Abstract
A positive correlation between cell size and DNA content has been recognized in many plant cell types. Conversely, misexpression of a dominant-negative cyclin-dependent kinase (CDK) or CDK inhibitor proteins (ICK/KRPs) in Arabidopsis and tobacco leaves has revealed that cell growth can be uncoupled from cell cycle progression and DNA content. However, cell growth also appears to be controlled in a non-cell-autonomous manner by organ size, making it difficult in a ubiquitous expression assay to judge the cell-autonomous function of putative cell growth regulators. Here, we investigated the function of the CDK inhibitor ICK1/KRP1 on cell growth and differentiation independent of any compensatory influence of an organ context using Arabidopsis trichomes as a model system. By analyzing cell size with respect to DNA content, we dissected cell growth in a DNA-dependent and a DNA-independent process. We further found that ICK1/KRP1 misexpression interfered with differentiation and induced cell death, linking cell cycle progression, differentiation, and cell death in plants. The function of ICK1/KRP1 in planta was found to be dependent on a C-terminal domain and regulated negatively by an N-terminal domain. Finally, we identified CDKA;1 and a D-type cyclin as possible targets of ICK1/KRP1 expression in vivo.
INTRODUCTION
In most species, the final size of an individual is controlled with astonishing precision. Two key parameters determine the growth of an organism (accumulation of mass): cell number and cell size. Although some control mechanisms for cell proliferation were discovered in the past (Doerner et al., 1996; Mizukami and Fischer, 2000; De Veylder et al., 2002), not much is known about cell growth in plants. One possible determinant of cell size is the amount of nuclear DNA, because in many species, a positive correlation has been found between cell size and DNA content (Nurse, 1985; Kondorosi et al., 2000; Gregory, 2001). A representative example of this correlation is found in Arabidopsis leaf hairs (trichomes). Wild-type trichomes undergo approximately four rounds of endoreduplication, leading to a DNA content of ∼32C (32-fold the DNA content of the haploid genome) per cell. In general, mutants with smaller trichomes were found to contain less DNA, whereas an increase in trichome cell size was correlated positively with additional endoreduplication rounds (Hulskamp et al., 1999).
Recent molecular data have revealed new aspects of cell growth control in plants. Misexpression of a dominant-negative CYCLIN-DEPENDENT KINASE (CDK) and of the CDK inhibitor proteins ICK/KRPs (INHIBITOR/INTERACTOR OF CYCLIN-DEPENDENT KINASES/KIP-RELATED PROTEINS) in Arabidopsis and tobacco leaves has resulted in a reduced cell division rate; the remaining cells were relatively large but contained only a small nucleus (Hemerly et al., 1995; Wang et al., 2000; De Veylder et al., 2001; Jasinski et al., 2002). This finding indicated that cell growth and cell cycle control can be uncoupled and suggested the existence of determinants of cell growth other than DNA amount. However, this DNA-independent increase in cell size is thought to represent a compensatory effect for a reduced number of cells to keep the proper leaf size (Hemerly et al., 1993; Doonan, 2000; De Veylder et al., 2001). Similar observations have been made in animals, in which cell expansion and cell division can compensate for each other to achieve a species-specific organ size (Day and Lawrence, 2000; Potter and Xu, 2001).
Non-cell-autonomous cell growth regulation controlled by the overall size of the organ hinders an evaluation of the cell-autonomous effects of ICK/KRP, leading us to wonder if ICK/KRP expression also results in a cell-autonomous uncoupling of DNA amount from cell size. To exclude any compensatory influence of an organ context, it is necessary to study gene function in single cells that do not contribute much to final leaf size. Therefore, we investigated the function of ICK1/KRP1 in cell growth and cell cycle progression in single-celled Arabidopsis trichomes.
By examining cell cycle progression in correlation with cell size in ICK1/KRP1-misexpressing trichomes, we were able to dissect two different growth mechanisms and found evidence for both DNA-dependent and DNA-independent growth regulation. The reduction of endoreduplication in trichomes was associated with an altered cell differentiation program that leads to trichomes with fewer branches. Strikingly, we found that trichomes that misexpressed ICK1/KRP1 died at later developmental stages. Thus, our data provide a new link between cell cycle progression, differentiation, and cell death in plants.
RESULTS
Misexpression of ICK1/KRP1 in Single-Celled Trichomes Reveals Two Growth Modes
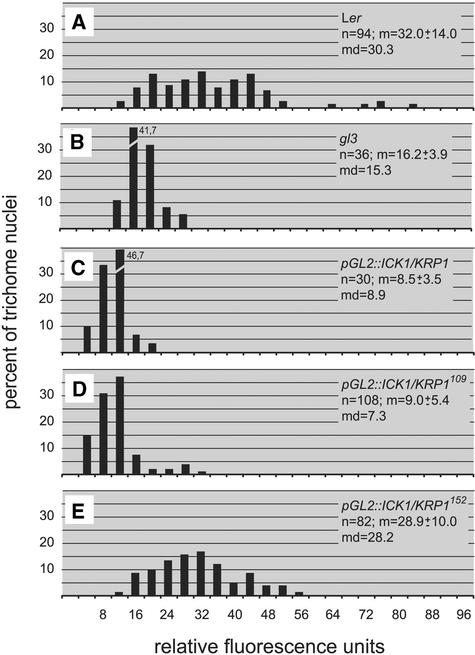
To analyze the function of the CDK inhibitor protein ICK1/KRP1 in a single-celled background, we expressed the coding sequence of ICK1/KRP1 in Arabidopsis trichomes using the GLABRA2 (GL2) promotor (Wang et al., 1997; Szymanski et al., 1998; De Veylder et al., 2001). Constitutive misexpression of CDK inhibitors under the control of the 35S promotor of Cauliflower mosaic virus (CaMV 35S) was reported to reduce endoreduplication levels in Arabidopsis leaves (Wang et al., 2000; De Veylder et al., 2001; Jasinski et al., 2002). Consistently, by measuring the fluorescence of 4′,6-diamidino-2-phenylindole (DAPI)–stained nuclei, we found that trichomes that misexpressed ICK1/KRP1 (pGL2:ICK1/KRP1) contained less DNA than wild-type trichomes (Figures 1A, 1B, 2A, and 2C). Although wild-type trichomes reached a DNA content of ∼30C (constituting approximately four endoreduplication rounds), pGL2:ICK1/KRP1 trichome nuclei had an average DNA content of ∼9C (corresponding to only approximately two rounds), clearly less than the trichome mutant glabra3 (gl3), with an average DNA content of 15C (approximately three endoreduplication rounds) (Figures 2B and 2C) (Hulskamp et al., 1994).
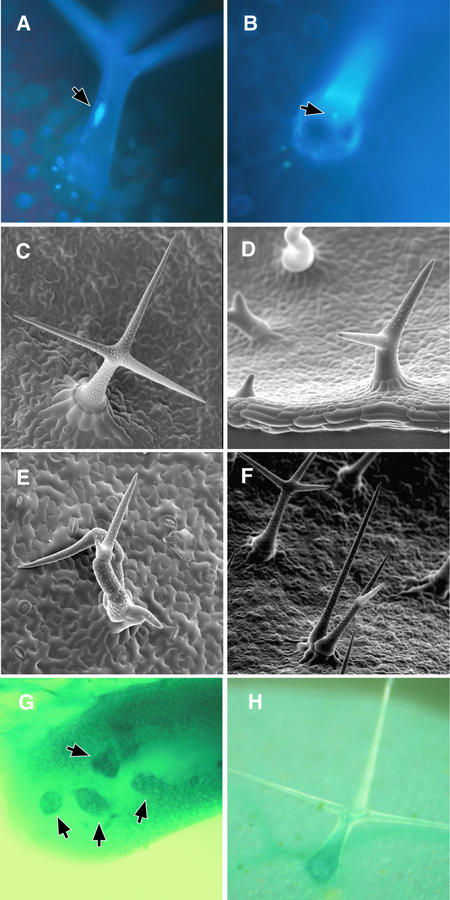
Figure 1.
Morphological Analysis.
(A) Light micrograph of a DAPI-stained wild-type trichome with a characteristically endoreduplicated nucleus (arrow).
(B) Light micrograph of a DAPI-stained pGL2:ICK1/KRP1 trichome with a much smaller nucleus (arrow) at the same magnification as in (A).
(C) Scanning electron micrograph of a mature wild-type trichome.
(D) Scanning electron micrograph of typical small and underbranched pGL2:ICK1/KRP1 trichomes at the same magnification as in (C).
(E) Scanning electron micrograph of clustered and multicellular pGL2:CYCD3;1 trichomes.
(F) Scanning electron micrograph of small but clustered and multicellular pGL2:CYCB1;2 pGL2:ICK1/KRP1 trichomes at the same magnification as in (E).
(G) and (H) Light micrographs of whole-mount GUS staining of the CDKA;1 reporter line pCDC2a:GUS.
(G) Strong expression of the CDKA;1 reporter in very young trichome cells (arrows).
