Abstract
We performed large-scale mRNA expression profiling using an Affymetrix GeneChip to study Arabidopsis responses to the bacterial pathogen Pseudomonas syringae. The interactions were compatible (virulent bacteria) or incompatible (avirulent bacteria), including a nonhost interaction and interactions mediated by two different avirulence gene–resistance (R) gene combinations. Approximately 2000 of the ∼8000 genes monitored showed reproducible significant expression level changes in at least one of the interactions. Analysis of biological variation suggested that the system behavior of the plant response in an incompatible interaction was robust but that of a compatible interaction was not. A large part of the difference between incompatible and compatible interactions can be explained quantitatively. Despite high similarity between responses mediated by the R genes RPS2 and RPM1 in wild-type plants, RPS2-mediated responses were strongly suppressed by the ndr1 mutation and the NahG transgene, whereas RPM1-mediated responses were not. This finding is consistent with the resistance phenotypes of these plants. We propose a simple quantitative model with a saturating response curve that approximates the overall behavior of this plant-pathogen system.
INTRODUCTION
Advances in genomic information and technologies have made it possible to study biological systems at a global level. mRNA expression profiling is one of the best established large-scale profiling technologies. One of the common uses of mRNA expression profiling is the identification of genes whose expression changes in association with a biological phenomenon of interest, suggesting that they are involved functionally. Another common use of expression profiling is the determination of large-scale expression phenotypes. If the state of a cell changes, it is very likely that the expression of some genes also changes. These genes may or may not be involved functionally in the change of the cell status. Therefore, an expression pattern of numerous genes can be used as a marker that represents a certain state of a cell, and similar expression patterns likely represent similar states.
GeneChip is an established oligomicroarray technology (Lipshutz et al., 1999). The Arabidopsis GeneChip used in this study represents ∼8000 genes, approximately one-third of the Arabidopsis genome (Zhu and Wang, 2000). Each gene is represented by at least one “probe set,” which is a set of 16 to 20 25-mer oligonucleotides. The presence of multiple probes per probe set allows statistical validation of the hybridization data for a particular probe set using a single array. Another advantage of an oligomicroarray for an organism with a substantial amount of genome information, such as Arabidopsis, is that probes can be designed to distinguish between members of gene families. The presence of mismatch probes improves background compensation (Lipshutz et al., 1999).
Strong disease resistance of plants often is conditioned by “gene-for-gene” interactions: when a pathogen has an avirulence (avr) gene and a plant has a corresponding resistance (R) gene, the plant can rapidly recognize the pathogen and effectively deploy defense responses (Hammond-Kosack and Jones, 1997; Dangl and Jones, 2001). When a plant is resistant, the interaction is called incompatible, and when a plant is susceptible, the interaction is called compatible. Understanding the recognition, signal transduction, and defense mechanisms involved in gene-for-gene resistance is not only of great scientific interest but also is important for controlling plant disease. Several genes show expression changes that are used as markers for incompatible interactions, but the magnitude and complexity of gene expression changes that occur during incompatible interactions have not been well documented at the genome level. Based on expression studies of several genes, it was proposed that the responses that occur in compatible interactions are accelerated in incompatible interactions (Lamb et al., 1992). It is not known whether this is true at the genome level.
The model plant-pathogen system consisting of Arabidopsis and the bacterial pathogen Pseudomonas syringae was developed to exploit the tractable genetics of both organisms. Early studies led to the isolation of the Arabidopsis R genes RPS2 and RPM1, which correspond to the P. syringae avr genes avrRpt2 and avrB (and avrRpm1), respectively (Bent et al., 1994; Mindrinos et al., 1994; Grant et al., 1995). Expression marker genes specific to RPS2- or RPM1-mediated incompatible interactions in Arabidopsis were reported (Reuber and Ausubel, 1996; Ritter and Dangl, 1996). However, it is not known to what extent RPS2- and RPM1-mediated responses differ at the genome level. Hypersensitive cell death is associated commonly with gene-for-gene resistance (Hammond-Kosack and Jones, 1997). A null mutation in the Arabidopsis NDR1 gene suppresses hypersensitive cell death in the RPS2-mediated incompatible interaction but not in the RPM1-mediated interaction. However, gene-for-gene resistance measured by in planta bacterial growth was abolished in both interactions (Century et al., 1995, 1997). The basis of this differential effect of the ndr1 mutation is not known. Plants carrying the NahG transgene, which encodes salicylate hydroxylase, cannot accumulate a high level of salicylic acid (SA) (Gaffney et al., 1993), which is an important signal molecule in responses to pathogen attack (Glazebrook, 2001). Arabidopsis plants carrying the NahG transgene are compromised in RPS2-mediated resistance (Delaney et al., 1994).
P. syringae pv phaseolicola NPS3121 (Psp) is a nonhost pathogen of Arabidopsis (Yu et al., 1993). In Arabidopsis, Psp elicits some defense responses, but not hypersensitive cell death, and NahG plants and nonhost1 (nho1) mutant plants allow Psp to grow to a limited extent (Lu et al., 2001). These observations suggest that active defense mechanisms make Psp a poor pathogen of Arabidopsis. It is not clear to what extent Arabidopsis responses to Psp resemble incompatible interactions.
Here, we report our GeneChip analysis of Arabidopsis–P. syringae interactions. Among ∼8000 genes whose expression was monitored, ∼2000 genes showed significant expression changes within 9 h after infection with bacterial strains. Our analysis indicates that a large part of the difference between compatible and incompatible responses can be explained by a quantitative difference in the behavior of a single signal transduction system. In addition, this quantitative model provides an explanation for the differential effects of ndr1 and NahG on RPS2- and RPM1-mediated responses.
RESULTS
Data Collection
We are particularly interested in early events during Arabidopsis–P. syringae interactions. Published mRNA expression profiling studies of plant–pathogen interactions have focused on relatively late events (Maleck et al., 2000; Schenk et al., 2000). We inoculated with a relatively high concentration of bacteria (2 × 107 colony-forming units [cfu]/mL) so that most leaf cells would have direct contact with bacteria immediately after inoculation. In one case, a lower concentration (1 × 106 cfu/mL) was used to obtain data for late responses in a compatible interaction. The combinations of Arabidopsis plants and P. syringae strains used are listed in Table 1. We used wild-type, ndr1-1 (Century et al., 1995), and NahG (Delaney et al., 1994) plants of the Columbia ecotype. We used virulent strains P. syringae pv tomato DC3000 (Pst) (Whalen et al., 1991) and P. syringae pv maculicola ES4326 (Psm) (Dong et al., 1991) and a nonhost strain, Psp (Yu et al., 1993). Strains carrying avrRpt2 (Innes et al., 1993) or avrB (Bisgrove et al., 1994) were used to study gene-for-gene interactions. For each combination of plant and bacterial strain (or water control), leaves were collected at 3, 6, and 9 h after inoculation, except when Psp was used (3 and 6 h). To reduce the impact of biological variation, for each experiment, each infection was conducted three times independently and equal amounts of RNA from each infection were pooled.
Table 1.
Experiments Performed
| Planta | Bacteriumb | Time Points (h) |
Number of Experiments |
Hypersensitive Cell Death? |
|---|---|---|---|---|
| Wild type | Water | 3, 6, 9, 30c | 1 | − |
| Pst | 3, 6, 9 | 2 | − | |
| Pst/avrRpt2 | 3, 6, 9 | 2 | + | |
| Pst/avrB | 3, 6, 9 | 2 | + | |
| Psm | 3, 6, 9 | 1 | − | |
| Psm/avrRpt2 | 3, 6, 9 | 1 | + | |
| Psp | 3, 6 | 1 | − | |
| Psm (low dose) | 30c | 1 | − | |
| NahG | Water | 3, 6, 9 | 1 | − |
| Pst | 3, 6, 9 | 1 | − | |
| Pst/avrRpt2 | 3, 6, 9 | 1 | + | |
| Pst/avrB | 3, 6, 9 | 1 | + | |
| Psp | 3, 6 | 1 | − | |
| ndr1-1 | Water | 3, 6, 9 | 1 | − |
| Pst | 3, 6, 9 | 1 | − | |
| Pst/avrRpt2 | 3, 6, 9 | 1 | − | |
| Pst/avrB | 3, 6, 9 | 1 | + |
All of the plants were in the Arabidopsis accession Columbia-0 background.
