Abstract
Objective: This study presents the results of an author co-citation analysis of the interdisciplinary field of medical informatics.
Methods: An author co-citation analysis was conducted for the years 1994 to 1998, using the fifty most-cited American College of Medical Informatics fellows as an author population. Co-citation data were calculated for every author pair, and multivariate analyses were performed to ultimately show the relationships among all authors. A multidimensional map was created, wherein each author is represented as a point, and the proximity of these points reflects the relationships of authors as perceived by multiple citers.
Results and Conclusion: The results from this analysis provide one perspective of the field of medical informatics and are used to suggest future research directions to address issues related to better understanding of communication and social networks in the field to inform better provision of information services.
INTRODUCTION
Medical informatics is an interdisciplinary field that draws from and contributes to a number of disciplines. It is a field that has a number of overlapping research foci within its boundaries and that often requires highly interactive collaboration among heterogeneous researchers. As a result, researchers and professionals in medical informatics may find it challenging to access and utilize the field's literature. Interdisciplinary fields such as medical informatics pose challenges to librarians and other information professionals who continually seek ways to reconcile relevant information sources with the needs of diverse user populations.
In support of this broad goal, library and information science (LIS) researchers and professionals have long benefited from understanding the scholarly communication structures and networks of the disciplines they serve. In LIS, the methodologies of bibliometrics have stood out as a compelling set of quantitative techniques used to understand the structure of disciplines. Bibliometrics seeks to quantitatively study the literatures of fields—primarily their bibliographies—to produce models of science, technology, and scholarship over time [1]. The intersection of bibliometrics and scholarly communication can be viewed as the application of the quantitative analyses of literature that enable qualitative assessments and interpretations of science and scholarly networks. The breadth of this intersection, of course, can be considered either narrowly or broadly. For instance, “some view the intersection narrowly, constituted only by the use of clustering methods to map relationships among disciplines or to identify scholarly communities” [2]. In a broader sense, others consider “any bibliometric study necessarily to concern scholarly communication and almost any quantitative analysis of scholarly communication to be bibliometric” [3]. Either of these might be oversimplifications, for it is the research questions, goals, and data that determine the information sought and the appropriate methods of interpretation.
This study uses the bibliometric method of author co-citation analysis (ACA), which has been a particularly compelling tool in LIS. ACA uses authors as the units of analysis and the co-citations of pairs of authors (the number of times they are cited together by a third party) as the variable that indicates their “distances” from each other. The underlying assumption of ACA is that the more two authors are cited together, the closer the relationship between them [4]. Two- or three-dimensional maps are produced using multidimensional scaling tools available in such statistical software packages as SPSS or SAS. Each point on the map represents an author, and the proximity of these points reflects the relationships of authors as perceived by multiple citers [5]. In effect, such maps can reveal clusters or networks of scientists in a given field. As White puts it: “This is a more rigorous grouping principle than that of typical subject indexing, because it depends not on perfunctory indication of content by nonspecialists, but on repeated statements of connectedness by citers with subject expertise” [6]. The presentation of these author clusters is interpreted as one perspective of the structure of a particular field.
The criticisms of this technique are not dissimilar from those of citation theory in general. An excellent encapsulation of such debates can be seen in White's chapter, “Author Co-Citation Analysis: Overview and Defense” [7], which, among other things, responds to an assault on this method written by Edge [8]. For instance, Edge claims that co-citation analysts infer relationships between two co-cited documents based on the fact that they are cited together. White's response is that it is not merely the occurrence of a single or few co-citations, but “the piling up of co-citations—the fact that their count over time exceeds a certain threshold—that indicates a relationship” [9]. More important, communication linkages or personal interaction are not simply assumed by co-citationists, but studies seem to show that strong ties are apparent, as reflected by the clusters. While authors are not necessarily engaged in interpersonal communication, it can be surmised that the relationship that holds “is generally a perceived similarity of subject matter or methodological approach in published and cited works” [emphasis in original] [10]. The following summarizes the crux of White's argument: “At most, citationists have, in effect, said to historians, ‘Here is some evidence from the literature of a connection you may have missed’” [11].
Although scarce, bibliometric studies of the field of medical informatics have mostly examined where to find the literature and have studied the field at the journal level as opposed to individual articles or groups of authors. For instance, one recent study of the medical informatics journal literature supports the fact that it is an interdisciplinary field [12] by examining journal intercitation relationships and journal co-citation patterns to analyze the field's structure. Several other studies have sought to understand medical informatics by analyzing its journal literature as well. More than a decade ago, Greenes and Siegel [13] attempted to characterize medical informatics and understand its most-valued journal titles by comparing the ISI Journal Citation Report (JCR) impact factor and immediacy index with a subjective analysis of American College of Medical Informatics (ACMI) fellows as reflected by survey responses. More recently, Sittig and Kaalaas-Sittig [14] developed a method for quantitatively ranking biomedical informatics serials based upon multiple citation analyses, library use statistics, expert opinion, and various selected distinguishing publication characteristics. A modified study was later conducted by Sittig [15] to look at medical informatics journal articles indexed in MEDLINE between 1990 and 1994. Again, the goal was to derive a core set of informatics serials that could be used by librarians in their selection of journal titles to add to their collections. Expanding upon the “core list” offered by Sittig, Vishwanatham [16] provided an objective list of journals that published medical informatics articles relevant to library and information science, particularly those not indexed in MEDLINE.