(H) CDKA;1 reporter expression in a differentiated mature trichome cell revealing that CDKA;1 is expressed throughout the lifetime of a trichome cell.
Figure 2.
Analysis of DNA Content.
Distribution of DNA contents given in relative fluorescence units (RFUs). The RFUs are calibrated with wild-type and gl3 trichome nuclei so that 2 RFUs represent ∼2C by defining the major peak in the wild-type trichomes as 32C and the major peak in gl3 as 16C in accordance with previously measured trichome nuclei (Schnittger et al., 1998; Szymanski and Marks, 1998; Walker et al., 2000). The sample size (n), the mean ± sd (m), and the median (md) are given.
(A) Wild-type Landsberg erecta (Ler).
(B) gl3.
(C) pGL2:ICK1/KRP1.
(D) pGL2:ICK1/KRP1109.
(E) pGL2:ICK1/KRP1152.
In contrast to what has been reported from the ubiquitous expression of ICK/KRPs, we found that not only the DNA amount but also the cell size was reduced in pGL2:ICK1/KRP1 trichomes (Figures 1C and 1D). To quantify this cell size reduction, we determined the area of DAPI-stained trichomes in optical cross-sections as a measure of cell size. Whereas wild-type trichomes reached an average of 13,000 μm2, ICK1/KRP1-expressing trichomes attained an area of only ∼8000 μm2 (Table 1). Interestingly, gl3 mutant trichomes covered a smaller area, ∼6000 μm2. Next, we determined the ratios of cell area to DNA content (Table 1). Assigning Landsberg erecta a relative value of 1, we observed that the size-to-DNA ratios for gl3 mutant trichomes also was 1. By contrast, ICK1/KRP1-misexpressing trichomes showed a ratio of ∼2, demonstrating that their cell size was much larger with respect to their small nuclei (Table 1). Thus, we found evidence for two growth mechanisms: a DNA-dependent mode setting up the approximate area in which growth could take place (responsible for the absolute reduction in ICK/KRP-misexpressing trichomes), and a DNA-independent mechanism (accountable for the relative increase in cell size with respect to DNA amount).
Table 1.
Trichome Cell Size
| Line | Trichome Areaa | No. of Trichomes | Area to DNA Content Ratiob |
|---|---|---|---|
| Landsberg erecta | 12,800 ± 3,200 (13,000) | 116 | 1.0 |
| g/3 | 7,300 ± 2,500 (6,000) | 103 | 0.9 |
| ICK1/KRP1 | 8,200 ± 1,900 (8,000) | 119 | 2.1 |
| ICK1/KRP1109 | 8,700 ± 2,300 (9,000) | 118 | 2.9 |
| ICK1/KRP1152 | 12,600 ± 2,900 (12,000) | 107 | 1.0 |
All trichomes on rosette leaves 3 or 4 were measured from at least two different plants per line. Data shown are average (μm2) ± sd (median).
Median trichome area and median DNA content as determined in this study (Figure 2) were used. The Landsberg erecta area-to-DNA ratio was set to 1.
In addition to cell size and DNA content, we found that branch number was reduced in ICK1/KRP1-misexpressing trichomes. Whereas wild-type trichomes were predominantly three branched (∼96%), the majority of ICK1/KRP1 trichomes were only two branched (∼70%), and a significant proportion were unbranched (12%) (Figures 1C and 1D, Table 2). In several trichome mutants, a similar correlation of trichome cell size, DNA amount, and branch number can be seen, which led to a previous model of branch initiation by DNA amount (Folkers et al., 1997). Because we presumably targeted cell cycle progression by ICK1/KRP1 expression directly, our data corroborate this hypothesis with molecular findings.
Table 2.
Trichome Branch Number
| No. of Branches (% per Leaf)b
|
|||||
|---|---|---|---|---|---|
| Linea | 1 | 2 | 3 | 4 | No. of Trichomes |
| Landsberg erecta | 0.0 ± 0.0 | 0.5 ± 1.5 | 95.8 ± 3.2 | 3.7 ± 3.5 | 261 |
| ICK1/KRP1 | 11.8 ± 5.9 | 70.1 ± 15.1 | 18.1 ± 12.1 | 0.0 ± 0.0 | 252 |
| ICK1/KRP1109 | 30.9 ± 14.5 | 62.1 ± 14.5 | 6.9 ± 8.6 | 0.0 ± 0.0 | 227 |
| ICK1/KRP1152 | 0.0 ± 0.0 | 0.0 ± 0.0 | 98.5 ± 6.8 | 1.5 ± 0.7 | 323 |
| Landsberg erecta × ICK1/KRP1109 | 6.9 ± 5.0 | 64.4 ± 13.1 | 28.6 ± 14.1 | 0.0 ± 0.0 | 377 |
| ICK1/KRP1109 × Landsberg erecta | 14.2 ± 9.2 | 71.9 ± 9.2 | 13.9 ± 9.4 | 0.0 ± 0.0 | 336 |
| GFP × ICK1/KRP1109 | 9.4 ± 6.9 | 85.2 ± 7.2 | 5.4 ± 4.4 | 0.0 ± 0.0 | 283 |
| ICK1/KRP1109 × GFP | 10.5 ± 8.1 | 80.4 ± 13.5 | 9.0 ± 7.7 | 0.0 ± 0.0 | 331 |
| CYCD4;1 | 0.0 ± 0.0 | 1.5 ± 1.8 | 98.4 ± 1.7 | 0.1 ± 0.2 | 730 |
| CYCD4;1 × ICK1/KRP1109 | 17.5 ± 11.8 | 69.6 ± 10.7 | 12.8 ± 8.4 | 0.0 ± 0.0 | 375 |
| ICK1/KRP1109 × CYCD4;1 | 14.1 ± 9.9 | 79.4 ± 9.3 | 6.5 ± 6.1 | 0.0 ± 0.0 | 340 |
| CYCD3;1 | NDc | ND | ND | ND | ND |
| ICK1/KRP1109 × CYCD3;1 | 0.0 ± 0.0 | 8.5 ± 5.7 | 91.1 ± 5.6 | 0.4 ± 0.6 | 662 |
| CYCD3;1 × ICK1/KRP1109 | 0.3 ± 0.5 | 2.6 ± 3.2 | 96.3 ± 3.4 | 0.8 ± 1.2 | 429 |
| CDKA;1 | 0.0 ± 0.0 | 0.4 ± 0.7 | 92.2 ± 4.8 | 7.4 ± 4.7 | 659 |
| CDKA;1 × ICK1/KRP1109 | 0.2 ± 0.5 | 7.4 ± 6.1 | 91.5 ± 5.6 | 0.9 ± 1.4 | 338 |
| ICK1/KRP1109 × CDKA;1 | 0.0 ± 0.0 | 5.5± 4.5 | 93.6 ± 4.4 | 0.8 ± 1.4 | 336 |
| CDKB1;1 | 0.0 ± 0.0 | 0.7 ± 1.1 | 97.4 ± 2.2 | 1.8 ± 1.8 | 714 |
| CDKB;1 × ICK1/KRP1109 | 4.0 ± 3.7 | 51.3 ± 10.8 | 44.7 ± 11.4 | 0.0 ± 0.0 | 376 |
| ICK1/KRP1109 × CDKB1;1 | 15.6 ± 12.6 | 70.4 ± 16.0 | 14.1 ± 9.5 | 0.0 ± 0.0 | 313 |
Single lines are homozygous; crosses refer to the F1 generation in which each construct is heterozygous.
All trichomes on rosette leaves 3 and 4 were counted from at least 10 plants per line. Data shown are averages ± sd. Branch numbers with the highest percentage are shown in boldface.
ND, Not determined because CYCD3;1-misexpressing trichomes develop multicellular trichomes each with several branching points.
ICK1/KRP1-Misexpressing Trichomes Undergo Cell Death
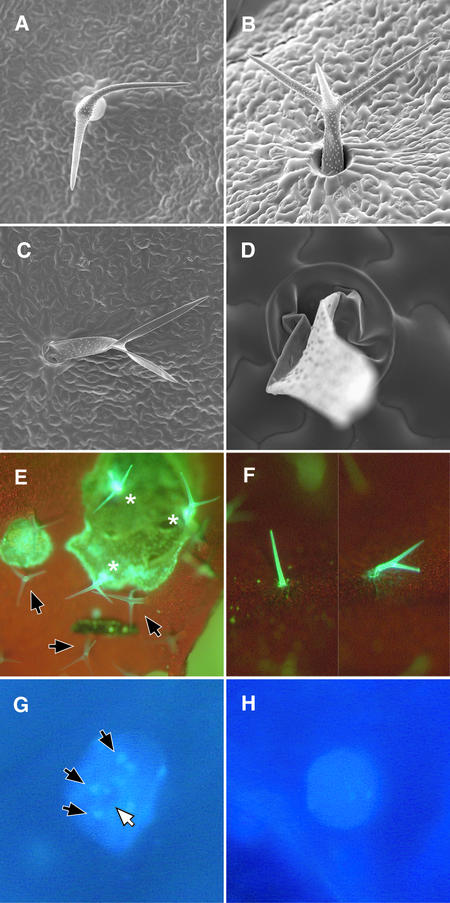
Reinspecting leaves at later developmental stages, we made a surprising discovery: pGL2:ICK1/KRP1 trichomes died. The first indication of a cell death process was a peculiar alteration of the cell morphology, including a bending and leaning of trichome branches (Figure 3A). This was followed by trichome collapse and finally full degradation, leaving only rudiments of a former trichome structure (Figures 3B to 3D). Concomitant with morphological alterations and typical of many cell death processes, the ICK1/KRP1 trichomes displayed a strong increase in yellow autofluorescence under blue light excitation, probably resulting from an accumulation of phenolic compounds (Figures 3E and 3F) (Kosslak et al., 1997; Takahashi et al., 1999; Hellmann et al., 2000).