Most strains were inoculated at 2 × 107 cfu/ml. “Low dose” indicates an inoculation at 106 cfu/mL.
10 mM MgCl2 was used instead of water for the bacterial suspension.
Data Analysis
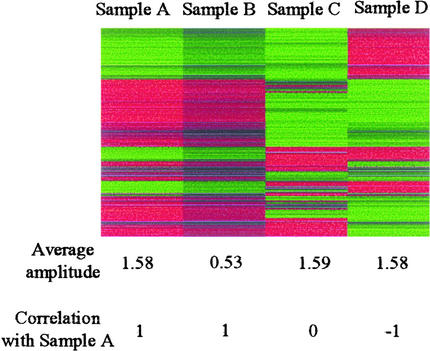
For each experimental set, the ratio of the expression level for each probe set to that in the corresponding water control was calculated, and probe sets that showed greater than twofold change in at least one sample were selected (see Methods for details). We considered the data from these probe sets to be informative data. The log2-transformed value of this ratio was used for complete linkage clustering (Eisen et al., 1998) and calculation of pair-wise correlation and average amplitude (see Methods for details). Results of the clustering analysis were visualized using TreeView (Eisen et al., 1998). As shown in Figure 1, the log2-transformed expression change value for each probe set is depicted by a thin colored horizontal line: if the expression value for the probe set in the sample is higher than that in the control, the line is red, and if it is lower, the line is green; the larger the absolute value of the change, the brighter the color of the line. The pattern made of red and green lines for each sample is called the expression change profile for the sample. The shape of the profile is defined by the pattern made of the relative log2-transformed expression change values for a sample: if all of the values in sample A are three times larger than those in sample B, the shapes of profiles for samples A and B are the same (Figure 1). The correlation is an index that represents the similarity in the shape of the profile. For example, if the shapes of the profiles are the same (e.g., samples A and B), unrelated (e.g., samples A and C), or totally opposite (e.g., samples A and D), the correlation value is 1, 0, or −1, respectively. The average amplitude of a profile is defined by the average of the absolute values of all log2-transformed expression change values in a sample. For example, the profile for sample A has a three times larger amplitude than that for sample B.
Figure 1.
Explanations of the Data Analysis Methods.
The values for each sample are generated artificially, and they are visualized by TreeView. The values in sample A are three times larger than the corresponding values in sample B. Therefore, the correlation between samples A and B is 1, which means that the shapes of the profiles are the same, and the average amplitude for sample A (1.58) is three times larger than that for sample B (0.53). The correlation between samples A and C is 0, which means that they do not share similarity. The values in sample D are the same as the corresponding values in sample A except that they have the opposite signs. Therefore, the correlation between them is −1, and the average amplitude is the same.
The hierarchical clustering method used in this study recognizes the similarity in the shapes of profiles but ignores the amplitudes of profiles because it uses the correlation as the similarity measure. Limitations of hierarchical clustering include its process dependency and one-dimensional result representation. Complete linkage clustering is defined as using the maximum distance between points in a cluster. Such a distance depends on which data points are already in a cluster, and this leads to process dependency of clustering. Different data points could be similar to a given data point in different ways (i.e., multidimensional). One-dimensional representation is not able to rationally handle such multidimensional relationships.
Some genes are represented by more than one probe set on the GeneChip, which explains why the GeneChip has ∼8700 probe sets corresponding to ∼8000 genes. Note that the data for some samples are used in more than one figure for easier comparison but that the set of selected probe sets and the order of the probe sets in each figure could be different.
Behavior of the Incompatible Interaction Is Robust
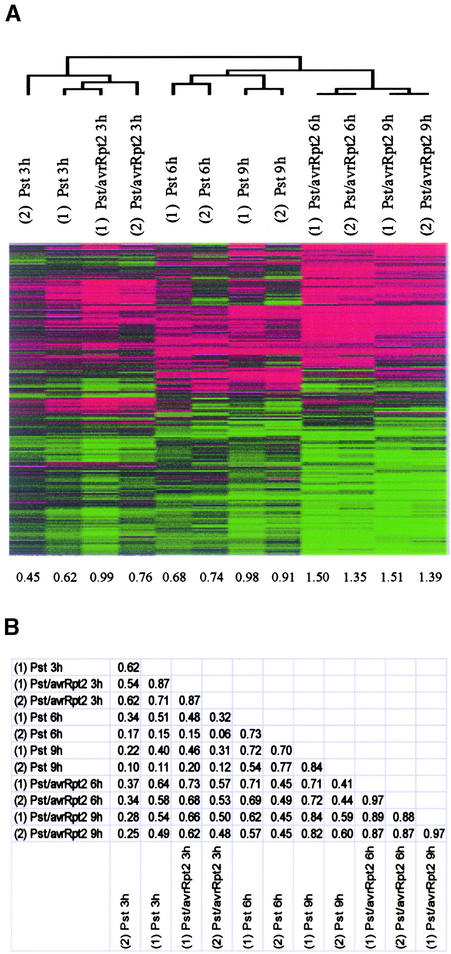
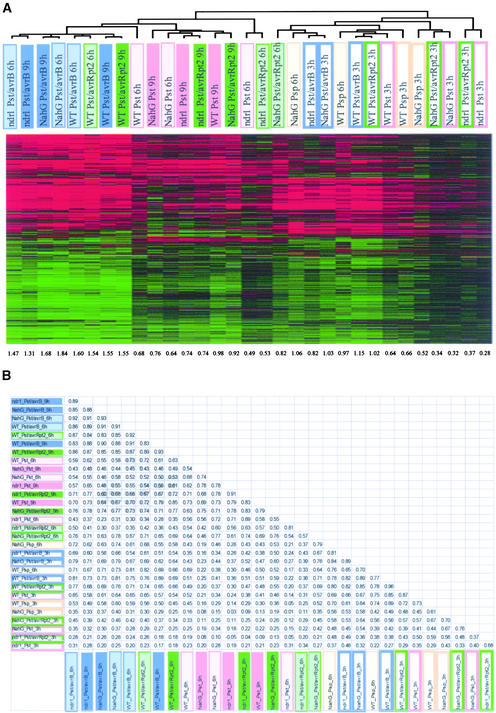
In expression profiles, two types of noise must be considered in interpreting the data: technical errors and biological variations. In our hands, the false-positive rate (percentages of probe sets that show greater than twofold difference between duplicated GeneChip hybridizations) was ∼0.2% (Zhu and Wang, 2000). This technical error corresponds to ∼16 genes among 8000 genes analyzed by the GeneChip. To reduce the impact of biological variation, we pooled RNA from triplicate samples as described above. For the samples indicated in Table 1 as being subjected to two experiments, two sets of pooled experiments were performed by different researchers ∼2 months apart to evaluate the level of biological variation in the results. These samples include a compatible interaction with Pst and an incompatible interaction with Pst/avrRpt2. Figure 2A shows the results of the hierarchical clustering analysis with 2725 selected probe sets representing 2338 genes. The corresponding samples from two different experiments cluster together except at 3 h (see the dendrogram in Figure 2A). Although (1) Pst/avrRpt2 3 h seems appreciably closer to (1) Pst 3 h than does (2) Pst/avrRpt2 3 h as a result of the process dependency of the clustering, the correlations between (1) Pst/avrRpt2 3 h and (1) Pst 3 h and between (1) Pst/avrRpt2 3 h and (2) Pst/avrRpt2 3 h are almost identical, as shown in Figure 2B. The major reason that part of the dendrogram for 3 h is disturbed is the similarity in the profile shapes between the compatible and incompatible interactions.
Figure 2.
Responses during an Incompatible Interaction Were More Robust Than Those during a Compatible Interaction.
