolic network modeling verified by chemostat cultivation. Biotechnol. Bioeng. 77:61-72. [DOI] [PubMed] [Google Scholar]
- 43.Szyperski, T. 1998. 13C-NMR, MS and metabolic flux balancing in biotechnological research. Q. Rev. Biophys. 31:41-106. [DOI] [PubMed] [Google Scholar]
- 44.Szyperski, T. 1995. Biosynthetically directed fractional 13C-labeling of proteinogenic amino acids: an efficient analytical tool to investigate intermediary metabolism. Eur. J. Biochem. 232:433-448. [DOI] [PubMed] [Google Scholar]
- 45.Szyperski, T., J. E. Bailey, and K. Wüthrich. 1996. Detecting and dissecting metabolic fluxes using biosynthetic fractional 13C labeling and two-dimensional NMR spectroscopy. Trends Biotechnol. 14:453-459. [Google Scholar]
- 46.Szyperski, T., R. W. Glaser, M. Hochuli, J. Fiaux, U. Sauer, J. E. Bailey, and K. Wüthrich. 1999. Bioreaction network topology and metabolic flux ratio analysis by biosynthetic fractional 13C-labeling and two-dimensional NMR spectroscopy. Metab. Eng. 1:189-197. [DOI] [PubMed] [Google Scholar]
- 47.van Dijken, J. P., and W. A. Scheffers. 1986. Redox balances in the metabolism of sugars by yeasts. FEMS Microbiol. Rev. 32:199-224. [Google Scholar]
- 48.van Roemund, C. W. T., E. H. Hettema, M. van der Berg, H. F. Tabak, and R. J. A. Wanders. 1999. Molecular characterization of carnithine-dependent transport of acetyl-CoA from peroxisomes to mitochondria in Saccharomyces cerevisiae and identification of a plasma membrane carnithine transporter, Agp2p. EMBO J. 18:5843-5852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van Urk, H., D. Schipper, G. J. Breedveld, P. R. Mak, W. A. Scheffers, and J. P. van Dijken. 1989. Localization and kinetics of pyruvate-metabolizing enzymes in relation to aerobic alcoholic fermentation in Saccharomyces cerevisiae. Biochim. Biophys. Acta 992:78-86. [DOI] [PubMed] [Google Scholar]
- 50.van Winden, W., P. Verheijen, and J. J. Heijnen. 2001. Possible pitfalls of flux calculations based on 13C-labeling. Metab. Eng. 3:151-162. [DOI] [PubMed] [Google Scholar]
- 51.Verduyn, C., E. Postma, W. A. Scheffers, and J. P. van Dijken. 1992. Effect of benzoic acid on metabolic fluxes in yeasts: a continuous-culture study on the regulation of respiration and alcoholic fermentation. Yeast 8:501-517. [DOI] [PubMed] [Google Scholar]
- 52.Wahlbom, C. F., A. Eliasson, and B. Hahn-Hägerdal. 2001. Intracellular fluxes in a recombinant xylose-utilizing Saccharomyces cerevisiae cultivated anaerobically at different dilution rates and feed concentrations. Biotechnol. Bioeng. 72:289-296. [DOI] [PubMed] [Google Scholar]
- 53.Walker, G. M. 1998. Yeast physiology and biotechnology. John Wiley & Sons, Ltd., Chichester, United Kingdom.
- 54.Wang, C. H., I. Stern, C. M. Gilmour, S. Klungsoyr, D. J. Reed, J. J. Bialy, B. E. Christensen, and V. H. Cheldelin. 1958. Comparative study of glucose catabolism by the radiorespirometric method. J. Bacteriol. 76:207-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wiechert, W. 2001. 13C metabolic flux analysis. Metab. Eng. 3:195-206. [DOI] [PubMed] [Google Scholar]
- 56.Wiechert, W., C. Siefke, A. A. de Graaf, and A. Marx. 1997. Bidirectional reaction steps in metabolic networks: II. Flux estimation and statistical analysis. Biotechnol. Bioeng. 55:118-135. [DOI] [PubMed] [Google Scholar]
- 57.Woldman, Y., and D. R. Appling. 2002. A general method for determining the contribution of split pathways in metabolite production in the yeast Saccharomyces cerevisiae. Metab. Eng. 4:170-181. [DOI] [PubMed] [Google Scholar]