Figure 3.
Analysis of Cell Death.
(A) to (D) Scanning electron micrographs of dying pGL2:ICK1/KRP1 trichomes. Cell death starts by bending of branches (A), followed by trichome collapse (B). Finally, trichomes become dismantled (C), leaving only rudiments behind (D).
(E) Light micrograph of mechanically ablated (stars) and living wild-type trichomes (arrows). Dead trichomes show strong yellow autofluorescence under blue light excitation.
(F) Light micrograph of a dead pGL2:ICK1/KRP1 trichome with strong yellow autofluorescence under blue light excitation.
(G) Light micrograph of a DAPI-stained wild-type trichome nucleus with brightly fluorescing chromocenters (black arrows mark the outermost three chromocenters) and the nucleolus (white arrow).
(H) Light micrograph of a DAPI-stained pGL2:ICK1/KRP1 trichome nucleus at the same magnification as in (G). Neither chromocenters nor a nucleolus can be recognized.
To further characterize this cell death, and in an attempt to distinguish programmed cell death from a rather unspecified necrotic cell death, we analyzed several cellular parameters, particularly the cytoskeleton and the nucleus, because they are known to undergo cell death–specific alterations. For an analysis of the cytoskeleton, we crossed ICK1/KRP1-misepxressing plants with plants expressing either a mitogen-activated protein–green fluorescent protein (GFP) or a TALIN-GFP fusion protein, which decorate the microtubules and microfilaments, respectively (Olson et al., 1995; Kost et al., 1998). Using both fusion proteins, we detected no early alteration of the cytoskeleton (data not shown). Also, the morphology of the nucleus and the integrity of the nuclear membrane were found to be undisturbed in cross-sections and by analysis of a β-glucuronidase (GUS) targeted to the nucleus, respectively (data not shown).
When the nuclear structure and the DNA were inspected in more detail using whole-mount DAPI staining, we discovered alterations from the wild type in the ICK1/KRP1 lines. In wild-type DAPI-stained trichome nuclei, there usually are 10 to 12 bright dots that appear; these might represent the chromocenters (Figure 3G, black arrows). Chromocenters comprise heterochromatin and coincide mostly with the centromeres (Nagl, 1976; Fransz et al., 2000). In the ICK1/KRP1-misexpressing trichomes, we observed that many of the nuclei contained a reduced number of chromocenters, including cells without chromocenters, implying that these structures were lost over time (Figure 3H). The decline in chromocenters was not attributable to reduced endoreduplication levels. This was evident because in the gl3 mutant trichomes and in less endoreduplicated wild-type cells (epidermal pavement cells and hypocotyl epidermis cells), the chromocenters were clearly visible (data not shown). The degradation of DNA is a hallmark of cells undergoing programmed cell death, and it has been reported that this degradation starts at heterochromatic regions of the genome (Compton, 1992; Dullea et al., 1999). However, we found no other signs of DNA degradation leading to the typical highly condensed nuclei of apoptotic cells. In addition to the loss of chromocenters, we discovered that the nucleoli disappeared in ICK1/KRP1 trichomes (Figures 3G, white arrow, and 3H). Also, the destruction of nucleoli has been seen during some variants of programmed cell death (Commean et al., 1985; Horky et al., 2001; Smetana, 2002).
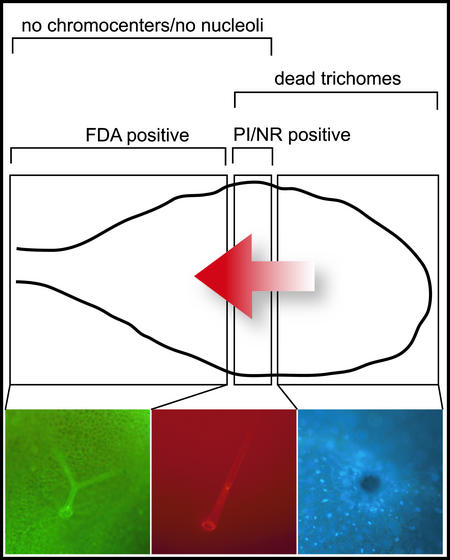
Next, we tried to resolve the temporal order of cell death processes. The vital dyes propidium iodine (PI) and neutral red (NR) mark dead cells because they can enter the cells only if the membrane integrity is disrupted and then stain DNA and the vacuole, respectively (Robinson et al., 2002). By contrast, fluorescein diacetate (FDA) freely crosses the membrane but fluoresces only if ATP is present; thus, it is a marker of living cells (Robinson et al., 2002). In Figure 4, a sketch of a representative leaf is shown. Trichome death started at the apical (oldest) part of the leaf, where the trichomes already were dismantled and could not be stained with any of the dyes. Moving toward the base of the leaf, we occasionally detected PI- and NR-positive cells that presumably were dying. At the base (youngest part), we found living trichomes, as indicated by FDA staining and the absence of PI-marked trichomes. Relating this pattern to the DAPI staining, we determined that in the basal part of the leaf, where cells were still alive, the alterations of the nuclei had started to indicate a defined temporal order of cell death processes, suggesting a form of programmed cell death (Figure 4).
Figure 4.
Temporal Order of Cell Death Processes.
Sketch of a rosette leaf of a pGL2:ICK1/KRP1 plant, with the basal area at left and the apical area at right. Trichome cell death starts at the apical (oldest) part of the leaf and moves toward the base; on older leaves, all trichomes are dead. In the very apical part, only rudiments of trichomes are left, for which no staining could be obtained (bottom right). Centrally, a few trichomes are stained with PI and NR, indicating dying trichomes that have lost their membrane integrity (bottom middle). More basally, trichomes are still alive, as judged by FDA staining and the absence of PI/NR-positive cells (bottom left). Already in living trichomes of the basal area, nuclear degradation processes have started (cf. Figures 3G and 3H).
The ICK1/KRP1 Phenotype Is Specific for the C-Terminal Part of the Protein and Is Regulated Negatively by an N-Terminal Domain
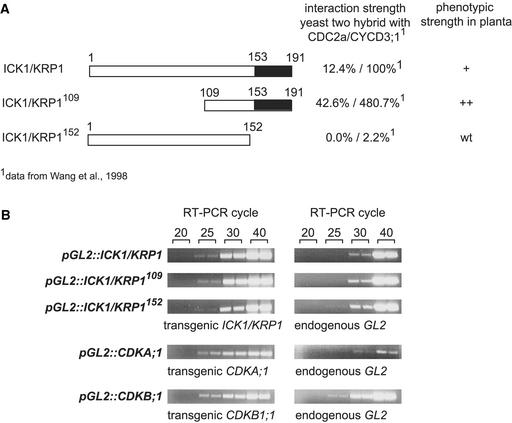
Is the ICK1/KRP1 trichome phenotype caused indirectly by a general overexpression of a protein, or is this phenotype related specifically to the nature of the ICK1/KRP1 protein? Wang et al. (1998) showed in a yeast two-hybrid interaction assay that ICK1/KRP1 consists of at least two functionally distinct domains: a C-terminal domain, which most likely includes two separate regions for CDKA;1 and CYCD3;1 binding, and a negative regulatory domain at the N terminus, which, if deleted, increases the physical interaction with CDKA;1 and CYCD3;1 (Figure 5A). Analogous to the constructs used in this two-hybrid assay, we created plants that misexpressed two truncated ICK1/KRP1 proteins. The first mutant protein contained the first 152 amino acids and thus lacked the CDKA;1- and CYCD3;1-interacting regions (designated ICK1/KRP1152). In the second ICK1/KRP1 mutant (designated ICK1/KRP1109), the first 108 amino acids were deleted, eliminating a putative negative regulatory domain (Figure 5A).
Figure 5.
Constructs and Expression Analysis.