(A) Comparison of the results from two experiments by hierarchical clustering. Wild-type Columbia plants were inoculated with Pst, Pst/avrRpt2, Pst/avrB, or water control, and tissue was harvested at 3, 6, and 9 h after inoculation. The samples from two experiments are indicated by (1) and (2) in the names of the samples. Data from 2725 probe sets, which correspond to 2338 genes, are shown. The dendrogram for the samples is shown at top. The numbers at bottom indicate the average amplitudes of the profiles.
(B) A table of pair-wise correlations among the samples shown in (A).
It is clear from the pair-wise correlations listed in Figure 2B that the results for the RPS2-mediated incompatible interaction show relatively small variation between two experiments, especially at 6 and 9 h (both correlations of 0.97). By contrast, the results for the compatible interaction show a fair amount of variation (correlations of 0.73 and 0.84 for 6 and 9 h, respectively). We think that the major reason for this effect is that the behavior of the compatible interaction as a biological system is not as robust as that of the incompatible interaction. This would mean that the level of biological variation observed in large-scale expression profiles depends heavily on an intrinsic characteristic of the biological system: the degree of robustness of the system.
A Large Part of the Difference between the Compatible and Incompatible Interactions Is Quantitative
The results shown in Figure 2A indicate that 1839 of ∼8000 genes monitored by the GeneChip changed expression level significantly (greater than twofold change) and reproducibly (in both experiments) within 9 h after infection with P. syringae. It is obvious that the incompatible interaction mediated by RPS2 led to more mRNA expression changes than the compatible interaction. During the incompatible interaction, the shapes of the profiles at 3 and 6 h after infection were quite different from each other, whereas those at 6 and 9 h were similar. The correlations between the 3- and 6-h time points were 0.73 and 0.53 in two experiments, and those between the 6- and 9-h time points were 0.89 and 0.87 (Figure 2B). This finding indicates that most qualitative mRNA expression changes in the infected tissue occur within 6 h after infection during the incompatible interaction.
Although the amplitudes of profiles were larger in the incompatible interaction than in the compatible interaction at all time points, the profile shapes between the compatible and incompatible interactions at the same time points were somewhat similar (Figure 2A). This trend of similarities at the same time points was much more prominent in experiment 1, especially at 3 h, than in experiment 2, but the trend still was recognizable even in experiment 2 (cf. the correlations listed in Figure 2B). This trend suggests that a large part of the difference between responses during the compatible and incompatible interactions is quantitative.
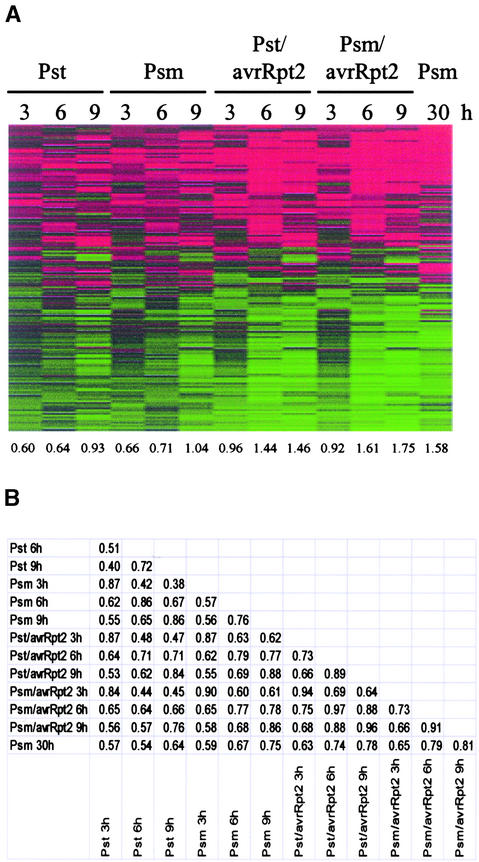
Figure 3 shows that the effects of bacterial strain background were very small in compatible and incompatible interactions. The results obtained with Pst and Psm were very similar at all three time points, as were the results obtained with Pst/avrRpt2 and Psm/avrRpt2.
Figure 3.
Responses during a Late Compatible Interaction Were Similar to Those during an Early Incompatible Interaction.
(A) Effects of different bacterial backgrounds and comparison of compatible and incompatible interactions at different time points (2971 probe sets, 2546 genes). Wild-type Columbia plants were inoculated with Pst, Psm, Pst/avrRpt2, Psm/avrRpt2, or water control, and tissue was harvested 3, 6, 9, or 30 h later. Note that for the 30-h sample, the bacterial dose used was 20 times lower than that used for the other samples. The GeneChip data were analyzed and visualized as in Figure 2A, except that the samples were not clustered.
(B) A table of pair-wise correlations for (A).
Late Compatible and Early Incompatible Interactions Show Similar Responses
It has been suggested that some responses observed at early stages of incompatible interactions occur at late stages of compatible interactions (Lamb et al., 1992), but this notion was based on observations of a limited number of responses. Using large-scale expression profiling, we tested this idea. Leaves were collected at 30 h after Psm inoculation for profiling of late responses in a compatible interaction. A lower dose was used because the high dose would have led to softening and necrosis of the leaf tissue before collection. As shown in Figure 3, the profile for the late compatible interaction was similar to those for incompatible interactions at 6 and 9 h (correlations of 0.79 and 0.81, respectively, with Psm background). Not only the shapes but also the amplitudes of the profiles were similar (1.61, 1.75, and 1.58 for Psm/avrRpt2 6 h, Psm/avrRpt2 9 h, and Psm 30 h, respectively). Thus, a broad range of defense responses in early incompatible interactions are very similar to those in late compatible interactions.
Responses during Incompatible Interactions Mediated by Two R Genes Are Very Similar
We compared the profiles for RPS2- and RPM1-mediated responses to determine to what extent these responses are similar. The Arabidopsis AIG1 and Eli3 genes were reported to be specific expression marker genes for RPS2- and RPM1-mediated responses, respectively (Reuber and Ausubel, 1996; Ritter and Dangl, 1996). In our hands, AIG1, Eli3-1, and Eli3-2 showed expression preferential to RPS2-, RPM1-, and RPM1-mediated responses, respectively (see supplemental data online). Figure 4 shows large-scale expression change profiles to compare these responses. At 3 h, the profiles of mRNA expression changes were very similar (correlation of 0.96). Note that this level of similarity is substantially larger than the level for the same 3-h Pst/avrRpt2 samples between two pooled experimental sets (correlation of 0.87; Figure 2B). The degree of difference increased slightly by 6 h (correlations of 0.92 and 0.93 at 6 and 9 h, respectively). In addition to the shapes of the profiles, the amplitudes of the profiles were also similar (see the values at the bottom of the figure). Thus, the overall responses mediated by RPS2 and RPM1 are very similar.
Figure 4.
RPS2- and RPM1-Mediated Responses Are Very Similar (2679 Probe Sets, 2302 Genes).
Wild-type Columbia plants were inoculated with Pst, Pst/avrRpt2, Pst/avrB, or water control, and tissue was harvested at 3, 6, or 9 h later. The GeneChip data were analyzed and visualized as in Figure 2A, except that the samples were not clustered.
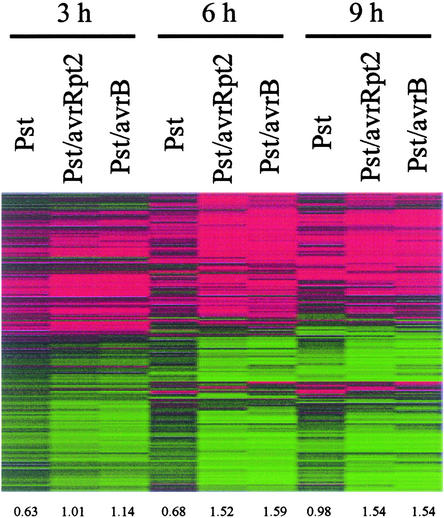
NahG and ndr1 Differentially Affect Responses during Incompatible Interactions Mediated by Two R Genes
The NahG transgene compromises RPS2-mediated resistance (Delaney et al., 1994), but some responses associated with the gene-for-gene resistance, including hypersensitive cell death, are not abolished in NahG transgenic plants. Inconsistent with a previous report (Rate et al., 1999), we reproducibly observed macroscopic hypersensitive cell death in NahG leaves inoculated with Pst/avrRpt2 (data not shown). Moreover, the minimum bacterial dose of Pst/avrRpt2 needed to induce macroscopic hypersensitive cell death was lower for NahG than for the wild type. This may be a result of faster bacterial growth in NahG plants (Delaney et al., 1994). The ndr1 mutation affects RPS2-mediated resistance more severely than RPM1-mediated resistance (Century et al., 1995).