Each of the above studies calls for further, more detailed bibliometric analyses of the interdisciplinary field of medical informatics. To date, no one has studied the relationships of core authors in medical informatics using ACA. This study seeks to present a representation of the structure of medical informatics research as derived by studying author co-citation patterns of core authors within the field. It is intended to be foundational research for further investigations of the field, so that librarians and other information professionals can better facilitate the information needs of this growing interdiscipline.
METHODS
Selection of authors.
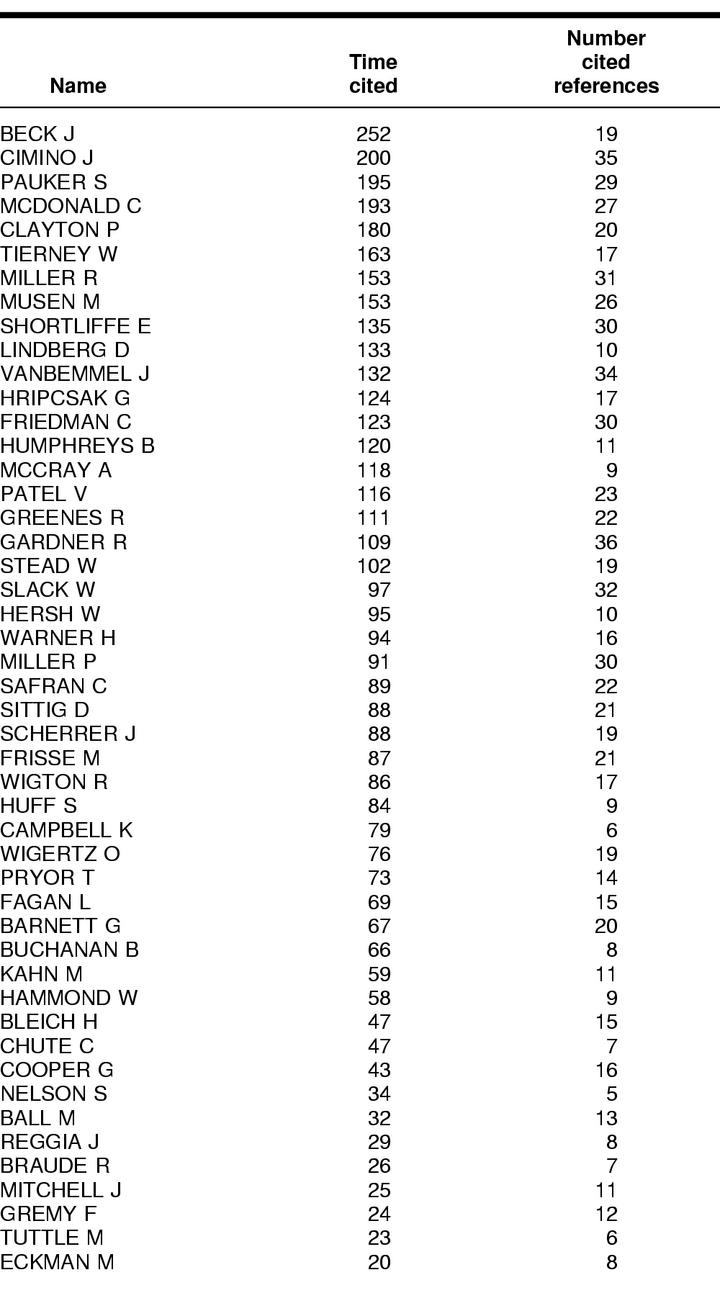
Determining a core author set poses an initial difficulty. As White states, co-cited author maps “are only as good as the analyst's choice of authors” [17]. There are no hard and fast rules, but the subjectivity inherent in the selection of authors should be limited. This study used American College of Medical Informatics (ACMI) fellows as a population from which to derive a core set of prominent medical informatics researchers. At the time these data were collected, 196 fellows had been elected into ACMI. These individuals ostensibly were elected into ACMI based on their significant contributions to the field. At the very least, ACMI fellows are well-recognized members in the field and are among some of the most prominent authors. A shorter list of fifty authors was determined after examining initial citation data retrieved from ISI files, as described below.
Retrieval of cocitation data.
Basic citation counts on ACMI fellows were collected from the ISI databases, Science Citation Index (SCI), and Social Science Citation Index (SSCI). These files were available via ISI's Web of Science (WoS) tool. Searches on each author's name were conducted to determine who were among the most-cited fellows. These data were used to shorten the list of authors to a manageable number; thus, the top fifty authors were selected based on the number of times cited and the number of cited references for the years 1994 to 1998.
It was determined that a five-year period was appropriate for the purposes of this study. First, this was not a longitudinal study examining the field over an extended period; otherwise, a much broader time span would have been merited and examined. On the other hand, too short a period would have limited the amount of citation and bibliographic information available, given the somewhat lengthy publication and indexing processes. The five-year period, while admittedly based on the author's own discretion, was meant to capture information on the most productive authors during this time. It also coincides with some of the most significant technological advances (e.g., the proliferation of the Web and Internet-based applications) that have affected the field. The resulting list of authors is shown in Table 1.