(A) ICK1/KRP1 constructs used in this analysis and their phenotypes in planta compared with a previous yeast two-hybrid analysis. At top is the full-length ICK1/KRP1 protein of 191 amino acids, with the CDKA;1 and CYCD3;1 interaction domain in the C-terminal part shown in black. In ICK1/KRP1109, the first 108 amino acids are deleted. ICK1/KRP1152 comprises amino acids 1 to 152, with the CDKA;1/CYCD3;1 interaction domain deleted. ICK1/KRP1109 has a stronger phenotype than the full-length ICK1/KRP1 protein (Table 2), demonstrating in planta the existence of a negative regulatory domain in the N-terminal part. ICK1/KRP1152 shows no deviation from wild-type (wt) plants, indicating that for ICK1/KRP1 function, the interaction domain with CDKA;1/CYCD3;1 is necessary.
(B) Semiquantitative RT-PCR showing the relative expression strength of the transgenic constructs pGL2:ICK1/KRP1, pGL2:ICK1/KRP1109, pGL2:ICK1/KRP1152, pGL2:CDKA;1, and pGL2:CDKB1;1 compared with endogenous GL2 expression. The numbers at top (20, 25, 30, and 40) indicate RT-PCR cycle number. All transgenes appear to be expressed similarly.
Transgenic plants that misexpress ICK1/KRP1152 were generated, and all of them showed no deviation from wild-type plants, as determined by DNA content, cell size, and branch number (Figure 2E, Tables 1 and 2). A comparison of expression levels by reverse transcriptase–mediated (RT) PCR confirmed that ICK1/KRP1152 was expressed similarly to the full-length ICK1/KRP1 gene (Figure 5B). This finding demonstrates that the ICK1/KRP1 phenotype was specific for the region that contains the interaction domains for CDKA;1 and CYCD3;1. Conversely, trichomes that express the N-terminally shortened protein showed an enhanced ICK1/KRP1 phenotype. The number of unbranched trichomes increased from 12 to 31%, and at the same time, the number of three-branched trichomes was reduced further from 18 to 7% (Table 2). Measurements of DNA content revealed an additional reduction in the amount of DNA (full-length ICK1/KRP1 had ∼9C, whereas ICK1/KRP1109 had ∼7C) (Figures 2C and 2D, Table 1). Thus, we confirmed in planta the existence of a negative regulatory domain in the N terminus of the ICK1/KRP1 protein.
In Vivo Targets of ICK1/KRP1 Action Are an A-Type CDK and Specifically One D-Type Cyclin, but Not a B-Type CDK or a Mitotic B-Type Cyclin
To find in planta targets of ICK1/KRP1, we set up a genetic interaction system using the strong trichome phenotype of pGL2:ICK1/KRP1109 plants. The effect of coexpressed candidate-interactor genes in trichomes was analyzed for any influence on branching pattern, cell size, and cell survival.
It has been reported that ICK1/KRP1 binds to D-type cyclins in yeast two-hybrid assays (Wang et al., 1998; De Veylder et al., 2001; Zhou et al., 2002a). Therefore, we crossed homozygous plants that express CYCD4;1 (CYCD2;2) in trichomes to our ICK1/KRP1109 plants. The expression of CYCD4;1 by itself has no influence on trichome morphology (Schnittger et al., 2002a). In the double transgenic plant pGL2:ICK1/KRP1109 pGL2:CYCD4;1, the same number of branches was observed with crosses of ICK1/KRP1109 to wild-type or pGL2:GFP:GUS:nls plants, which suggests that CYCD4;1 and ICK1/KRP1 do not interact in planta (Table 2).
Next, we introduced plants that misexpress CYCD3;1 in trichomes into the ICK1/KRP1109 line. The sole CYCD3;1 expression in trichomes results in multicellular trichomes and, because of divisions that occur before outgrowth, also in clusters of directly neighboring trichomes (Figure 1E, Table 3) (Schnittger et al., 2002a). In the F1 generation of pGL2:ICK1/KRP1109 crossed to pGL2:CYCD3;1, we found only unicellular trichomes, which were equally spaced, fully branched, and grown out to the wild-type cell size and which did not undergo cell death (Tables 2 and 3). The mutual rescue of the ICK1/KRP1109 and CYCD3;1 phenotypes shows a functional interaction of the two proteins in planta. A similar result was obtained recently by Jasinski et al. (2002), who rescued Arabidopsis plants that misexpressed the tobacco CDK inhibitor NtKIS1a by coexpressing the Arabidopsis CYCD3;1 under the control of the CaMV 35S promotor.
Table 3.
Trichome Cluster Frequency
| Line | Cluster Frequency (% per leaf)a |
No. of TISb |
|---|---|---|
| Landsberg erecta | 0.2 ± 0.9 | 409 |
| CYCD3;1 × Landsberg erecta | 94.4 ± 4.4 | 804 |
| CYCD3;1 × ICK1/KRP1109 | 1.4 ± 2.3 | 423 |
| CYCB1;2 × Landsberg erecta | 13.5 ± 7.9 | 649 |
| CYCB1;2 × ICK1/KRP1109 | 19.3 ± 9.4 | 378 |
Twenty rosette leaves (number 3 or 4) were counted from at least 10 plants per line. Data shown are averages ± sd.
TIS, trichome initiation site, the place where one or more directly neighboring (clustered) trichomes are found.
Because CYCD3;1 misexpression in trichomes induces DNA replication and cell division, CYCD3;1 could function at both the G1-S and G2-M transition points (Schnittger et al., 2002a). To define the time of action for ICK1/KRP1 more closely, we crossed the ICK1/KRP1109 line to plants that mis-expressed in trichomes the mitotic B-type cyclin CYCB1;2. As seen for CYCD3;1, the misexpression of CYCB1;2 also caused multicellular trichomes and clustering, presumably directly and exclusively, promoting the entry into mitosis (Schnittger et al., 2002b). The offspring of the ICK1/KRP1109 × CYCB1;2 cross showed an additive phenotype in which small and multicellular trichomes in clusters were formed (Figure 1F, Table 3). This finding indicates that ICK1/KRP1 does not block the mitosis-promoting activity but rather functions on a cyclin D complex, perhaps at the G1-S transition.
A further candidate for a target of ICK1/KRP1 action is CDKA;1 (CDC2a), which also was identified as an interacting protein with ICK1/KRP1 (Wang et al., 1998; De Veylder et al., 2001; Zhou et al., 2002a). To test for a CDKA;1–ICK1/KRP1 genetic interaction, we generated transgenic plants that misexpress CDKA;1 in trichomes and compared their phenotype with that of the double transgenic plants. CDKA;1 misexpression resulted in no alteration of trichome morphology in the transgenic plants analyzed (Table 2, Figure 5B, and data not shown). We next crossed three pGL2:CDKA;1 lines to the ICK1/KRP1109 plants. The offspring of these reciprocal crosses showed a complete rescue of the ICK1/KRP1109 phenotype, including the branch-number and cell-survival defects (Table 2 and data not shown). To test for the specificity of the interaction between ICK1/KRP1 and CDKA;1, we expressed another CDK, CDKB1;1, in ICK1/KRP1109-containing trichomes. In a yeast two-hybrid assay, CDKB1;1 failed to interact with ICK1/KRP1 (De Veylder et al., 2001; Zhou et al., 2002a). From >30 transgenic pGL2:CDKB1;1 plants generated, none showed a change in trichome morphology (Table 2 and data not shown). In crosses with pGL2:CDKB1;1 plants, we found that despite a strong expression of the transgene, there was no rescue of the ICK1/KRP1109 trichome phenotype, indicating that ICK1/KRP1 interacts specifically with CDKA;1 in vivo (Figure 5B, Table 2).
Could the two in vivo interactors, CYCD3;1 and CDKA;1, also be the targets of ICK1/KRP1 misexpression in trichomes? CYCD3;1 was shown previously not to be expressed in trichomes; thus, it can be excluded (Schnittger et al., 2002a). To determine whether CDKA;1 is expressed in trichomes, we used a CDKA;1 promotor-reporter line that has been shown to reflect the mRNA pattern of CDKA;1 (Hemerly et al., 1993; Jacqmard et al., 1999). Consistent with the results of Imajuku et al. (2001), we found that CDKA;1 is expressed throughout the development of a trichome cell, suggesting a downregulation of CDKA;1 kinase activity as one likely reason for the ICK1/KRP1-misexpression phenotype in trichomes (Figures 1G and 1H).
DISCUSSION
What determines cell size, and how is cell size connected to the developmental program of a cell? On the one hand, DNA amount has been recognized as one putative determinant of cellular size, because a positive correlation can be found in many plant species (Nurse, 1985; Kondorosi et al., 2000; Gregory, 2001). On the other hand, the misexpression of CDK inhibitors and dominant-negative CDKs in Arabidopsis and tobacco leaves has resulted in a reduction of cell cycle progressions, leading to fewer cells with smaller (less endoreduplicated) nuclei. These fewer cells were larger than the wild-type cells, demonstrating that cell cycle progression and cell growth can be uncoupled, which raises questions about the impact of DNA amount as a cell size regulator (Hemerly et al., 1993; Wang et al., 2000; De Veylder et al., 2001). However, especially from work in animal systems, it is known that cell size also is regulated from an organ size–derived signal that is thought to maintain organ size by compensatively controlling cell division and cell expansion. For instance, overexpression of the Drosophila E2F transcription factor resulted in an increase in imaginal disc cells, but the overall disc size was not altered as a result of a concomitant reduction in cell size (Neufeld et al., 1998). Similarly, the increase of cell size in plants that misexpress CDK inhibitors and dominant-negative CDKs has been regarded as a compensative mechanism for the reduced number of cells (Hemerly et al., 1993; Doonan, 2000; De Veylder et al., 2001).