We tested the effects of NahG and ndr1 on large-scale expression profiles. Whereas the shapes and amplitudes of the profiles for RPS2- and RPM1-mediated responses in wild-type plants were very similar, Figure 5 demonstrates that the effect of NahG or ndr1 was much stronger on RPS2-mediated responses than on RPM1-mediated responses. Most RPS2-mediated responses were suppressed strongly in NahG and ndr1 plants. At 3 h, the amplitudes of expression changes in NahG and ndr1 plants infected with Pst/avrRpt2 was much smaller than that in the wild type (0.34, 0.37, and 1.02 for NahG, ndr1, and the wild type, respectively). At 6 and 9 h, the profiles from NahG and ndr1 plants infected with Pst/avrRpt2 were quite different from those from the wild type in both shape and amplitude. The correlations at 6 and 9 h were 0.71 and 0.77, respectively, between NahG and the wild type and 0.42 and 0.72 between ndr1 and the wild type. The amplitudes at 6 and 9 h were 0.82 and 0.92, respectively, for NahG, 0.53 and 0.74 for ndr1, and 1.54 and 1.55 for the wild type. Generally, they were more similar to the profiles from wild-type plants infected with Pst. The correlations at 6 and 9 h were 0.64 (which is the only exception for this argument) and 0.79, respectively, between NahG Pst/avrRpt2 and wild-type Pst and 0.54 and 0.83 between ndr1 Pst/avrRpt2 and wild-type Pst. The amplitudes at 6 and 9 h were 0.68 and 0.98, respectively, for wild-type Pst. The effects of NahG and ndr1 on RPM1-mediated responses generally were small. The correlations at 3, 6, and 9 h were 0.89, 0.91, and 0.90, respectively, between NahG and the wild type and 0.82, 0.86, and 0.88 between ndr1 and the wild type. The amplitudes at 3, 6, and 9 h were 1.03, 1.84, and 1.68, respectively, for NahG, 0.82, 1.47, and 1.31 for ndr1, and 1.15, 1.60, and 1.55 for the wild type. Both NahG and ndr1 also suppressed responses during the compatible interaction with Pst at all three time points. The amplitudes at 3, 6, and 9 h were 0.32, 0.64, and 0.76, respectively, for NahG, 0.28, 0.49, and 0.74 for ndr1, and 0.64, 0.68, and 0.98 for the wild type.
Figure 5.
Differential Effects of ndr1 and NahG on RPS2- and RPM1-Mediated Responses.
(A) RPS2-mediated responses are strongly suppressed by the ndr1 mutation and the NahG transgene, but RPM1-mediated responses are not (2679 probe sets: the same probe sets as in Figure 4, but the order of the probe sets is different). Wild-type (WT), ndr1, and NahG plants were inoculated with Pst, Pst/avrRpt2, Pst/avrB, Psp, or water control, and tissue was harvested at 3, 6, or 9 h later. ndr1 plants were not inoculated with Psp. The GeneChip data were analyzed and visualized as in Figure 2A.
(B) A table of pair-wise correlations for (A).
Samples are color coded: Pst, Pst/avrRpt2, Pst/avrB, and Psp are pink, green, blue, and peach, respectively. The time points 3, 6, and 9 h are indicated by boxes with no fill color, boxes with faint fill color, and solid-colored boxes, respectively. Wild-type, NahG, and ndr1 plants are not distinguished by color.
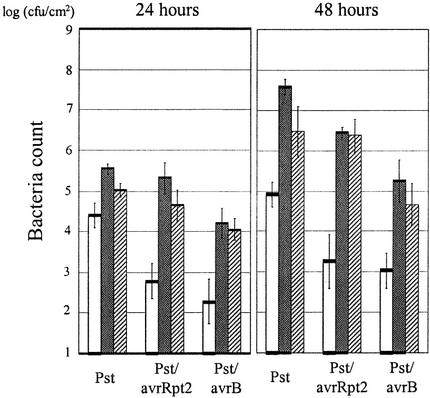
As shown in Figure 6, we measured in planta bacterial growth in these plants to compare it with the expression profile data. There was a good correlation between how much the expression profile was affected and how much more bacterial growth was allowed in these plants. For example, although the titer of Pst/avrB was ∼60 times higher in ndr1 plants than in wild-type plants at 48 h, that of Pst/avrRpt2 was ∼1000 times higher. This finding is consistent with the observation that ndr1 affects RPS2-mediated expression responses much more strongly than RPM1-mediated responses. As reported previously (Century et al., 1995; Shapiro and Zhang, 2001), the virulent strain Pst grew better in ndr1 plants than in wild-type plants within 2 days. This observation is consistent with the profile change in ndr1 infected with Pst.
Figure 6.
Quantitative Effects of NahG and ndr1 on in Planta Bacterial Growth.
Wild-type (white bar), NahG (gray bar), and ndr1 (stippled bar) plants were inoculated with Pst, Pst/avrRpt2, or Pst/avrB, and bacterial titer was measured at 24 and 48 h later. At 0 h, the bacterial counts of Pst, Pst/avrRpt2, and Pst/avrB were 2.49 ± 0.33, 2.10 ± 0.46, and 2.08 ± 0.43 log (cfu/cm2), respectively. Each data point represents the mean and standard deviation of six replicates. This experiment was repeated with similar results.
A Nonhost Pathogen Weakly Induces Responses Somewhat Similar to Those in Incompatible Interactions
Psp is a nonhost pathogen of Arabidopsis, whereas it is a genuine pathogen of bean (Lindgren et al., 1986; Yu et al., 1993). Unlike many bacterial pathogens on nonhost plants, Psp does not induce hypersensitive cell death on Arabidopsis, although it does induce expression of PR1 and GST1 (Lu et al., 2001). As shown in Figure 5, the profile shape of expression changes in Psp-infected plants at 3 h was somewhat similar to those in Pst-, Pst/avrRpt2-, and Pst/avrB-infected plants at 3 h (correlations of 0.73, 0.72, and 0.69, respectively), and the profile amplitude of Psp-infected plants was as small as that of Pst-infected plants (0.66 for Psp and 0.64 for Pst). The profile shape of Psp-infected plants at 6 h was similar to those of Pst/avrRpt2-infected plants at both 3 and 6 h (correlations of 0.78 and 0.82, respectively). These observations suggest that Psp induces defense mechanisms somewhat similar to those induced by RPS2-mediated resistance, but at a slower late.
NahG plants allow a limited level of Psp growth (Lu et al., 2001). At 3 h, the profile amplitude for NahG plants infected with Psp was very small (0.52; Figure 5A). At 6 h, the profile shape of NahG plants infected with Psp was more similar to the 3-h incompatible interaction profiles than to the 6-h profiles in wild-type plants. The correlations of NahG plants at 6 h after Psp infection with wild-type plants infected with Pst/avrRpt2 and Pst/avrB were 0.80 and 0.78, respectively, at 3 h and 0.68 and 0.61 at 6 h. NahG may slow the response to Psp or inhibit the progress of defense responses beyond a certain point.
DISCUSSION
Robust Behavior of an Incompatible Interaction as a Biological System
In this report, we emphasize the use of large-scale mRNA expression profiling to characterize the biological states of Arabidopsis cells. Monitoring the expression of thousands of genes simultaneously allowed us to use large-scale profiles to characterize biological states. One interesting aspect revealed by expression profiles is that biological variations in the profile were much smaller in an incompatible interaction than in a compatible interaction. Because the profiles for the incompatible interaction in two experiments were very similar, it is unlikely that the large variation between two experiments for the compatible interaction was attributable to experimental errors. Instead, this observation suggests that responses during an incompatible interaction are not much affected by various conditions that are difficult to control, such as soil quality, including microbial environments and moisture level, light quality/quantity as a result of lamp age (2 months difference in this case), etc. In other words, the responses with smaller biological variations are likely to be correlated with robust system behavior. It appears that plant responses in an incompatible interaction are more robust than those in a compatible interaction.