Table 1 List of top fifty American College of Medical Informatics (ACMI) authors based on times cited (as of May 1999)
Once this core set of authors was identified, various bibliographic data were downloaded. It was necessary to limit searches to avoid retrieving a large number of false hits; that is, where references for authors with similar or the same last names and initials were retrieved but were not the authors of interest. To minimize these instances, as well as to ensure the retrieval of references related to medical informatics, searches were limited to articles that appeared in any of the eighteen journals listed under “medical informatics” in ISI's 1997 SCI Journal Citation Reports. Reviewing the titles from this list, they appeared to be an excellent representation of journals in the field, so no titles were added nor were any other adjustments made. There were a few reasons for this. First, ISI's impact factor afforded some objective standard by which to select journal titles, and, second, based on this researcher's personal experience in the field, these titles appeared to represent a broad range of literature in medical informatics and were, for the most part, common titles. Using these also meant that searches could be restricted to the SCI database, because all of these titles are indexed there. Searches included each author's name, with truncation following the first initial (i.e., CIMINO J*). Using a Boolean “AND,” authors were combined with the eighteen journal titles, which were strung together with an “OR” operator.
Traditionally, author co-citation data are collected during an ISI search session by using a Boolean “AND” for every possible author pair at the time of the search. This task can be formidable if performed manually because the number of iterations equals N(N–1)/2, where N equals the number of authors. For this study, all the necessary bibliographic data for each individual author were collected from WoS for further processing. The data gathered were bibliographic citations (excluding abstracts) for every article that cited at least one of these core authors' articles published in informatics journals within the five-year period. Also, bibliographic citations for each of the authors' cited references (the actual article being cited) were downloaded. All of the citations were imported into EndNote libraries using filters designed for downloading ISI WoS information and subsequently exported into a Microsoft Access database for additional processing. All duplicates were removed to account for instances of co-authorship, so that redundant cocitation credit would not be given.
The co-citation counts for each author pair were derived using a program created by Jim Ries, a former predoctoral medical informatics fellow at the University of Missouri–Columbia, Department of Health Management and Informatics. Essentially, this program searched the citation field of each bibliographic record, counting the number of times two authors were cited together. The result was the basis of all future analyses used in the ACA portion of this project.
Cluster analysis, factor analysis, multidimensional scaling.
Three types of multivariate analysis—cluster analysis, factor analysis, and multidimensional scaling—were performed using the raw co-citation matrix described above as the raw data set. Descriptions of these methods follows.
Cluster analysis
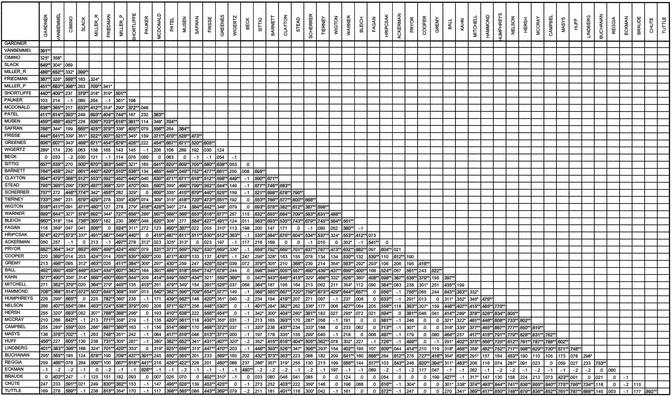
The first multivariate analysis, cluster analysis, requires conversion of the raw data matrix to a correlation matrix. The functions available in SPSS create this conversion automatically as a by-product of the ANALYZE>CLASSIFY>HIERARCHICAL CLUSTER . . . selection using the software's graphical user interface. Pearson product-moment correlations are usually computed to measure the extent to which two variables have a linear relationship. McCain [18] and others [19] subscribe to Pearson correlation as being the predominant measure of similarity used in ACA. Use of Pearson correlations between variables allows for a similarity between authors to be determined not solely on their raw co-citations, but in terms of their “co-citation profiles.” To quote McCain [20]: “Two authors who are always cited highly with certain third authors, but infrequently with others, will have a high positive correlation and can be said to be perceived as related or ‘similar’ in some sense by the citing population”; also, the correlation coefficient removes differences in “scale” that can occur between authors with similar profiles but where one is more frequently cited than another. Correlation coefficients for each author pair are presented in a matrix form with author names appearing in rows and columns in equal order. Given issues related to treatment of diagonal values, this study has used McCain's [21] technique of treating diagonal values as missing data and calculating the co-cited author correlations accordingly (which has shown little difference in the results of other studies).
Although there are a variety of cluster analysis techniques [22, 23], the primary method in ACA for cluster formation has been hierarchical agglomerative clustering. Hierarchical agglomerative clustering refers to methods where each object (or individual) starts out as its own cluster. Each subsequent step is meant to combine closest clusters until a single cluster of all objects remains. The specific hierarchical agglomerative method chosen for this study is furthest neighbor (also known as complete linkage), which uses, as described above, Pearson correlations as the measure of similarity, and uses cluster criteria based on maximum distances between objects [24].
To graphically display the clustering results, a dendrogram plot was selected. A representation of the clustering process for the data, it initially shows each individual author and then successive groupings of authors to the point where a single cluster remains. “Cutting” the dendrogram, or drawing a vertical line through a given point on the dendrogram, is done to make some determination of what the best number of clusters might be, although different analysts can interpret this differently. The goal is not to find the perfect level of cluster membership but to show an approximation to spur further discussion.