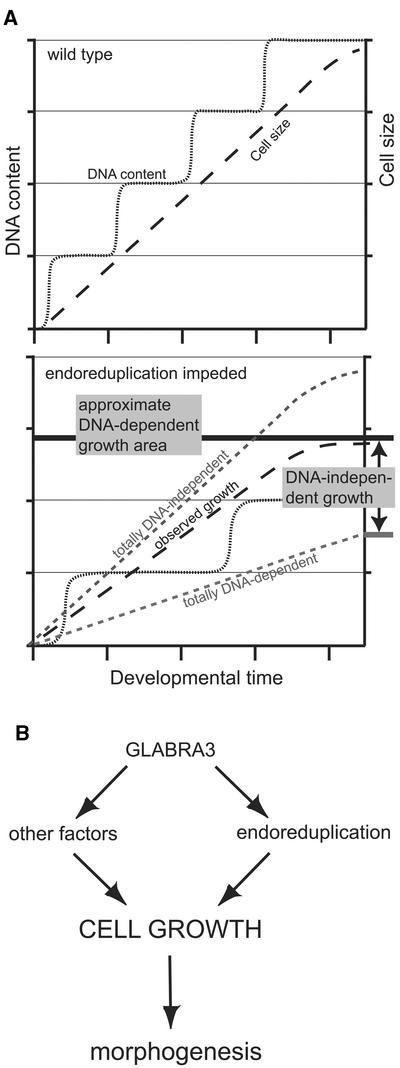
In an attempt to minimize the presumed compensatory influence of an organ context, we used Arabidopsis trichomes to analyze the effect of one CDK inhibitor, ICK1/KRP1, on cell growth. Trichomes do not contribute much to the final leaf size, because on the fully expanded first leaf pair in Landsberg erecta, only approximately nine trichomes are formed per leaf (Larkin et al., 1996). Thus, a reduction in trichome size is not expected to lead to additional organ growth. The use of this single-cell system enabled us to reveal two growth mechanisms and their relative contributions for trichome growth (Figure 6A). First, a reduction in endoreduplication levels was associated with a reduced overall cell size, supporting a role for DNA amount as a regulator of cell growth, at least for trichomes. This finding also showed that ICK1/KRP1 is not a positive regulator of cell growth. On the other hand, comparing the ratios of cell size and DNA content, we also saw the influence of a DNA-independent growth mechanism, because trichomes could grow in a certain range without an increase in DNA content, leading cell autonomously to an uncoupling of DNA amount from cell growth (Figure 6A). Therefore, in 35S:ICK/KRP plants, the postulated organ size checkpoint–derived signal might strongly promote a DNA-independent expansion program, eclipsing/overpowering a primary influence of DNA content.
Figure 6.
Model of Growth Control in Trichomes.
(A) Scheme of the two observed growth control mechanisms. In the wild type, a given ratio of nuclear size to cell size is observed (top). If endoreduplication is reduced, as in the ICK1/KRP1-misexpressing cells, two extreme scenarios are possible (bottom, dashed lines). Totally DNA-independent growth would lead to the same cell size as in wild-type trichomes; this was not observed. Alternatively, totally DNA-dependent growth would lead to cells with the same cell size-to-nuclear size ratio; this also was not observed. Instead, both control mechanisms seemed to be involved. A DNA amount–dependent mechanism sets up the approximate area of cell growth; this area might vary between different cell types. Within this range, a DNA-independent growth mechanism is responsible for the final cell size. However, this DNA-independent growth can overpower the DNA-dependent mechanism. For further discussion, see the text.
(B) Proposed model for cell growth regulation and its relationship to cell morphology in trichomes. Besides other factors, one determinant of cell growth is nuclear size. In turn, cell growth controls cell morphology. GL3 regulates not only endoreduplication but also other determinants of cell growth. Thus, gl3 mutants have smaller cells but show the same ratio of cell size to nuclear size as the wild type. For further discussion, see the text.
The relative contributions of the two growth modes to the final cell size might vary among different cell types and tissues. Because of their highly elaborate structure, Arabidopsis trichomes could be very sensitive to changes in DNA amount, whereas epidermal cells could grow comparatively larger. This notion is in agreement with the observation that the transgenic expression of the Arabidopsis ICK1/KRP1 gene in petals of Brassica plants resulted in a diminished organ size, mostly as a result of a reduced cell number with very little change in cell size (Zhou et al., 2002b).
Another reason for the different observations regarding absolute cell size might be the expression strength of the transgene. The GL2 promotor used in this study is expressed much more strongly in trichomes than the CaMV 35S promotor used in previous studies on ICK1/KRP1 (A. Schnittger, unpublished data).
The finding that ICK1/KRP1-misexpressing trichomes had reduced DNA contents but comparatively large cell sizes sheds new light on the function of the GL3 gene. Formerly, gl3 was described as an endoreduplication mutant (Hulskamp et al., 1994). If only DNA amplification were affected in the gl3 mutant, we would have expected a similar uncoupling of cell size and DNA amount as in the ICK1/KRP1-misexpressing trichomes. But because in gl3 both cell size and DNA content are reduced, gl3 should be regarded as a general growth mutant (Figure 6B). GL3 encodes bHLH transcription factor, and interestingly, overexpression of GL3 in trichomes leads to larger trichomes with more branches and a higher DNA amount (Payne et al., 2000; S. Schellmann and M. Hülskamp, unpublished data). Thus, the previous model of branch initiation by DNA amount should be expanded with a potential role for cell size on the differentiation pathway (Figure 6B).
What are the targets of ICK1/KRP1 cell growth control/cell cycle progression in trichomes? We found that the two targets of ICK1/KRP1 from the yeast two-hybrid analysis, CYCD3;1 and CDKA;1, also interacted functionally with ICK1/KRP1 in planta. CYCD3;1 seems to be expressed in young and meristematic tissues; thus, it is tempting to speculate that ICK1/KRP1 might have a function in balancing this cyclin in proliferating tissues (Riou-Khamlichi et al., 1999; Schnittger et al., 2002a). An expression analysis of ICK/KRPs at the cellular level in meristematic tissues will be a first step in answering this question. CYCD3;1 was not expressed in Arabidopsis trichomes; therefore, it appears not to represent a target of ICK1/KRP1 in the pGL2:ICK1/KRP1 lines investigated here (Schnittger et al., 2002a). However, because the subfamily of D3-type cyclins contains two more members in Arabidopsis, these genes are additional candidates for interactors with ICK1/KRP1 in trichomes (Vandepoele et al., 2002). In addition, Zhou et al. (2002a) and De Veylder et al. (2001) found that ICK/KRPs bind to CYCD1 and CYCD2 in yeast two-hybrid assays, adding two more candidates to the list of in planta targets of ICK1/KRP1. However, for another D-type cyclin, CYCD4;1 (CYCD2;2), which was not tested in a two-hybrid assay, we did not obtain a rescue by crossing plants expressing this cyclin to ICK1/KRP1-misexpressing plants, suggesting that ICK1/KRP1 does not interact with this D-type cyclin. ICK1/KRP1 also appeared not to interact with a B-type cyclin, because in our experiments, double transgenic plants that misexpress both ICK1/KRP1 and CYCB1;2 displayed an additive phenotype with small and multicellular trichomes. This finding suggests that the time of action for ICK1/KRP1 is restricted to the G1-S transition.
In animals as well, CDK inhibitor proteins (Cip/Kip family) function at the G1-S transition (Sherr and Roberts, 1999). However, in animals, the Cip/Kip proteins are not only negative factors but also serve in certain cell types as assembly factors for CDK4/6 with cyclin D; thus, they also have a positive function for the entry into G1-phase (Sherr and Roberts, 1999). In animals, the function of Cip/Kip appears to be concentration dependent; at low stoichiometries, the assembly function takes place, whereas at higher concentrations of Cip/Kip, CDK4/6 Rb-kinase activity is blocked (LaBaer et al., 1997). In plants, only a negative function for cell cycle progres-sion has been found for ICK/KRPs. However, both the CaMV 35S and GL2 promotors are expressed quite strongly; thus, only one aspect of a subtle regulation mechanism might have been captured.
