Abstract
The hypersensitive response (HR) in plants is a programmed cell death that is commonly associated with disease resistance. A novel mutation in Arabidopsis, hlm1, which causes aberrant regulation of cell death, manifested by a lesion-mimic phenotype and an altered HR, segregated as a single recessive allele. Broad-spectrum defense mechanisms remained functional or were constitutive in the mutant plants, which also exhibited increased resistance to a virulent strain of Pseudomonas syringae pv tomato. In response to avirulent strains of the same pathogen, the hlm1 mutant showed differential abilities to restrict bacterial growth, depending on the avirulence gene expressed by the pathogen. The HLM1 gene encodes a cyclic nucleotide–gated channel, CNGC4. Preliminary study of the HLM1/CNGC4 gene pro-duct in Xenopus oocytes (inside-out patch-clamp technique) showed that CNGC4 is permeable to both K+ and Na+ and is activated by both cGMP and cAMP. HLM1 gene expression is induced in response to pathogen infection and some pathogen-related signals. Thus, HLM1 might constitute a common downstream component of the signaling pathways leading to HR/resistance.
INTRODUCTION
In response to pathogen attack, plants have developed complex signaling and defense mechanisms to protect themselves. One of the most efficient and immediate resistance reactions is the hypersensitive response (HR), which is characterized by the rapid death of the cells directly in contact with, or in close proximity to, the pathogen. The HR is thought to confine the pathogen by stopping it from spreading from the site of the attempted infection and to trigger local and systemic signaling for the activation of defenses in noninfected cells. However, in many cases, the importance of this defense response in plant resistance has not been proven, and our understanding of the molecular mechanisms underlying the HR remains limited (Richberg et al., 1998).
Data suggest that the HR is a form of programmed cell death (Greenberg, 1997), because its initiation depends on active plant metabolism (Keen et al., 1981; Levine et al., 1996) and can be triggered by elicitor molecules from bacteria or fungi. In addition, a number of plant mutants develop spontaneous necrotic lesions in the absence of any pathogen attack, and these lines also show other hallmarks of the HR, such as the expression of pathogenesis-related genes and/or enhanced resistance to pathogen infection (Greenberg et al., 1993, 1994; Dietrich et al., 1994). Genetic analysis of these lesion-mimic mutants has permitted the identification of certain actors and/or regulators of this program, including the LSD1 gene, which encodes a putative zinc finger protein (Dietrich et al., 1997).
Some of the earliest signaling events detected during the HR include ion fluxes, influx of calcium and H+, and efflux of K+ and Cl− (Atkinson et al., 1990). Among these ions, calcium acts as an important second messenger in the activation of resistance responses (Grant et al., 2000), in particular the HR. Calcium channel blockers inhibit the HR in tobacco and soybean systems (Atkinson et al., 1990; He et al., 1993; Levine et al., 1996; Sasabe et al., 2000). In addition, calcium influx and the transient increase in cytosolic calcium levels after elicitor treatment are necessary for the induction of the oxidative burst, one of the signaling events associated with the HR (Jabs et al., 1997). Thus, a number of studies have implicated ion fluxes and calcium in cell death signaling. Recently, the phenotypes of the two Arabidopsis lesion-mimic mutants dnd1 and cpn1, both of which exhibit increased disease resistance to pathogens, were shown to be caused by mutations in genes that encode calcium-related proteins: CNGC2, a cation channel that can conduct calcium; and a copine, a calcium-dependent, phospholipid binding protein (Clough et al., 2000; Jambunathan et al., 2001). Interestingly, although both of these mutations cause spontaneous lesions and increase pathogen resistance, they have opposing effects on the HR in response to the same pathogen (Pseudomonas syringae): the HR is accelerated and enhanced in cpn1, but it is almost completely abolished in dnd1. This observation challenges current belief about the role of the hypersensitive cell death in resistance to pathogens. From the study of dnd1, it has been suggested that cell death can be uncoupled from resistance. However, this mutant displays spontaneous cell death and, as a result, constitutively active defenses. Thus, it is difficult to establish clearly the role of the hypersensitive cell death in this context. By contrast, the accelerated HR in cpn1 supports the role of CPN1 as a negative regulator of HR cell death and the existence of a link between HR and resistance.