Differences between Responses during Compatible and Incompatible Interactions Are Quantitative and Kinetic
What are the differences in responses between compatible and incompatible interactions? We observed quantitative and kinetic differences. During the first 9 h, a large part of the difference between compatible and incompatible interactions was quantitative. The profile shapes were similar in both interactions at corresponding time points. Because the amplitude of expression changes during a compatible interaction is small, the sensitivity, accuracy, and broad spectrum of large-scale mRNA expression profiling was crucial to detect this similarity. This similarity of profile shapes was most evident at 3 h. One possible explanation for why the degree of similarity declined at later times is that effects of quantitative differences at 3 h were amplified over time and resulted in larger differences at later times. It also is possible that the difference may be attributable to the effects of virulence factors. It is conceivable that virulence factors delivered by Pst become more effective at suppressing plant defense responses at later hours. These explanations are not mutually exclusive. It is tempting to speculate that although the plant signal transduction mechanisms are largely shared between compatible and incompatible interactions, strong signaling very soon after infection enables plants to overcome the effects of virulence factors during incompatible interactions.
There also was a kinetic difference between compatible and incompatible interactions. After 9 h, the profile amplitude appeared increased in a compatible interaction. Although we need to be cautious in interpreting these results because of the difference in the inoculation dose, the shape and the amplitude of the profile at 30 h were similar to those of incompatible interactions at 6 and 9 h. The similarity in the profile shapes and amplitudes of early incompatible and late compatible interactions demonstrates that the notion of a kinetic difference between compatible and incompatible interactions generally is true on a large scale. Quantitative and kinetic differences often are related closely to each other. For example, if the induced expression of a certain gene is reduced, it takes more time for the protein encoded by that gene to accumulate; consequently, the response mediated by the protein shows slower kinetics.
In summary, our analysis suggests that plant responses in compatible and incompatible interactions are qualitatively similar but quantitatively different soon after infection and that the amplitude of the responses in the compatible interaction increases later. These observations suggest that signal transduction mechanisms involved in compatible and incompatible interactions are largely shared.
RPS2- and RPM1-Mediated Responses Are Very Similar
RPS2- and RPM1-mediated responses were very similar on the large scale represented by expression profiles. This observation strongly suggests that the major signal transduction mechanisms used in RPS2- and RPM1-mediated responses are shared. Specific or preferential expression of marker genes, such as AIG1 and Eli3 (Reuber and Ausubel, 1996; Ritter and Dangl, 1996), may be under the control of some minor, specific signal transduction mechanisms. Alternatively, their expression may be highly sensitive to subtle quantitative or kinetic differences in signaling through shared mechanisms.
The 6-h time points revealed small differences between the profiles of RPS2- and RPM1-mediated responses. These differences may reflect kinetic differences in the responses. RPS2-mediated responses generally are slower than RPM1-mediated responses, as illustrated by different onset times of hypersensitive cell death (Ritter and Dangl, 1996; Shapiro and Zhang, 2001). Alternatively, the profile differences may be attributable to differences in the virulence functions of AvrRpt2 and AvrB (Chen et al., 2000; Nimchuk et al., 2000; Guttman and Greenberg, 2001). These two possibilities are not mutually exclusive. The virulence function of AvrRpt2 might be the cause that slows down the RPS2-mediated responses.
How Do ndr1 and NahG Affect R Gene–Mediated Responses?
Despite the overall similarities in the shapes and amplitudes of the mRNA profiles of RPS2- and RPM1-mediated responses, RPS2-mediated responses were suppressed strongly by the ndr1 mutation and the NahG transgene, whereas RPM1-mediated responses were not affected as strongly. Century et al. (1995) reported that ndr1 abolishes hypersensitive cell death and gene-for-gene specific inhibition of bacterial growth in RPS2-mediated resistance, whereas it abolishes only gene-for-gene specific inhibition of bacterial growth in RPM1-mediated resistance. In our hands, RPM1-mediated inhibition of bacterial growth was only partially affected in ndr1 plants (Figure 6). Note that ndr1 also affected Pst growth and that a major part of the increase in Pst/avrB growth in ndr1 resulted from the effect on nonspecific resistance rather than from the effect on RPM1-mediated resistance. We conclude that NDR1 is not an essential factor for RPM1-mediated resistance but a quantitative factor. Its quantitative effect could appear stronger depending on the experimental conditions and the detection range of the assay used. The quantitative effect of the ndr1 mutation on RPM1-mediated responses also was described by others (Shapiro and Zhang, 2001; Tornero et al., 2002).
In agreement with the report by Century et al. (1995), we observed that Pst and Pst/avrRpt2 grow similarly in ndr1 plants. Therefore, it is reasonable to conclude that NDR1 is required for RPS2-mediated resistance. However, the correlations in profiles between ndr1 plants infected with Pst/avrRpt2 and wild-type plants infected with Pst/avrRpt2 were higher than those between ndr1 plants infected with Pst and wild-type plants infected with Pst/avrRpt2 at all three time points. The correlations at 3, 6, and 9 h were 0.43, 0.42, and 0.72, respectively, between ndr1 Pst/avrRpt2 and wild-type Pst/avrRpt2 and 0.29, 0.34, and 0.61 between ndr1 Pst and wild-type Pst/avrRpt2 (Figure 5B). This finding indicates that ndr1 does not totally abolish the avrRpt2-dependent responses and strongly suggests that the effect of ndr1 on RPS2-mediated responses is quantitative. Previously, based on the results of a transient expression assay for the resistance response, we suggested that NDR1 could be a quantitative factor for RPS2-mediated resistance (Tao et al., 2000). Hypersensitive cell death can be elicited by a very high inoculum of P. syringae pv glycinea carrying avrRpt2 in ndr1 plants (Shapiro and Zhang, 2001). This finding is consistent with the notion of NDR1 as a quantitative factor for RPS2-mediated responses.
The ndr1 mutation increased the growth of Pst, at least during the first 2 days (Figure 6) (Shapiro and Zhang, 2001). The ndr1 mutation also affected plant responses after infection with Pst (Figure 5A). Therefore, the function of NDR1 is not restricted to gene-for-gene resistance.
The behavior of NahG plants in response to the bacterial infections is similar to that of ndr1 plants in both expression change profiles and bacterial growth. It is conceivable that the function affected by the NahG transgene in early stages of interactions is related closely to that affected by the ndr1 mutation. NahG encodes an SA hydroxylase, and NahG transgenic Arabidopsis plants accumulate very little SA compared with wild-type plants (Lawton et al., 1995). The ndr1 plant also has a defect in SA accumulation (Shapiro and Zhang, 2001). Therefore, the simplest interpretation of these findings is that SA plays a quantitative role in RPS2- and RPM1-mediated resistance. If we assume that in this case SA functions as a signaling molecule, as it does in systemic acquired resistance (Ryals et al., 1996), most genes responding in RPM1-mediated resistance must be regulated in an SA-independent manner. This notion is difficult to reconcile with the fact that an almost identical set of genes are strongly affected by NahG or ndr1 when elicited through RPS2-mediated mechanisms. These genes appear to be regulated in an SA-dependent manner in this case.
Another role of SA could be the potentiation of R gene–mediated responses in a very early step of signaling at a low concentration (Shirasu et al., 1997). It is conceivable that the signal flow for the RPM1-mediated responses is so strong that it does not need additional potentiation by a low level of SA for regulation of the majority of genes. However, the signal flow for the RPS2-mediated responses is weaker and requires potentiation by a low level of SA. This view also is consistent with the kinetics of the RPS2-mediated responses mentioned above. Shapiro and Zhang (2001) discussed the role of SA in potentiation to explain the differential responses of ndr1 plants in RPS2- and RPM1-mediated signaling. It will be interesting to determine whether other classes of SA pathway–related mutants, such as eds5, sid2 (Nawrath and Metraux, 1999), pad4 (Zhou et al., 1998), and npr1/nim1 (Cao et al., 1994; Delaney et al., 1995), have similar effects on RPS2- and RPM1-mediated resistance. The induction of EDS5 mRNA expression by Pst/avrRpt2 is suppressed strongly in NahG, ndr1, and pad4 plants but not in sid2 and npr1 plants (Nawrath et al., 2002). If this expression pattern of EDS5 is representative of the general trend we observed using large-scale mRNA profiling, a deficiency in SA accumulation alone cannot explain the trend because sid2 plants accumulate a very low level of SA (Nawrath and Metraux, 1999).