At the sequence level, only the C terminus of ICK1/KRP1 is related to the N-terminal part of p21Cip1 and p27Kip1. This region was shown for both animal and plant proteins to bind to CDKs (Polyak et al., 1994; Toyoshima and Hunter, 1994; Wang et al., 1998). The CYCD3;1 binding domain might reside immediately before the CDK interaction domain in the plant CDK inhibitor protein (Wang et al., 2000). Indeed, when we expressed a C-terminally truncated plant ICK1/KRP1 in which both domains were eliminated, no effect was obtained. By contrast, our misexpression of an N-terminally truncated ICK1/KRP1 protein resulted in an enhanced trichome phenotype. The nature of this negative regulatory domain remains obscure. Post-translationally, animal p27Kip1 is regulated by localization and degradation. p27Kip1 is phosphorylated by CDK2, creating a binding site for Skp2-containing E3-ubiquitin ligase, and upon SCF-mediated ubiquitylation, it is degraded via the proteasome (Carrano et al., 1999; Montagnoli et al., 1999; Tsvetkov et al., 1999). However, we found no consensus CDK phosphorylation site (S/T-P-X-K/R) in the ICK1/KRP1 sequence that could mark the protein for degradation. We also found no obvious transport signature that could have been eliminated by truncation of the protein. It remains a challenge to assign functions for the N terminus of ICK1/KRP1.
In animals, p27Kip1 also is a key regulator of cell survival (Lloyd et al., 1999; Philipp-Staheli et al., 2001, and references therein). In animal systems, cell cycle progression seems to be connected inherently with cell survival; by contrast, evidence for a similar connection in plants is scarce and indirect at best. Here, we report that ICK1/KRP1 misexpression in plants led to cell death, unraveling potential parallels in animals. The question now is what kind of cell death is executed. Trichomes could die nonspecifically, perhaps as a result of the observed altered cell size-to-DNA ratio. In this scenario, trichomes would not contain enough DNA to support the large cell, resulting in a classic case of necrosis. Arguing against this possibility, we found evidence that the pGL2:ICK1/KRP1 trichomes initiated a programmed cell death program; as in living cells, the nuclear structure started to change (i.e., the chromocenters and the nucleolus disappeared). Additional experiments will be necessary to analyze the context of this cell death, including the determination of whether a ubiquitin-mediated protein-degradation program is involved. Because we have identified CDKA;1 as a likely target of the ICK1/KRP1 action described here, cell death might be dependent on the activity of this kinase. However, attempts to phenocopy ICK1/KRP1-induced cell death by silencing the CDKA;1 gene in trichomes, which would support this model, have failed to date (A. Schnittger, unpublished data). ICK1/KRP1 expression has been found to increase in aging leaves; concomitantly, a reduction in CDK activity was observed (Wang et al., 1998). Our collective results now offer a link between ICK1/KRP1 expression and cell death during leaf senescence.
METHODS
Plant Material, Growth Conditions, and Plant Transformation
Arabidopsis thaliana plants were grown under long-day conditions (16 h of light, 8 h of darkness) between 18 and 25°C under standard greenhouse conditions. The Arabidopsis ecotype Landsberg erecta was used as a wild-type control. For pGL2:ICK1/KRP1, pGL2:ICK1/KRP1152, pGL2:ICK1/KRP1109, pGL2:CDKA;1, pGL2:CDKB1;1, and pGL2:GFP:GUS:nls, we generated at least 20 transgenic plants and selected homozygous lines in the T3 generation. For ICK1/KRP1 and ICK1/KRP1109, >50% of the transgenic plants showed a phenotype. For further analysis, a representative reference line with strong transgene expression was chosen. The pGL2:CYCB1;1, pGL2: CYCB1;2, and pGL2:CYCD3;1 expression lines have been described previously (Schnittger et al., 2002a, 2002b). Transgenic plants were generated as described previously (Schnittger et al., 2002b).
Expression Constructs
To achieve trichome expression of all constructs described in this article, the plant transformation vector pBI101.1pGL2 containing a 2.1-kb HindIII-NheI fragment from the 5′ upstream region of the GL2 gene was used (a gift from David Marks, University of Minnesota, Twin Cities) (Szymanski et al., 1998). To generate the pGL2:ICK1/KRP1 construct, the ICK1/KRP1 cDNA was amplified with the gene-specific primers 5′-CCCCGGGATGGTGAGAAAATATAGAAAAGC-3′ and 5′-GCGAGCTCTCACTCTAACTTTACCCATTCG-3′, containing SmaI and SacI restriction sites, from a cDNA library generated from Landsberg erecta flowers and young siliques (a gift from Markus Grebe and Marika Kientz, Zentrum für Molekularbiologie der Pflanzen, University of Tübingen, Germany). This cDNA was subcloned in pBS (pART70), excised with SmaI and SacI, and subcloned into SmaI-SacI–digested pBI101.1pGL2 to generate pART71. The truncated ICK1/KRP1 version ICK1/KRP1152 was generated via PCR with the primers 5′-CCCCGGGATGGTGAGAAAATATAGAAAAGC-3′ and 5′-TGAGCTCTTAAATTTCCGATTCCGTTGGCAT-3′, containing SmaI and SacI sites, respectively, using pART70 as a template. This PCR product was subcloned in pGEMT, sequenced, excised with SmaI and SacI, and subcloned into SmaI-SacI–digested pBI101.1pGL2. ICK1/KRP1109 was generated using the primers 5′-GGATCCACAATGGAATTTGAATCGGCGGTTAAAGAA-3′ and 5′-CGAGCTCTCACT-CTAACTTTACCCATTCG-3′, containing BamHI and SacI sites, respectively, using pART70 as a template. This PCR product was subcloned in pGEMT, sequenced, excised with SmaI and SacI, and subcloned into BamHI-SacI–digested pBI101.1pGL2. To generate the pGL2:CDKA;1 construct, the CDKA;1 cDNA was excised from pCDC2aAT (a gift from Dirk Inzé) (Ferreira et al., 1991) with XhoI and SacI, treated with Klenow fragment to fill in the recessed 3′ overhang, and inserted into SmaI-SacI–digested pBI101.1pGL2 to yield plasmid pART63. To generate the pGL2:CDKB1;1 construct, the CDKB1;1 cDNA was excised from pGEM2bF (a gift from Dirk Inzé, Gent University, Gent, Belgium) (Segers et al., 1996) with BamHI and XbaI, treated with Klenow fragment to fill in the recessed 3′ overhang, and inserted into BamHI-Ecl136II–cleaved pBI101.1pGL2 to yield plasmid pART66.
Unless stated otherwise, all manipulations were performed using standard molecular methods (Sambrook et al., 1989; Ausubel, 1994).
Crosses of Transgenic Lines
Only homozygous T3 plants, as determined by their segregation ratios, were used for crosses, and F1 generation lines were analyzed directly as double transgenic plants. The F2 generation was checked to ensure the segregation of the respective single-transgene phenotypes.
Reverse Transcriptase–Mediated PCR Analysis
RNA was prepared with Dynabeads (Dynal, Oslo, Norway). This RNA was treated with DNase I to ensure the removal of genomic DNA. Reverse transcriptase–mediated (RT) PCR was performed with the TITAN One Tube RT-PCR mix (Roche Diagnostics, Mannheim, Germany). The 5′ primer used was designed against the 5′ untranslated region of the GL2 gene, which is included in the GL2 promotor fragment used in vector construction, whereas the 3′ primers were designed against ICK1/KRP1, CDKA;1, CDKB1;1, and GL2 genes. A total of 20 μL of RT-PCR products, after 20, 25, 30, and 40 cycles, were separated on agarose gels and visualized by UV excitation of ethidium bromide–stained DNA. All RT expression analysis was performed in duplicate and repeated at least once in an independent experiment.
Microscopy and Measurement of Trichome Areas
Light microscopy was performed with an Axiophot microscope (Zeiss, Jena, Germany) equipped with differential interference contrast (Nomarski) and epifluorescence optics. The DISKUS software package (Carl H. Hilgers-Technisches Büro, Königswinter, Germany; version 4.25.7) was used to calculate the trichome area in optical sections. Cryo-scanning electron microscopy was performed as described by Rumbolz et al. (1999).
DNA Measurements
Trichome nuclei were measured as described by Schnittger et al. (1998).
Vitality/Death and 4′,6-Diamidino-2-Phenylindole Stainings
Staining with the vital dye fluorescein diacetate was used to detect living cells (Robinson et al., 2002). A stock solution (5 mg/mL) was prepared in acetone and diluted with PBST (137 mM NaCl, 3.7 mM KCl, 4.3 mM Na2HPO4, 1.4 mM KH2PO4, and 0.1% Tween 20, pH ∼7.3) to a final concentration of 10 pg/mL. Whole leaves or seedlings were vacuum-infiltrated for 15 min, incubated for 15 min, and washed in PBST. Cells were examined under epifluorescence light using the appropriate filter set (excitation filter, 450 to 490 nm; dichroic mirror, 510 nm; barrier filter, 520 nm). The vital dyes neutral red (NR) and propidium iodine (PI) were used to test for the loss of membrane integrity and thus to detect dead or dying cells (Robinson et al., 2002). For NR, a stock solution of 1 mg/mL in water was diluted for staining to a final concentration of 100 μg/mL in PBST. Whole leaves or seedlings were vacuum-infiltrated for 15 min, incubated in this staining solution for 15 min, and washed in PBST. Accumulation of NR within the vacuole was observed under bright light. For PI, a stock solution of 1 mg/mL in water was diluted for staining to a final concentration of 10 μg/mL in PBST. Additionally, a combination of the vital stains fluorescein diacetate and PI was used at the concentrations given above. Whole leaves were vacuum-infiltrated in each case for 15 min, incubated in the staining solution for 15 min, and washed in PBST. Accumulation of PI within the nucleus was observed under epifluorescence light using the appropriate filter set (excitation filter, 535 to 550 nm; dichroic mirror, 565 nm; barrier filter, 590 nm). 4′,6-Diamidino-2-phenylindole staining was performed as described by Schnittger et al. (1998).