Here, we describe a new Arabidopsis lesion-mimic mutant, named hlm1 for HR-like lesion mimic, that shows constitutive expression of defense genes and salicylic acid (SA) production but that is impaired in its HR to different bacterial pathogens. It also differs from the wild type in its resistance to avirulent Pseudomonas syringae pv tomato strains containing the avrRps4 or avrRpm1 gene. HLM1 was found to encode a member of the CNGC (cyclic nucleotide–gated channel) ion channel family (Schuurink et al., 1998; Köhler et al., 1999; Maser et al., 2001) and is upregulated in response to pathogen infection.
RESULTS
hlm1 Is a Lesion-Mimic Mutant and Shows Constitutive Expression of Defenses
We screened a population of Arabidopsis mutagenized by T-DNA insertion (Bechtold et al., 1993) for alterations of the HR as described previously (Godard et al., 2000) or for their ability to display spontaneous lesions on the leaves. The hlm1 mutant displayed clear developmental and growth abnormalities, as shown in Figure 1A. hlm1 plants had a more compact and reduced stature, and their leaves were smaller and thicker than wild-type leaves, with shorter petioles. Leaves of the mutant line displayed small necrotic lesions that were not visible to the naked eye until 3 weeks after germination but that could be observed with a microscope (Figure 1B). These lesions usually occurred randomly in the leaf, but they were more numerous at the margins and tip, leading to a chlorotic, senescence-like phenotype at the tips of leaves after 3 to 4 weeks of growth (Figure 1A). These lesions occurred in hlm1 plants in the absence of the pathogen and appeared consistently in all mutant progeny plants. Evans blue staining revealed the presence of numerous microscopic lesions consisting of clusters of a few dead cells (Figure 1C). No lesions or dead cells were observed in the wild-type plants (data not shown). Remarkably, hlm1 plants accumulated autofluorescent phenolic compounds around the lesions (Figure 1D), a hallmark of plant defense responses that also is observed in some lesion-mimic mutants. Because the phenotype of some lesion-mimic mutants depends on environmental conditions, we monitored the kinetics of appearance and/or the number of lesions under varying photoperiods, humidity conditions, and light intensities. Although some differences were seen, no strict conditionality was observed (data not shown).
Figure 1.
Lesion-Mimic Phenotypes Associated with the hlm1 Mutation.
(A) Four-week-old plants photographed at the same distance. The three mutant alleles exhibit similar lesions and alterations in growth and morphology compared with Ws-4.
(B) Spontaneous lesion formation in hlm1 mutant plants. A representative leaf of a 3-week-old plant, and the lesions observed with a microscope (Zeiss Axiophot), are shown. Bars = 100 μm.
(C) Staining of leaves with Evans blue reveals the presence of numerous microscopic lesions consisting of clusters of a few dead cells.
(D) Observation of leaves with a fluorescence microscope (Zeiss Axiophot using an FT510 filter) shows autofluorescence corresponding to the lesions observed in (B).
Because lesion-mimic mutants are altered in genes that might play a role in the hypersensitive cell death program, we examined whether hlm1 plants show constitutive expression of defense genes. hlm1 plants had constitutively high levels of defense gene transcripts, at all developmental stages, for the PR1, GST2, and CHIB genes (Figure 2A). In wild-type plants, GST2 and CHIB expression was low but increased with the age of the plants, whereas PR1 expression was not detectable. By contrast, PDF1-2 was found to be partially repressed in the mutant. This result, together with the accumulation of PR1 transcripts in hlm1, prompted us to measure the level of SA in mutant plants, because PR1 expression is largely dependent on SA and SA signaling and PDF1-2 expression depends on ethylene/jasmonate signaling, another pathway leading to defense (Glazebrook, 2001). SA levels were found to be much higher in the mutant than in the wild type (Figure 2B).