The fact that ndr1 and NahG also affect plant responses to Pst suggests that the presumed potentiation of signaling by SA occurs in the signaling mechanism involved in basic resistance. This also supports the notion that signaling mechanisms are largely shared between compatible and incompatible interactions. Shirasu et al. (1997) observed such a potentiation with exogenously added SA in an incompatible interaction but not in a compatible interaction. This finding could be explained by assuming that the potentiation of signaling in compatible interactions already is saturated with the wild-type level of SA and that the effect can be observed only when the SA level is reduced from the wild-type level.
Is Inducible Defense Responsible for One Form of Nonhost Resistance?
It is likely that different mechanisms are responsible for nonhost resistance to different pathogens. In the case of Arabidopsis nonhost resistance against Psp, it is associated with inducible defense (Lu et al., 2001). The shapes of the expression profiles are similar to those for early incompatible interactions. Unlike many other bacterial pathogens on nonhosts, Psp does not induce hypersensitive cell death (Yu et al., 1993). This is similar to the extreme resistance mediated by the potato Rx gene against Potato virus X (Bendahmane et al., 1999). It has been proposed that Rx-mediated resistance is so effective at a very early step that the resistance response does not reach the step at which hypersensitive cell death is induced. Such step-wise progress of inducible defense might be more general. It is conceivable that the very early inducible defense against Psp is judged to be successful by the plant cell and that inducible defense does not proceed to a step that induces hypersensitive cell death. This model is consistent with the observation that the expression change induced by Psp inoculation seems to have a slower kinetics than incompatible interactions.
Quantitative Nature of the Biological System
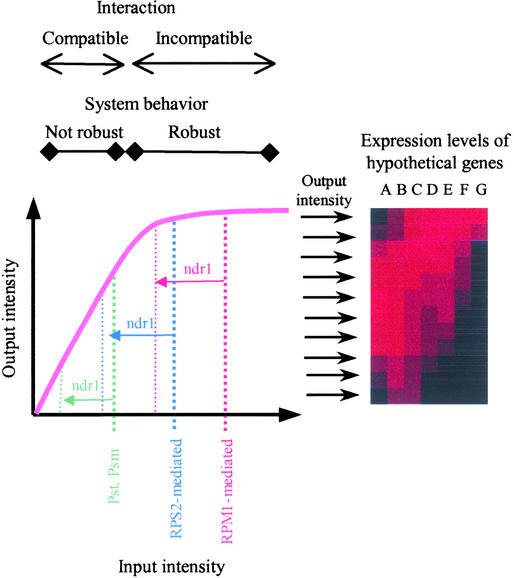
A large part of the difference between compatible and incompatible interactions is quantitative. NahG and ndr1 affect RPS2- and RPM1-mediated resistance differently and quantitatively. These observations led us to hypothesize that the quantitative nature of this plant-pathogen system is its fundamental characteristic. Figure 7 illustrates a quantitative model to explain our observations. In this model, a common signal transduction mechanism with the depicted input-output response curve is postulated. We assume that early signaling events that occur before this common mechanism are likely different for compatible and incompatible interactions and for RPS2- and RPM1-mediated resistance (e.g., different signal perception mechanisms and different R proteins). For the sake of simplicity, a time factor is not incorporated in this model. Consider that this model describes a particular time point.
Figure 7.
A Quantitative Signaling Model for Compatible and Incompatible Pathogen Interactions.
A common signal transduction mechanism with the depicted saturating response curve between its input and output signal intensities is postulated. For simplicity, a time factor is not included in this model. The difference between incompatible (RPM1-mediated and RPS2-mediated) and compatible (Pst and Psm) interactions is defined by the difference in the range of the input signal intensity. The robustness of the system behavior is defined by how sensitive the output intensity is to changes in the input intensity. How gene expression pattern changes according to the output intensity is illustrated at right. The relative expression levels of hypothetical genes A to G are shown by the brightness of red shading. The relative expression levels of these genes change according to the output intensities of the common mechanism. This illustrates the situation in which both the average amplitude of expression and the overall expression pattern gradually change when the output intensity changes. The effect of ndr1 is hypothesized to nonspecifically reduce the input intensity.
On the right side of the figure are hypothetical genes whose expression levels are indicated by red shading. These genes illustrate what kind of expression profiles are expected according to the output level of the common signal transduction mechanism. Genes A and B are hypothesized to have the highest expression levels at medium output intensities of the common mechanism. Such patterns were hypothesized to explain the expression patterns of a small number of genes that are induced by Pst in wild-type, NahG, and ndr1 plants and that are induced by Pst/avrRpt2 in NahG and ndr1 plants but not in wild-type plants (see supplemental data online). The response curve has a typical saturation curve shape, so that the output level is relatively stable to a change in the input level when the input level is high but not when it is low. Incompatible interactions generate a high level of the input signal; thus, the responses during incompatible interactions are insensitive to a small input signal change and therefore are robust. On the other hand, the compatible interaction generates a medium level of input signal, which corresponds to the middle of the slope of the response curve; therefore, the responses during compatible interactions are sensitive to a small input signal change and not robust.
The ndr1 mutation generally reduces the input signal intensity. This notion is based on the discussion above that the ndr1 mutation may affect the potentiation of a very early signaling step. RPM1-mediated signal has such a high input signal intensity in wild-type plants that the reduced input level in ndr1 plants is still high enough to result in the high output level. By contrast, because RPS2-mediated signal in wild-type plants is close to the edge of the plateau, the reduced input signal intensity in ndr1 plants decreases the output signal intensity drastically. The effect of the ndr1 mutation is nonspecific, and it also reduces the intensity of the input signal generated by a compatible interaction. This explains the ndr1 effect on responses to Pst. The effect of NahG may be considered to be similar to that of ndr1. It seems that the effect of ndr1 is stronger than that of NahG, judging from the expression profile differences at 6 h (Figure 5). This quantitative difference is consistent with the effects of ndr1 and NahG on hypersensitive cell death elicited by Pst/avrRpt2.
Recently, the rar1 mutation was reported to have quantitative effects on RPS2- and RPM1-mediated responses (Muskett et al., 2002; Tornero et al., 2002). However, the rar1 mutation affects RPM1-mediated responses more strongly than RPS2-mediated responses, which is opposite from the effects of ndr1 (Tornero et al., 2002). The rar1 and ndr1 mutations seem to affect a similar signaling pathway. How can we reconcile this apparent discrepancy? In rar1 plants, RPM1 was not detectable (Tornero et al., 2002). RAR1 may be involved in the stabilization of R proteins in a differential manner. Although RPM1 protein accumulation was strongly affected in rar1 plants, RPS2 protein accumulation might be affected only slightly in rar1 plants. If we assume the existence of such a regulatory mechanism that is different from the one for NDR1 (potentiation of an early signaling process), it is easy to explain the difference in the effects of the rar1 and ndr1 mutations.