β-Glucuronidase Assays
Whole-mount β-glucuronidase staining was performed as described by Sessions et al. (1999).
Computer Work
Photographs were processed using Adobe Photoshop 6.0 and Adobe Ilustrator 9.0 (Mountain View, CA).
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes.
Acknowledgments
We are grateful to Richard Guggenheim and his team from the Raster-Elektronen-Mikroskopie–Labor Universität Basel, especially to Marcel Düggelin, for their help with the scanning electron microscopy analysis. We thank Elmon Schmelzer and Rolf-Dieter Hirtz from the Central Microscopy of the Max-Planck-Institut für Züchtungsforschung in Cologne for advice and help with microscopy devices. We are grateful to Charles N. David and the Department of Zoology at the Universität München for the use of the cytophotometry device. We thank Dirk Inzé and his group for providing the DNA and seeds used in this analysis. We also thank Maren Heese, Michael Lenhard, Seth Davis, and Tom Beeckman for critical reading and helpful comments on the manuscript. This work was supported by grants from the Deutsche Forschungsgemeinschaft to M.H.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.008342.
References
- Ausubel, F.M. (1994). Current Protocols in Molecular Biology. (New York: John Wiley & Sons).
- Carrano, A.C., Eytan, E., Hershko, A., and Pagano, M. (1999). SKP2 is required for ubiquitin-mediated degradation of the CDK inhibitor p27. Nat. Cell Biol. 1, 193–199. [DOI] [PubMed] [Google Scholar]
- Commean, V.L., Pappelis, G.A., Pappelis, A.J., and Bemiller, J.N. (1985). Changes in DNA and in nuclear and nucleolar dry mass and area in senescing parenchyma cells of corn cob and stalk tissues. Mech. Ageing Dev. 29, 205–213. [DOI] [PubMed] [Google Scholar]
- Compton, M.M. (1992). A biochemical hallmark of apoptosis: Internucleosomal degradation of the genome. Cancer Metastasis Rev. 11, 105–119. [DOI] [PubMed] [Google Scholar]
- Day, S.J., and Lawrence, P.A. (2000). Measuring dimensions: The regulation of size and shape. Development 127, 2977–2987. [DOI] [PubMed] [Google Scholar]
- De Veylder, L., Beeckman, T., Beemster, G.T., de Almeida Engler, J., Ormenese, S., Maes, S., Naudts, M., Van Der Schueren, E., Jacqmard, A., Engler, G., and Inze, D. (2002). Control of proliferation, endoreduplication and differentiation by the Arabidopsis E2Fa-DPa transcription factor. EMBO J. 21, 1360–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Veylder, L., Beeckman, T., Beemster, G.T., Krols, L., Terras, F., Landrieu, I., van der Schueren, E., Maes, S., Naudts, M., and Inze, D. (2001). Functional analysis of cyclin-dependent kinase inhibitors of Arabidopsis. Plant Cell 13, 1653–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerner, P., Jorgensen, J.E., You, R., Steppuhn, J., and Lamb, C. (1996). Control of root growth and development by cyclin expression. Nature 380, 520–523. [DOI] [PubMed] [Google Scholar]
- Doonan, J. (2000). Social controls on cell proliferation in plants. Curr. Opin. Plant Biol. 3, 482–487. [DOI] [PubMed] [Google Scholar]
- Dullea, R.G., Robinson, J.F., and Bedford, J.S. (1999). Nonrandom degradation of DNA in human leukemic cells during radiation-induced apoptosis. Cancer Res. 59, 3712–3718. [PubMed] [Google Scholar]
- Ferreira, P.C., Hemerly, A.S., Villarroel, R., Van Montagu, M., and Inze, D. (1991). The Arabidopsis functional homolog of the p34cdc2 protein kinase. Plant Cell 3, 531–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folkers, U., Berger, J., and Hulskamp, M. (1997). Cell morphogenesis of trichomes in Arabidopsis: Differential control of primary and secondary branching by branch initiation regulators and cell growth. Development 124, 3779–3786. [DOI] [PubMed] [Google Scholar]
- Fransz, P.F., Armstrong, S., de Jong, J.H., Parnell, L.D., van Drunen, C., Dean, C., Zabel, P., Bisseling, T., and Jones, G.H. (2000). Integrated cytogenetic map of chromosome arm 4S of A. thaliana: Structural organization of heterochromatic knob and centromere region. Cell 100, 367–376. [DOI] [PubMed] [Google Scholar]
- Gregory, T.R. (2001). Coincidence, coevolution, or causation? DNA content, cell size, and the C-value enigma. Biol. Rev. Camb. Philos. Soc. 76, 65–101. [DOI] [PubMed] [Google Scholar]
- Hellmann, H., Funck, D., Rentsch, D., and Frommer, W.B. (2000). Hypersensitivity of an Arabidopsis sugar signaling mutant toward exogenous proline application. Plant Physiol. 122, 357–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemerly, A., Engler, J., Bergounioux, C., Van Montagu, M., Engler, G., Inze, D., and Ferreira, P. (1995). Dominant negative mutants of the Cdc2 kinase uncouple cell division from iterative plant development. EMBO J. 14, 3925–3936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemerly, A.S., Ferreira, P., de Almeida Engler, J., Van Montagu, M., Engler, G., and Inze, D. (1993). cdc2a expression in Arabidopsis is linked with competence for cell division. Plant Cell 5, 1711–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horky, M., Wurzer, G., Kotala, V., Anton, M., Vojtesek, B., Vacha, J., and Wesierska-Gadek, J. (2001). Segregation of nucleolar components coincides with caspase-3 activation in cisplatin-treated HeLa cells. J. Cell Sci. 114, 663–670. [DOI] [PubMed] [Google Scholar]
- Hulskamp, M., Misra, S., and Jurgens, G. (1994). Genetic dissection of trichome cell development in Arabidopsis. Cell 76, 555–566. [DOI] [PubMed] [Google Scholar]
- Hulskamp, M., Schnittger, A., and Folkers, U. (1999). Pattern formation and cell differentiation: Trichomes in Arabidopsis as a genetic model system. Int. Rev. Cytol. 186, 147–178. [DOI] [PubMed] [Google Scholar]
- Imajuku, Y., Ohashi, Y., Aoyama, T., Goto, K., and Oka, A. (2001). An upstream region of the Arabidopsis thaliana CDKA;1 (CDC2aAt) gene directs transcription during trichome development. Plant Mol. Biol. 46, 205–213. [DOI] [PubMed] [Google Scholar]
- Jacqmard, A., De Veylder, L., Segers, G., de Almeida Engler, J., Bernier, G., Van Montagu, M., and Inze, D. (1999). Expression of CKS1At in Arabidopsis thaliana indicates a role for the protein in both the mitotic and the endoreduplication cycle. Planta 207, 496–504. [DOI] [PubMed] [Google Scholar]
- Jasinski, S., Riou-Khamlichi, C., Roche, O., Perennes, C., Bergounioux, C., and Glab, N. (2002). The CDK inhibitor NtKIS1a is involved in plant development, endoreduplication and restores normal development of cyclin D3;1-overexpressing plants. J. Cell Sci. 115, 973–982. [DOI] [PubMed] [Google Scholar]
- Kondorosi, E., Roudier, F., and Gendreau, E. (2000). Plant cell-size control: Growing by ploidy? Curr. Opin. Plant Biol. 3, 488–492. [DOI] [PubMed] [Google Scholar]
- Kosslak, R.M., Chamberlin, M.A., Palmer, R.G., and Bowen, B.A. (1997). Programmed cell death in the root cortex of soybean root necrosis mutants. Plant J. 11, 729–745. [DOI] [PubMed] [Google Scholar]
- Kost, B., Spielhofer, P., and Chua, N.H. (1998). A GFP-mouse talin fusion protein labels plant actin filaments in vivo and visualizes the actin cytoskeleton in growing pollen tubes. Plant J. 16, 393–401. [DOI] [PubMed] [Google Scholar]
- LaBaer, J., Garrett, M.D., Stevenson, L.F., Slingerland, J.M., Sandhu, C., Chou, H.S., Fattaey, A., and Harlow, E. (1997). New functional activities for the p21 family of CDK inhibitors. Genes Dev. 11, 847–862. [DOI] [PubMed] [Google Scholar]
- Larkin, J.C., Young, N., Prigge, M., and Marks, M.D. (1996). The control of trichome spacing and number in Arabidopsis. Development 122, 997–1005. [DOI] [PubMed] [Google Scholar]
- Lloyd, R.V., Erickson, L.A., Jin, L., Kulig, E., Qian, X., Cheville, J.C., and Scheithauer, B.W. (1999). p27kip1: A multifunctional cyclin-dependent kinase inhibitor with prognostic significance in human cancers. Am. J. Pathol. 154, 313–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizukami, Y., and Fischer, R.L. (2000). Plant organ size control: AINTEGUMENTA regulates growth and cell numbers during organogenesis. Proc. Natl. Acad. Sci. USA 97, 942–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montagnoli, A., Fiore, F., Eytan, E., Carrano, A.C., Draetta, G.F., Hershko, A., and Pagano, M. (1999). Ubiquitination of p27 is regulated by Cdk-dependent phosphorylation and trimeric complex formation. Genes Dev. 13, 1181–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagl, W. (1976). Zellkern und Zellzyklen: Molekularbiologie, Organisation und Entwicklungsphysiologie der Desoxyribonucleinsaeure und des Chromatins. (Stuttgart, Germany: Ulmer).