Factor analysis
Like cluster analysis, factor analysis techniques also are used to identify groups of related variables. Unlike cluster analysis, however, which is more ad hoc, factor analysis has an underlying theoretical model [25]. Factor analysis seeks to study correlations among a number of interrelated variables and to group them into a few highly descriptive factors. In ACA research, the type of factor analysis known as “principal component analysis” has been used to complement clustering techniques. This method attempts to “explain” the interrelationships observed among the variables through the creation of a much smaller number of “derived” variables or factors [26]; it is, in effect, a data reduction method. In ACA, this translates into determining how much each author “loads” on a particular factor. Authors can contribute to more than one factor, which is not the case in clustering or in scaling techniques, although authors usually load most heavily on a single factor, with author loadings of 0.7 or greater as likely to be the most useful for interpretation [27]. Determining the number of factors to be extracted from the data can vary depending on the interpretation sought, but usually the most descriptive information occurs in the first several factors. The eigenvalue refers to the amount of variance accounted for by a factor [28], or the sum of the squared loadings on the factor. This is usually a set as a default of > 1, which means that the program will only extract eigenvalues for factors greater than one. Studying the “scree plot” produced by statistical packages such as SPSS is commonly done to determine the number of factors with the most explanatory power. Rotation is also commonly used to interpret results, with varimax rotation being the most popular in ACA. This is a type of orthogonal rotation that seeks to enable easier interpretation of the results by separating the factor loadings. In effect, maximizing the sum of variances of factors is done to show variables as loading either high or low on factors, that is, closer to one or zero with less gradation in between.
This study used the same raw co-citation matrix used in the other analyses as the initial data and the routines used to again create a correlation matrix for the analysis. Using SPSS, the following were selected: ANALYZE>DATA REDUCTION>FACTOR . . . As stated, principal components analysis was the method used, with a varimax rotation and the default extraction of eigenvalues over one.
Multidimensional scaling
The final method used for the ACA was multidimensional scaling, again using the same correlation matrix as created by the other methods. The purpose of this method is to further elucidate the hierarchical agglomerative clustering and factor analysis results. Multidimensional scaling allows for the creation of a graphical display of the similarity of authors, which can be used for further discussion and, in general, provides a richer display of the clustering results. Essentially, “the closeness of author points on such maps is algorithmically related to their similarity as perceived by citers” [29]. Boundaries can be drawn around groups of related authors based on the clusters derived from the cluster analysis (depending where one cuts the dendrogram) to find further support for interpretation from the results of factor analysis.
Using SPSS, the following selections were made: ANALYZE>SCALE>MULTIDIMENSIONAL SCALING . . . Again, a correlation matrix was created based upon the initial raw co-citation matrix used as input. As the measure of dissimilarity, the Euclidean distance was used, although other options are available (e.g., Chebychev, Block, or Minkowski). A result of the calculations performed with programs such as SPSS are the stress measure and R-square, which are used as indicators of the “goodness of fit.” Generally speaking, a stress value that is close to zero and an R-square (percentage of total variance) that is close to 1 are indications that the data fit the model well. Increasing the number of dimensions usually results in more ideal stress values and R-squares; however, fewer dimensions are desirous because they are considered easier to interpret. In ACA, because cocitation data is inherently “noisy” [30], stress measures below 0.2 are generally acceptable, especially in cases where the R-square is high (that is, close to or greater than 0.90). Interpretation is impressionistic and almost solely based on the graphical representation of the data, although it does offer an enhancement of the results of other analyses.
ACA interpretation
The results of the above quantitative techniques offer a picture of the field of medical informatics as derived from an unobtrusive examination of co-citation patterns. Visualization of the relationships among authors in a two-dimensional space comes from a combined analysis of multidimensional scaling, cluster analysis, and factor analysis (principal components analysis). Thus, maps were created wherein authors who appear most similar appear in closer proximity to one another, while those who are less similar appear further apart. To further illustrate the findings represented through the map, boundaries were drawn around clusters of authors. Such drawings are largely impressionistic and are based upon the individual analyst's interpretation of the data. However, supporting evidence for drawing boundaries comes from the principal components analysis, where groupings of authors are identified based on how they load on each factor, as well as the cluster analysis results.
McCain [31] suggests other means for supporting interpretations of author maps, such as consultation with experts, text-based methods of validating results, and other forms of comparison. As will be discussed in the next section, interpretation for this project comes from studying language use as well as experiences of the researcher. This also informs how the axes, or different areas on the map, are labeled to give the display more informational value and, thus, readers a better understanding of how the results were interpreted.
Limitations
The structure of medical informatics as presented through an ACA is limited in a few important ways. First, the data cover only a short period of time in the history of medical informatics: five years. Thus, one cannot make conclusions similar to the kind of longitudinal ACA study that White and McCain [32] did on the field of information science. The period is also not up to the minute, and the time that has elapsed since the data were collected might mean that there have been some changes in focus of these authors. It is, in essence, a snapshot that is restricted in its ability to comprehensively portray the field.
Another limitation comes from the somewhat subjective selection of the core author set. While ACMI fellows seemed a reasonable population to draw from, some may argue that election into ACMI is not a true reflection of prominence in the field. However, the researcher has made the assumption—largely based on personal experience in the field and consultation with others—that ACMI fellows are indeed representative and key members of medical informatics. This assumption is supported to some degree by Greenes and Siegel's [33] decision to use ACMI fellows as experts in their study.
RESULTS
Author co-citation analysis.