We are aware of shortcomings of the quantitative model caused by oversimplification, especially when the model includes NahG plants. For example, when we compare the pair-wise correlations among responses of wild-type plants to Pst/avrRpt2, those of wild-type plants to Pst, and those of NahG plants to Pst/avrRpt2 at 6 h, they are 0.72 between wild-type Pst and wild-type Pst/avrRpt2, 0.71 between wild-type Pst/avrRpt2 and NahG Pst/avrRpt2, and 0.64 between wild-type Pst and NahG Pst/avrRpt2 (Figure 5B). These values are inconsistent with the prediction according to the model, which is that the correlation between wild-type Pst and NahG Pst/avrRpt2 should not be the smallest among the three. In addition, the similarity among these three pair-wise correlation values suggests that a model to explain these data points requires more than one degree of freedom. Note that our quantitative model assumes only one degree of freedom. It is possible that NahG has an additional effect, which contributes to an additional degree of freedom, relative to ndr1. Such an additional effect may come from a reduced level of SA as a signaling molecule instead of a potentiation factor. When in planta bacterial growth was measured, all of the bacteria tested grew better in NahG than in ndr1 plants. Although the ndr1 mutation has a stronger effect on early expression changes, the presumed additional factor associated with NahG may explain this effect on long-term bacterial growth. The observation that SA accumulates slowly in ndr1 plants (Shapiro and Zhang, 2001) is consistent with the notion that the additional factor is SA acting as a signaling molecule. It is clear that we need more sophisticated analysis than the comparison of correlation values to handle systems with more than one degree of freedom and to identify the additional factors involved.
Concluding Remarks
The sensitivity, accuracy, and broad spectrum of large-scale mRNA expression profiling as a phenotyping method revealed the quantitative nature of plant responses to pathogens. We demonstrated that apparently specific biological phenomena, such as compatible and incompatible interactions, may not necessarily be attributable to largely distinct molecular mechanisms; rather, they may share many of the same molecular mechanisms. The availability of large-scale profiling data collected under various conditions (including from organisms with different genotypes) and sophisticated analyses of the data will bring us much better views of the topology and dynamics of biological systems.
METHODS
Plants and Bacteria
Arabidopsis thaliana plants were grown at 22°C with 70% RH under ∼130 μmol·m−2·s−1 light with a 12-h-light/12-h-dark cycle in controlled-environment chambers. Approximately 4-week old plants were used. The plants and bacterial strains used are listed in Table 1.
Sample Preparation and Data Collection
Leaves of plants were hand-infiltrated with a water suspension of a bacterial strain at a dose of 2 × 107 colony-forming units (cfu)/mL (except one case in which 106 cfu/mL was used; Table 1) using a needleless syringe (Katagiri et al., 2002). The leaf tissue was harvested at the times indicated in Table 1 and immediately frozen in liquid nitrogen. RNA was prepared from frozen tissue using RNAwiz (Ambion, Austin, TX). Equal amounts of RNA from triplicate samples were pooled before the cRNA labeling reaction. Quality control of RNA, complementary RNA labeling, hybridization to a GeneChip, and collection of data from the hybridized GeneChip were performed as described previously (Zhu et al., 2001). The AtGenome1 Array (Affymetrix, Santa Clara, CA) was used as the GeneChip.
Data Analysis
Unless indicated otherwise, analysis of mRNA expression profile data was performed as follows. After hybridization, expression values from each pooled sample were normalized globally to the average value of 100. When an expression value was <5, it was adjusted to 5. The ratio of expression values for each probe set between a particular sample and the corresponding water-infiltrated control (same plant genotype and same time point) was calculated. For each comparison, the probe sets that showed greater than twofold difference from the control in at least one sample were selected. In this ratio calculation, we did not select probe sets unless the larger value of the two expression values was >50. The selected expression ratio values were log2 transformed and subjected to complete linkage hierarchical clustering analysis with uncentered correlations using Cluster (Eisen et al., 1998). Both “genes” (probe sets in this case) and “arrays” (samples) were clustered in Figures 2A and 5A, and only genes were clustered in Figures 3A and 4. The results of the hierarchical clustering analysis were visualized using TreeView (Eisen et al., 1998), in which increased and decreased expression for each probe set are depicted by red and green horizontal lines, respectively. The correlation for each pair of samples also was calculated. The correlation (normalized dot product) R is defined as
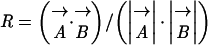 |
(1) |
where
| → |
| A |
and
| → |
| B |
are vectors that represent a pair of sample data points in the linear space with dimensions equal to the number of the probe sets with the log2-transformed ratio values. The absolute values of the log2-transformed expression ratio values of the selected probe sets for each data set were averaged to represent the average amplitudes of the profiles shown in Figures 2A, 3A, 4, and 5A. All of the expression data (after the global normalization) used to generate the figures are provided as supplemental data online.
In Planta Bacterial Growth Measurement
Plants were inoculated as described above, except that a dose of 105 cfu/mL was used. Viable bacteria from the leaf were counted at 24 and 48 h after inoculation, as described previously (Katagiri et al., 2002).
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes.
Supplementary Material
Acknowledgments
We thank Brain Staskawicz for ndr1-1 seeds, Bob Dietrich for NahG seeds, and Willem Rensink for critical reading of the manuscript.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.007591.
Footnotes
Online version contains Web-only data.
References
- Bendahmane, A., Kanyuka, K., and Baulcombe, D.C. (1999). The Rx gene from potato controls separate virus resistance and cell death responses. Plant Cell 11, 781–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bent, A.F., Kunkel, B.N., Dahlbeck, D., Brown, K.L., Schmidt, R., Giraudat, J., Leung, J., and Staskawicz, B.J. (1994). RPS2 of Arabidopsis thaliana: A leucine-rich repeat class of plant disease resistance genes. Science 265, 1856–1860. [DOI] [PubMed] [Google Scholar]
- Bisgrove, S.R., Simonich, M.T., Smith, N.M., Sattler, A., and Innes, R.W. (1994). A disease resistance gene in Arabidopsis with specificity for two different pathogen avirulence genes. Plant Cell 6, 927–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, H., Bowling, S.A., Gordon, S., and Dong, X. (1994). Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell 6, 1583–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Century, K.S., Holub, E.B., and Staskawicz, B.J. (1995). NDR1, a locus of Arabidopsis thaliana that is required for disease resistance to both a bacterial and a fungal pathogen. Proc. Natl. Acad. Sci. USA 92, 6597–6601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Century, K.S., Shapiro, A.D., Repetti, P.P., Dahlbeck, D., Holub, E., and Staskawicz, B.J. (1997). NDR1, a pathogen-induced component required for Arabidopsis disease resistance. Science 278, 1963–1965. [DOI] [PubMed] [Google Scholar]
- Chen, Z., Kloek, A.P., Boch, J., Katagiri, F., and Kunkel, B.N. (2000). The Pseudomonas syringae avrRpt2 gene product promotes pathogen virulence from inside plant cells. Mol. Plant-Microbe Interact. 13, 1312–1321. [DOI] [PubMed] [Google Scholar]
- Dangl, J.L., and Jones, J.D.G. (2001). Plant pathogens and integrated defence responses to infection. Nature 411, 826–833. [DOI] [PubMed] [Google Scholar]
- Delaney, T.P., Friedrich, L., and Ryals, J.A. (1995). Arabidopsis signal transduction mutant defective in chemically and biologically induced disease resistance. Proc. Natl. Acad. Sci. USA 92, 6602–6606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney, T.P., Uknes, S., Vernooij, B., Friedrich, L., Weymann, K., Negrotto, D., Gaffney, T., Gut-Rella, M., Kessmann, H., Ward, E., and Ryals, J. (1994). A central role of salicylic acid in plant disease resistance. Science 266, 1247–1250. [DOI] [PubMed] [Google Scholar]