- Neufeld, T.P., de la Cruz, A.F., Johnston, L.A., and Edgar, B.A. (1998). Coordination of growth and cell division in the Drosophila wing. Cell 93, 1183–1193. [DOI] [PubMed] [Google Scholar]
- Nurse, P. (1985). The genetic control of cell volume. In The Evolution of Genome Size, T. Cavalier-Smith, ed (Chichester, UK: John Wiley & Sons), pp. 185–196.
- Olson, K.R., McIntosh, J.R., and Olmsted, J.B. (1995). Analysis of MAP 4 function in living cells using green fluorescent protein (GFP) chimeras. J. Cell Biol. 130, 639–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne, C.T., Zhang, F., and Lloyd, A.M. (2000). GL3 encodes a bHLH protein that regulates trichome development in Arabidopsis through interaction with GL1 and TTG1. Genetics 156, 1349–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philipp-Staheli, J., Payne, S.R., and Kemp, C.J. (2001). p27(Kip1): Regulation and function of a haploinsufficient tumor suppressor and its misregulation in cancer. Exp. Cell Res. 264, 148–168. [DOI] [PubMed] [Google Scholar]
- Polyak, K., Lee, M.H., Erdjument-Bromage, H., Koff, A., Roberts, J.M., Tempst, P., and Massague, J. (1994). Cloning of p27Kip1, a cyclin-dependent kinase inhibitor and a potential mediator of extracellular antimitogenic signals. Cell 78, 59–66. [DOI] [PubMed] [Google Scholar]
- Potter, C.J., and Xu, T. (2001). Mechanisms of size control. Curr. Opin. Genet. Dev. 11, 279–286. [DOI] [PubMed] [Google Scholar]
- Riou-Khamlichi, C., Huntley, R., Jacqmard, A., and Murray, J.A. (1999). Cytokinin activation of Arabidopsis cell division through a D-type cyclin. Science 283, 1541–1544. [DOI] [PubMed] [Google Scholar]
- Robinson, J.P., Darzynkiewicz, Z., Dean, P.N., Hibbs, A.R., Orfao, A., Rabinovitch, P.S., and Wheeless, L.L. (2002). Current Protocols in Cytometry. (New York: John Wiley & Sons).
- Rumbolz, J., Kassemeyer, H.-H., Steinmetz, V., Deising, H.B., Mendgen, K., Mathys, D., Wirtz, S., and Guggenheim, R. (1999). Differentiation of infection structures of the powdery mildew fungus Uncinula necator and adhesion to the host cuticle. Can. J. Bot. 78, 409–421. [Google Scholar]
- Sambrook, J., Fritsch, E., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Schnittger, A., Jurgens, G., and Hulskamp, M. (1998). Tissue layer and organ specificity of trichome formation are regulated by GLABRA1 and TRIPTYCHON in Arabidopsis. Development 125, 2283–2289. [DOI] [PubMed] [Google Scholar]
- Schnittger, A., Schobinger, U., Bouyer, D., Weinl, C., Stierhof, Y.D., and Hulskamp, M. (2002. a). Ectopic D-type cyclin expression induces not only DNA replication but also cell division in Arabidopsis trichomes. Proc. Natl. Acad. Sci. USA 99, 6410–6415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnittger, A., Schobinger, U., Stierhof, Y.D., and Hulskamp, M. (2002. b). Ectopic B-type cyclin expression induces mitotic cycles in endoreduplicating Arabidopsis trichomes. Curr. Biol. 12, 415–420. [DOI] [PubMed] [Google Scholar]
- Segers, G., Gadisseur, I., Bergounioux, C., de Almeida Engler, J., Jacqmard, A., Van Montagu, M., and Inze, D. (1996). The Arabidopsis cyclin-dependent kinase gene cdc2bAt is preferentially expressed during S and G2 phases of the cell cycle. Plant J. 10, 601–612. [DOI] [PubMed] [Google Scholar]
- Sessions, A., Weigel, D., and Yanofsky, M.F. (1999). The Arabidopsis thaliana MERISTEM LAYER 1 promoter specifies epidermal expression in meristems and young primordia. Plant J. 20, 259–263. [DOI] [PubMed] [Google Scholar]
- Sherr, C.J., and Roberts, J.M. (1999). CDK inhibitors: Positive and negative regulators of G1-phase progression. Genes Dev. 13, 1501–1512. [DOI] [PubMed] [Google Scholar]
- Smetana, K. (2002). Structural features of nucleoli in blood, leukemic, lymphoma and myeloma cells. Eur. J. Histochem. 46, 125–132. [DOI] [PubMed] [Google Scholar]
- Szymanski, D.B., Jilk, R.A., Pollock, S.M., and Marks, M.D. (1998). Control of GL2 expression in Arabidopsis leaves and trichomes. Development 125, 1161–1171. [DOI] [PubMed] [Google Scholar]
- Szymanski, D.B., and Marks, M.D. (1998). GLABROUS1 overexpression and TRIPTYCHON alter the cell cycle and trichome cell fate in Arabidopsis. Plant Cell 10, 2047–2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, A., Kawasaki, T., Henmi, K., Shi, I.K., Kodama, O., Satoh, H., and Shimamoto, K. (1999). Lesion mimic mutants of rice with alterations in early signaling events of defense. Plant J. 17, 535–545. [DOI] [PubMed] [Google Scholar]
- Toyoshima, H., and Hunter, T. (1994). p27, a novel inhibitor of G1 cyclin-Cdk protein kinase activity, is related to p21. Cell 78, 67–74. [DOI] [PubMed] [Google Scholar]
- Tsvetkov, L.M., Yeh, K.H., Lee, S.J., Sun, H., and Zhang, H. (1999). p27(Kip1) ubiquitination and degradation is regulated by the SCF(Skp2) complex through phosphorylated Thr187 in p27. Curr. Biol. 9, 661–664. [DOI] [PubMed] [Google Scholar]
- Vandepoele, K., Raes, J., De Veylder, L., Rouze, P., Rombauts, S., and Inze, D. (2002). Genome-wide analysis of core cell cycle genes in Arabidopsis. Plant Cell 14, 903–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker, J.D., Oppenheimer, D.G., Concienne, J., and Larkin, J.C. (2000). SIAMESE, a gene controlling the endoreduplication cell cycle in Arabidopsis thaliana trichomes. Development 127, 3931–3940. [DOI] [PubMed] [Google Scholar]
- Wang, H., Fowke, L.C., and Crosby, W.L. (1997). A plant cyclin-dependent kinase inhibitor gene. Nature 386, 451–452. [DOI] [PubMed] [Google Scholar]
- Wang, H., Qi, Q., Schorr, P., Cutler, A.J., Crosby, W.L., and Fowke, L.C. (1998). ICK1, a cyclin-dependent protein kinase inhibitor from Arabidopsis thaliana interacts with both Cdc2a and CycD3, and its expression is induced by abscisic acid. Plant J. 15, 501–510. [DOI] [PubMed] [Google Scholar]
- Wang, H., Zhou, Y., Gilmer, S., Whitwill, S., and Fowke, L.C. (2000). Expression of the plant cyclin-dependent kinase inhibitor ICK1 affects cell division, plant growth and morphology. Plant J. 24, 613–623. [DOI] [PubMed] [Google Scholar]
- Zhou, Y., Fowke, L.C., and Wang, H. (2002. a). Plant CDK inhibitors: Studies of interactions with cell cycle regulators in the yeast two-hybrid system and functional comparison in transgenic Arabidopsis plants. Plant Cell Rep. 20, 967–975. [Google Scholar]
- Zhou, Y., Wang, H., Gilmer, S., Whitwill, S., Keller, W., and Fowke, L.C. (2002. b). Control of petal and pollen development by the plant cyclin-dependent kinase inhibitor ICK1 in transgenic Brassica plants. Planta 215, 248–257. [DOI] [PubMed] [Google Scholar]