As stated, the ACA was conducted based on co-citation frequencies for the top fifty ACMI fellows, according to times cited, for the period 1994 to 1998. The raw co-citation matrix derived from data collected from the ISI Science Citation Index database via WoS is comprised of every author as both a case and a variable, with the cell values indicating the number of times each author has been co-cited with every other author. The raw co-citation data were used for the analyses conducted for this portion of the project. Descriptive statistics for these data were calculated. The range of co-citations was from 3 to 97, and the highest mean co-citation count was 17.76 (both for McDonald).
Cluster analysis
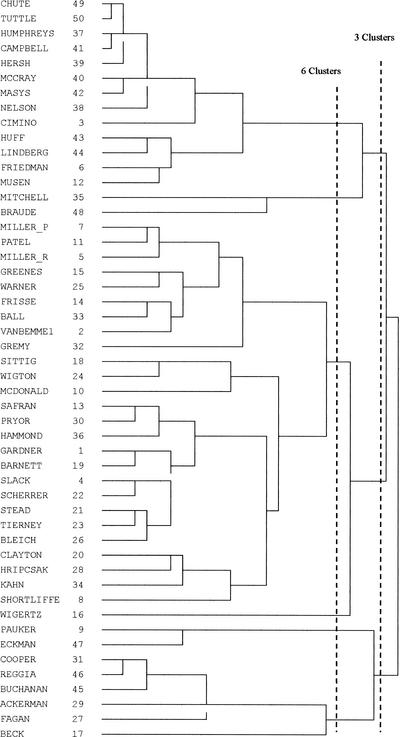
The results of the cluster analysis include a Pearson correlation matrix (Figure 1) and a dendrogram depicting the complete linkage results, shown in Figure 2. “Cutting” the dendrogram in the manner shown in Figure 2 reveals both a six-cluster and three-cluster solution. The six-cluster solution reveals two large groups and four smaller groups. Cluster 1 is the largest, showing twenty-six members. Given the fact that there are three very small clusters (two clusters with two members and one with only one member), and because these authors are those with the lowest overall mean co-citation counts, cutting the dendrogram to show a three cluster solution is more appropriate. That is, cutting up one level (to the right), a three-cluster solution shows that Mitchell and Braude join cluster 2, cluster 1 gains Wigertz, and cluster 3 gets Pauker and Eckman. This seems to tidy up the groupings better. In addition, as will be seen below in the multidimensional mapping of these authors, this works out to be a clearer representation because the smaller clusters tend to be intermingled with the larger clusters in other solutions. Also, this level of clustering appears to be supported and further elucidated through factor analysis.
Figure 1.
Pearson correlation matrix * Correlation is significant at the 0.05 level (2-tailed).** Correlation is significant at the 0.01 level (2-tailed).
Figure 2.
Dendrogram of complete linkage clustering This dendrogram shows the clustering process. Each author starts out as being the sole member in his or her cluster; moving right, cluster membership increases to a final, single cluster solution. The dotted lines represent either a 3 or 6 cluster solution.
Factor analysis
A factor analysis using authors-as-variables was also conducted to provide another perspective on how authors in this field might be grouped. Extraction was performed using principal components analysis with the default of eigenvalues greater than 1. A varimax rotation was used to simplify how the data could be viewed or interpreted, and missing data were handled with means substitution.
The discussion of factor analysis in the above “Methods” section shows that, similar to cluster analysis, factor analysis techniques are used to identify groups of related variables. Because the goal is to help interpret the interrelationships among a number of variables, it is considered, in effect, a data reduction method. For this study, use of this method helps in determining how much each author loads on a particular factor. What these “factors” are, in a qualitative sense, is largely explained by the individual researcher's interpretation of these and other data.
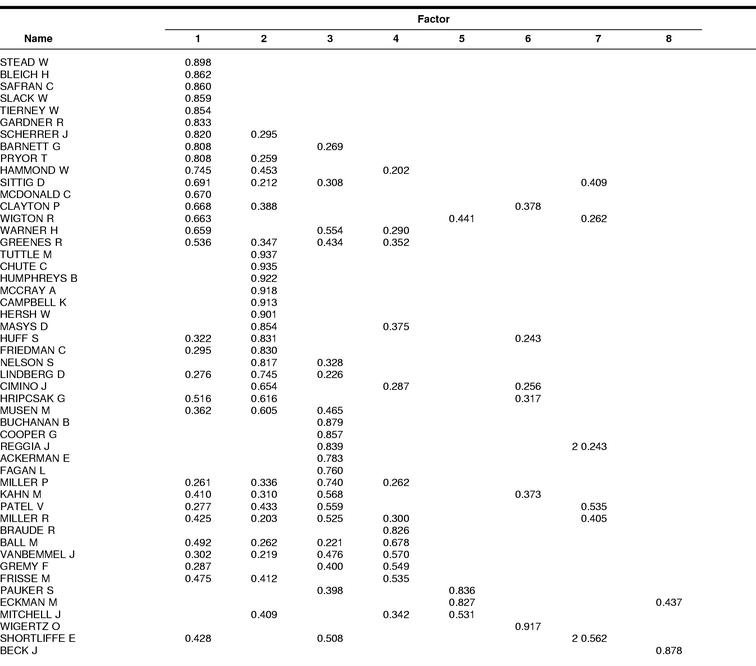
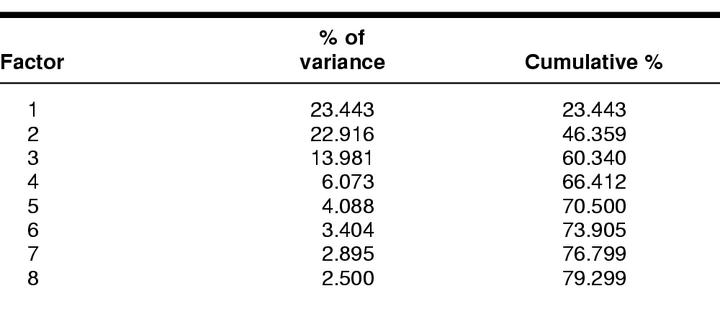
Table 2 shows the rotated solution of the factor analysis done for this study. A total of eight factors were extracted and explain 79.3% of the total variance. It is from the first three or four factors, however, that the greatest amount of this total variance is accounted for (Table 3); the first component accounts for 22.44% of the variance; the second, 22.91%; the third, 13.98%; and the fourth, 6.07%.