- Dong, X., Mindrinos, M., Davis, K.R., and Ausubel, F.M. (1991). Induction of Arabidopsis defense genes by virulent and avirulent Pseudomonas syringae strains and by a cloned avirulence gene. Plant Cell 3, 61–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen, M.B., Spellman, P.T., Brown, P.O., and Botstein, D. (1998). Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95, 14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaffney, T., Friedrich, L., Vernooij, B., Negrotto, D., Nye, G., Uknes, S., Ward, E., Kessmann, H., and Ryals, J. (1993). Requirement of salicylic acid for the induction of systemic acquired resistance. Science 261, 754–756. [DOI] [PubMed] [Google Scholar]
- Glazebrook, J. (2001). Genes controlling expression of defense responses in Arabidopsis: 2001 status. Curr. Opin. Plant Biol. 4, 301–308. [DOI] [PubMed] [Google Scholar]
- Grant, M.R., Godiard, L., Straube, E., Ashfield, T., Lewald, J., Sattler, A., Innes, R.W., and Dangl, J.L. (1995). Structure of the Arabidopsis RPM1 gene which enables dual-specificity disease resistance. Science 269, 843–846. [DOI] [PubMed] [Google Scholar]
- Guttman, D.S., and Greenberg, J.T. (2001). Functional analysis of the type III effectors AvrRpt2 and AvrRpm1 of Pseudomonas syringae with the use of a single-copy genomic integration system. Mol. Plant-Microbe Interact. 14, 145–155. [DOI] [PubMed] [Google Scholar]
- Hammond-Kosack, K.E., and Jones, J.D.G. (1997). Plant disease resistance genes. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48, 575–607. [DOI] [PubMed] [Google Scholar]
- Innes, R.W., Bent, A.F., Kunkel, B.N., Bisgrove, S.R., and Staskawicz, B.J. (1993). Molecular analysis of avirulence gene avrRpt2 and identification of a putative regulatory sequence common to all known Pseudomonas syringae avirulence genes. J. Bacteriol. 175, 4859–4869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katagiri, F., Thilmony, R.L., and He, S.Y. (2002). The Arabidopsis thaliana-Pseudomonas syringae interaction. In The Arabidopsis Book, C.R. Somerville and E.M. Meyerowitz, eds (Rockville, MD: American Society of Plant Biologists), doi/10.1199/tab.0039, http://www.aspb.org/publications/arabidopsis. [DOI] [PMC free article] [PubMed]
- Lamb, C.J., Ryals, J.A., Ward, E.R., and Dixon, R.A. (1992). Emerging strategies for enhancing crop resistance to microbial pathogens. Biotechnology 10, 1436–1445. [DOI] [PubMed] [Google Scholar]
- Lawton, K.A., Weymann, K., Friedrich, L., Vernooij, B., Uknes, S., and Ryals, J. (1995). Systemic acquired resistance in Arabidopsis requires salicylic acid but not ethylene. Mol. Plant-Microbe Interact. 8, 863–870. [DOI] [PubMed] [Google Scholar]
- Lindgren, P.B., Peet, R.C., and Panopoulos, N.J. (1986). Gene cluster of Pseudomonas syringae pv. phaseolicola controls pathogenicity on bean and hypersensitivity on non-host plants. J. Bacteriol. 168, 512–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipshutz, R.J., Fodor, S.P., Gingeras, T.R., and Lockhart, D.J. (1999). High density synthetic oligonucleotide arrays. Nat. Genet. 21, 20–24. [DOI] [PubMed] [Google Scholar]
- Lu, M., Tang, X., and Zhou, J.-M. (2001). Arabidopsis NHO1 is required for general resistance against Pseudomonas bacteria. Plant Cell 13, 437–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maleck, K., Levine, A., Eulgem, T., Morgan, A., Schmid, J., Lawton, K.A., Dangl, J.L., and Dietrich, R.A. (2000). The transcriptome of Arabidopsis thaliana during systemic acquired resistance. Nat. Genet. 26, 403–410. [DOI] [PubMed] [Google Scholar]
- Mindrinos, M., Katagiri, F., Yu, G.-L., and Ausubel, F.M. (1994). The Arabidopsis thaliana disease resistance gene RPS2 encodes a protein containing a nucleotide-binding site and leucine-rich repeats. Cell 78, 1089–1099. [DOI] [PubMed] [Google Scholar]
- Muskett, P.R., Kahn, K., Austin, M.J., Moisan, L.J., Sadanandom, A., Shirasu, K., Jones, J.D.G., and Parker, J.E. (2002). Arabidopsis RAR1 exerts rate-limiting control of R gene–mediated defenses against multiple pathogens. Plant Cell 14, 979–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrath, C., Heck, S., Parinthawong, N., and Metraux, J.P. (2002). EDS5, an essential component of salicylic acid–dependent signaling for disease resistance in Arabidopsis, is a member of the MATE transporter family. Plant Cell 14, 275–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrath, C., and Metraux, J.P. (1999). Salicylic acid induction-deficient mutants of Arabidopsis express PR-2 and PR-5 and accumulate high levels of camalexin after pathogen inoculation. Plant Cell 11, 1393–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimchuk, Z., Marois, E., Kjemtrup, S., Leister, R.T., Katagiri, F., and Dangl, J.L. (2000). Eukaryotic fatty acylation drives plasma membrane targeting and enhances function of several type III effector proteins from Pseudomonas syringae. Cell 101, 353–363. [DOI] [PubMed] [Google Scholar]
- Rate, D.N., Cuenca, J.V., Bowman, G.R., Guttman, D.S., and Greenberg, J.T. (1999). The gain-of-function Arabidopsis acd6 mutant reveals novel regulation and function of the salicylic acid signaling pathway in controlling cell death, defenses, and cell growth. Plant Cell 11, 1695–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuber, T.L., and Ausubel, F.M. (1996). Isolation of Arabidopsis genes that differentiate between resistance responses mediated by the RPS2 and RPM1 resistance genes. Plant Cell 8, 241–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritter, C., and Dangl, J.L. (1996). Interference between two specific pathogen recognition events mediated by distinct plant disease resistance genes. Plant Cell 8, 251–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryals, J.A., Newenschwander, U.H., Willits, M.G., Molina, A., Steiner, H.-Y., and Hunt, M.D. (1996). Systemic acquired resistance. Plant Cell 8, 1809–1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenk, P.M., Kazan, K., Wilson, I., Anderson, J.P., Richmond, T., Somerville, S.C., and Manners, J.M. (2000). Coordinated plant defense responses in Arabidopsis revealed by microarray analysis. Proc. Natl. Acad. Sci. USA 97, 11655–11660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro, A.D., and Zhang, C. (2001). The role of NDR1 in avirulence gene-directed signaling and control of programmed cell death in Arabidopsis. Plant Physiol. 127, 1089–1101. [PMC free article] [PubMed] [Google Scholar]
- Shirasu, K., Nakajima, H., Rajasekhar, V.K., Dixon, R.A., and Lamb, C. (1997). Salicylic acid potentiates an agonist-dependent gain control that amplifies pathogen signals in the activation of defense mechanisms. Plant Cell 9, 261–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao, Y., Yuan, F., Leister, R.T., Ausubel, F.M., and Katagiri, F. (2000). Mutational analysis of the Arabidopsis nucleotide binding site-leucine-rich repeat resistance gene RPS2. Plant Cell 12, 2541–2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tornero, P., Merritt, P., Sasanandom, A., Shirasu, K., Innes, R.W., and Dangl, J.L. (2002). RAR1 and NDR1 contribute quantitatively to disease resistance in Arabidopsis, and their relative contributions are dependent on the R gene assayed. Plant Cell 14, 1005–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whalen, M.C., Innes, R.W., Bent, A.F., and Staskawicz, B.J. (1991). Identification of Pseudomonas syringae pathogens of Arabidopsis and a bacterial locus determining avirulence on both Arabidopsis and soybean. Plant Cell 3, 49–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, G.-L., Katagiri, F., and Ausubel, F.M. (1993). Arabidopsis mutations at the RPS2 locus result in loss of resistance to Pseudomonas syringae strains expressing the avirulence gene avrRpt2. Mol. Plant-Microbe Interact. 6, 434–443. [DOI] [PubMed] [Google Scholar]
- Zhou, N., Tootle, T.L., Tsui, F., Klessig, D.F., and Glazebrook, J. (1998). PAD4 functions upstream from salicylic acid to control defense responses in Arabidopsis. Plant Cell 10, 1021–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, T., Budworth, P., Han, B., Brown, D., Chang, H.-S., Zou, G., and Wang, X. (2001). Toward elucidating the global gene expression patterns of developing Arabidopsis: Parallel analysis of 8300 genes by high-density oligonucleotide probe array. Plant Physiol. Biochem. 39, 221–242. [Google Scholar]
- Zhu, T., and Wang, X. (2000). Large-scale profiling of the Arabidopsis transcriptome. Plant Physiol. 124, 1472–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.