Table 2 Rotated solution matrix
Table 3 Variance accounted for by eight factors
Comparing the cluster memberships with the factor analysis, many similarities are seen. Namely, that the first two factors and the two largest clusters seem to be more or less the same in terms of author membership. There are a couple exceptions but, as should be noted, authors can and do load on more than one factor in many cases. For instance, Greenes shows up as 0.536 on factor 1, 0.434 on factor 3, 0.352 on factor 4, and 0.347 on factor 2. This is more revealing than the clustering, where an author is forced to be in one cluster or another. The advantage of adding this method to the analyses, then, is that more insight is afforded into the degree of membership a single author might have in more than a single grouping.
Multidimensional scaling
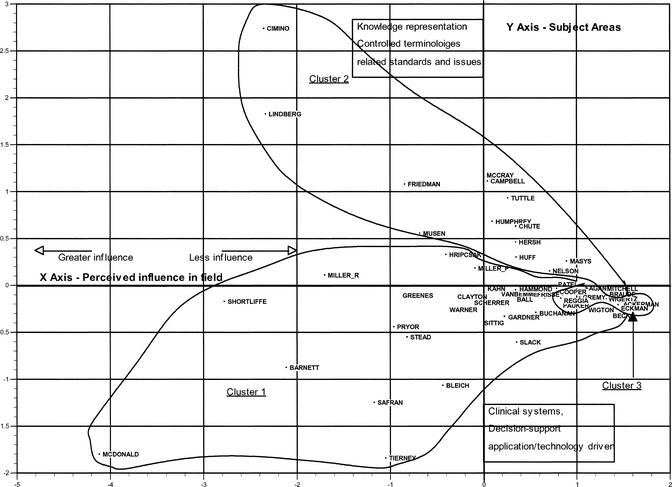
With the multidimensional scaling, the relationships of authors can be studied graphically. Distances were created from the raw co-citation matrix using the Euclidean distance measure, as described earlier. The relationships of authors in a two-dimensional space are represented in Figure 3. The stress measure was 0.1160 and the R-square was 0.962. Lines were drawn around each cluster based on the information from the cluster analysis.
Figure 3.
Multidimensional scaling two-dimensional map of medical informatics authors
Examining this map at a gross level, it shows a small, compressed group of points on the center X axis to the right side of the chart and more evenly distributed points above and below that axis but left of the Y axis. The lines drawn around each of the three clusters of authors show that the first two clusters essentially are those larger, albeit more dispersed, groupings of authors above and below the center X axis: cluster 1 below it and cluster 2 above. These are oblong-shaped, with the bottom right of the top cluster and the top right of the bottom cluster seemingly gravitating toward the less discrete clustering of points on the center right X axis.
In general, the mapping is representative of the data used to create it. For instance, Cimino, who appears as the topmost point on the map, is plotted far away from Tierney, located on the bottom. The Pearson correlation matrix in Figure 1 shows they have a low correlation with one another, only 0.231. Conversely, McCray and Campbell, who appear toward the top right side of the map, are highly correlated (0.819) and are thus plotted in close proximity to each other. This illustration also is supported by the memberships as derived through the factor analysis. As mentioned previously, virtually all of the members loading most heavily on factor 1 appear in cluster 1, and likewise for cluster 2.
For a clearer interpretation of this display, the X and Y axes can be labeled in the following way (again, this is open to reader interpretation but is one impression of the field based on the data). First, the X axis suggests a continuum of “perceived influence on the field.” That is, those toward the left part of the X axis are authors who have the highest mean co-citation counts. Anyone familiar with the medical informatics literature will quickly recognize most of these names, such as Cimino, Lindberg, Friedman, Shortliffe, McDonald, and so on. Working toward the right along the X axis, the mean co-citation counts are increasingly lower until, ultimately, the furthest right grouping of authors have mean co-citation counts less than two. This could suggest that the prominent authors are likely to have been co-cited with more individuals, perhaps many of those within their clusters and even on the low-mean-co-citation side of the map; therefore, they are not “close” to any single author on the right center of the map but more equally related to all or most of them. To restate, this reflects the idea that more prominent authors are cited more frequently and so have more opportunities for co-citation.
Looking up and down the Y axis shows representative authors from different “subject areas.” For instance, judging from an examination of the titles of works published by Cimino and his “neighbors,” during the years 1994 to 1998 (the same time period covered by the ACA), it becomes clear that a large part of his work has been focused on medical terminology or standards in knowledge representation issues. Tierney and his neighbors, on the other hand, seem to have focused on subjects related to decision support applications or, generally, clinical information systems of one form or another.
Given these broad level labels of the axes, the multidimensional scaling map can be “read” more easily. Looking at authors in the top left quadrant shows that these authors are prominent (are highly co-cited and, therefore, seem important contributors to the knowledgebase of the field) in the subject areas they seem most likely to address such as controlled medical terminology issues (based on terms from the titles of their articles and the subject headings assigned to them). In the bottom left quadrant also are a number of influential authors whose articles (and subject headings assigned to them) seem to address subject areas such as clinical information systems, artificial intelligence, decision support systems (in various forms), and, generally, more technology and application-related topics. In addition, on the left side of the Y axis, a few of the high-mean-co-citation count authors appear close to the X axis. This might suggest more of a mixed research agenda. For instance, Shortliffe seems to work in a number of areas covering both vocabulary and system issues equally. The top and bottom right quadrants are similar, in terms of subjects being addressed, to their equivalents on the left of the Y axis; however, these authors may be considered less influential based on mean co-citation rates. Yet, the far right of the X axis, at the zero point on the Y axis, shows the least co-cited authors, leading to the realization that it is more difficult to distinguish the subjects they address in their articles. This view of the graph suggests that a more focused or well-developed research agenda is related in some way to higher co-citation rates or being co-cited with a larger number of people.
DISCUSSION
This bibliometric study of the field of medical informatics provides LIS professionals a perspective that heretofore has not been afforded. As such, it can be one of several tools used to help individuals access and visualize scholarly communication within the field. For instance, while those familiar with the medical informatics community and its literature will already know that, say, McCray and Campbell work in similar areas and are often cited together, those who are not well oriented with the field, particularly new researchers or those information professionals assisting them, could find such information useful. Thus, the type of graph provided here, based on quantitative data and informed qualitative interpretations, could be of great assistance. Other basic uses of this study, or similar studies, are that it helps identify the most productive and prominent authors in the field, the amount they are cited, the amount they are co-cited with other informatics authors, and the authors who appear to work in similar subject areas. From this, one may better locate literature produced in particular areas of medical informatics through such methods as citation- or author-based retrieval.
FUTURE DIRECTIONS
Information professionals in general, and medical librarians in particular, are facing significant challenges as new interdisciplines emerge. Medical librarians increasingly are being asked to inform the development of new databases, tools (e.g., controlled vocabularies), services, and systems for providing quality access to information for this variety of researchers who are crossing disciplinary boundaries. Therefore, information professionals must meet these challenges by developing their understanding of scholarly communication issues of such researchers and by utilizing a variety of tools for developing collections and providing access to information resources.
This study is meant as foundational research to inspire further explorations into the interdisciplinary field of medical informatics. Future studies are needed to understand more clearly how information is actually communicated within the field and what this means for those interested in making the medical informatics literature more accessible both to informaticians and researchers outside the field. To this end, the evidence presented here could be followed up with an extended analysis of language use by informaticians. That is, it is a reasonable assumption that impediments to knowledge sharing are based in language. For instance, in ideal situations, if someone works in a particular research area of medical informatics (e.g., controlled medical terminologies), a common language to discuss concepts in this area might be expected, to enable an equal understanding of, and access to, a shared knowledge. But, establishing a consistent, consensual understanding of even a single research area can be difficult. As Cimino stated in his discussion on the challenges to developing controlled medical terminologies, “there is no common language by which we can communicate our ideas . . . although we are often talking about the same thing, we do so in confusing dialects, with seemingly interchangeable phrases” [34]. The type of unimpeded understanding he seems to call for would require common language use and disambiguated communication among researchers. Initial results shown in a study by Andrews [35] suggest that there are indeed areas of apparent ambiguous and inconsistent language use in the field, and further, more in-depth studies are needed.
ACA techniques and more advanced language analyses could also be applied to look not only within medical informatics, but also to see how it is linked to other disciplines. That is, given that this field is interdisciplinary, the impact of heterogeneous disciplinary perspectives, methods, language, and other factors might be identified to examine where impediments to knowledge sharing might exist across boundaries.
In general, modifications to the methods used here can likely be joined with other methodologies to study scholarly communication in this field. At the very least, a better understanding of the information needs of informaticians, and the people using the knowledge they create, might emerge.
Footnotes
* Based on the author's dissertation completed at the University of Missouri–Columbia, School of Information Science and Learning Technologies, Fall 2000.
REFERENCES
- White HD, McCain KW. Bibliometrics. In: Williams ME, ed. Annual review of information science and technology. v. 24. Amsterdam, Netherlands: Elsevier Publications; 1989:119–86. [Google Scholar]
- Borgman CL. ed. Scholarly communication and bibliometrics. Newbury Park, CA: Sage Publications, 1990:14. [Google Scholar]
- Borgman CL. ed. Scholarly communication and bibliometrics. Newbury Park, CA: Sage Publications, 1990:14. [Google Scholar]
- White HD, Griffith BC. Author cocitation: a literature measure of intellectual structure. J Am Soc Info Sci. 1981 May; 32(3):163–72. [Google Scholar]
- White HD. Author co-citation analysis: overview and defense. In: Borgman CL, ed. Scholarly communication. Newbury Park, CA: Sage Publications, 1990:84–106. [Google Scholar]
- White HD, McCain KW. Visualizing a discipline: an author co-citation analysis of information science, 1972–1995. J Am Soc Info Sci. 1998 Apr; 49(4):327–55. [Google Scholar]
- White HD. Author co-citation analysis: overview and defense. In: Borgman CL, ed. Scholarly communication. Newbury Park, CA: Sage Publications, 1990:84–106. [Google Scholar]
- Edge DO.. Why I am not a co-citationist. Society for Social Studies of Science Newsletter. 1977;2:13–9. [Google Scholar]
- White HD. Author co-citation analysis: overview and defense. In: Borgman CL, ed. Scholarly communication. Newbury Park, CA: Sage Publications, 1990: 96. [Google Scholar]
- White HD. Author co-citation analysis: overview and defense. In: Borgman CL, ed. Scholarly communication. Newbury Park, CA: Sage Publications, 1990: 96. [Google Scholar]
- White HD. Author co-citation analysis: overview and defense. In: Borgman CL, ed. Scholarly communication. Newbury Park, CA: Sage Publications, 1990: 94. [Google Scholar]
- Morris TA, McCain KW. The structure of medical informatics journal literature. J Am Med Inform Assoc. 1998 Sep–Oct; 5(5):448–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenes RA, Siegel ER. Characterization of an emerging field: approaches to defining the literature and disciplinary boundaries of medical informatics. In: Stead WW, ed. Eleventh Annual Symposium on Computer Applications in Medical Care. Washington, DC: Institute of Electrical and Electronics Engineers, 1987 Nov 1–4:411–5. [Google Scholar]
- Sittig DF, Kaalaas-Sittig J.. A citation analysis of medical informatics journals. Medinfo. 1995;8:1452–6. [PubMed] [Google Scholar]
- Sittig DF. Identifying a core set of medical informatics serials: an analysis using the MEDLINE database. Bull Med Libr Assoc. 1996 Apr; 84(2):200–4. [PMC free article] [PubMed] [Google Scholar]
- Vishwanatham R. Citation analysis in journal rankings: medical informatics in the libary and information science literature. Bull Med Libr Assoc. 1998 Oct; 86(4):518–22. [PMC free article] [PubMed] [Google Scholar]
- White HD. Author co-citation analysis: overview and defense. In: Borgman CL, ed. Scholarly communication. Newbury Park, CA: Sage Publications, 1990: 99. [Google Scholar]
- McCain KW. Mapping authors in intellectual space: a technical overview. J Am Soc Info Sci. 1990 Sep; 41(6):433–43. [Google Scholar]
- White HD, McCain KW. Visualizing a discipline: an author co-citation analysis of information science, 1972–1995. J Am Soc Info Sci. 1998 Apr; 49(4):327–55. [Google Scholar]
- McCain KW. Mapping authors in intellectual space: a technical overview. J Am Soc Info Sci. 1990 Sep; 41(6):436. [Google Scholar]
- McCain KW. Mapping authors in intellectual space: a technical overview. J Am Soc Info Sci. 1990 Sep; 41(6):433. [Google Scholar]
- Hair JF, Anderson RE, Tatham RL, and Black WC. Multivariate data analysis. 4th ed. Upper Saddle River, NJ: Prentice-Hall, 1995. [Google Scholar]
- SPSS. SPSS base 10.0 applications guide. Chicago, IL: SPSS, 1999. [Google Scholar]
- Hair JF, Anderson RE, Tatham RL, and Black WC. Multivariate data analysis. 4th ed. Upper Saddle River, NJ: Prentice-Hall, 1995: 439. [Google Scholar]
- SPSS. SPSS base 10.0 applications guide. Chicago, IL: SPSS, 1999: 304. [Google Scholar]
- McCain KW. Mapping authors in intellectual space: a technical overview. J Am Soc Info Sci. 1990 Sep; 41(6):433. [Google Scholar]
- McCain KW. Mapping authors in intellectual space: a technical overview. J Am Soc Info Sci. 1990 Sep; 41(6):433. [Google Scholar]
- Hair JF, Anderson RE, Tatham RL, and Black WC. Multivariate data analysis. 4th ed. Upper Saddle River, NJ: Prentice-Hall, 1995: 365. [Google Scholar]
- White HD, McCain KW. Visualizing a discipline: an author co-citation analysis of information science, 1972–1995. J Am Soc Info Sci. 1998 Apr; 49(4):331. [Google Scholar]
- McCain KW. Mapping authors in intellectual space: a technical overview. J Am Soc Info Sci. 1990 Sep; 41(6):433. [Google Scholar]
- McCain KW. Mapping authors in intellectual space: a technical overview. J Am Soc Info Sci. 1990 Sep; 41(6):433. [Google Scholar]
- White HD, McCain KW. Visualizing a discipline: an author co-citation analysis of information science, 1972–1995. J Am Soc Info Sci. 1998 Apr; 49(4):331. [Google Scholar]
- Greenes RA, Siegel ER. Characterization of an emerging field: approaches to defining the literature and disciplinary boundaries of medical informatics. In: Stead WW, ed. Eleventh Annual Symposium on Computer Applications in Medical Care. Washington, DC: Institute of Electrical and Electronics Engineers, 1987 Nov 1–4:411–5. [Google Scholar]
- Cimino JJ. Editorial: the concepts of language and the language of concepts. Meth Info Med. 1998 Nov; 37(4–5):311. [PubMed] [Google Scholar]
- Andrews JE. A bibliometric investigation of medical informatics: a communicative action perspective. [doctoral dissertation]. University of Missouri–Columbia, Columbia, MO, 2000. [Google Scholar]