Figure 2.
Defense Gene Expression and SA Levels in Wild-Type and hlm1 Plants.
(A) Transcript levels of the PR1 (Pathogenesis-Related1), CHIB (Chitinase B), GST2 (Glutathione Sulfo-Transferase2), and PDF1-2 genes in wild-type (Ws-4) and hlm1 plants as determined by RNA gel blot analysis at 14, 21, and 28 days after sowing.
(B) Total SA levels in wild-type (Ws-4) (hatched bars) and hlm1 (closed bars) plants. Leaves were harvested from plants grown on soil under short-day conditions. SA measurements and standard errors are derived from four replicates. F.W., fresh weight.
hlm1 Is Impaired in the HR and in Resistance to Pathogens
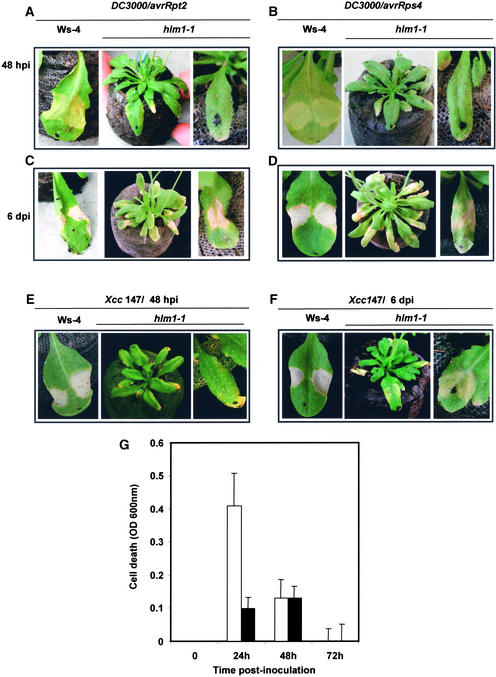
Because hlm1 shows spontaneous necrotic lesions and constitutive expression of defense markers, the wild-type gene could play a role in the control of cell death signaling in the context of the HR. To investigate this possibility further, the ability of hlm1 plants to develop an HR was examined by inoculation with different avirulent Pseudomonas strains and with Xanthomonas campestris pv campestris. hlm1 and wild-type (Wassilewskija [Ws-4]) leaves were infiltrated with two different concentrations (106 and 5 × 107 colony-forming units [cfu]/mL) of Pseudomonas bearing the avrRpm1 (data not shown), avrRpt2, or avrRps4 avirulence gene, and the appearance of the HR was monitored over time (Figure 3). Interestingly, the mutant plants did not exhibit any visible sign of the HR at 24 and 48 h after inoculation (0% of inoculated leaves at 106 cfu/mL; 10 to 27% of leaves, depending on the strain, at 5 × 107 cfu/mL), whereas the infiltrated regions of wild-type leaves collapsed (100% of inoculated leaves regardless of the strain or the inoculum) (Figures 3A and 3B). However, 6 days after inoculation, a diffuse chlorosis was observed at the inoculation site in 80% of the mutant leaves in response to Pseudomonas (avrRps4) (Figure 3D), whereas HR-like symptoms were visible in response to Pseudomonas (avrRpt2) (Figure 3C). An avirulent strain of Xanthomonas gave a response similar to that of Pseudomonas (avrRps4) (Figures 3E and 3F). Different resistance genes control responsiveness to these bacterial strains, suggesting that hlm1 is disrupted in a common component of the plant defense system leading to HR.
Figure 3.
HR Phenotypes of hlm1 Plants in Response to Diverse Avirulent Pathogens.
(A) to (D) Leaves from wild-type (Ws-4) and hlm1 plants after inoculation with Pseudomonas DC3000 containing the avrRpt2 gene ([A] and [C]) or the avrRps4 gene ([B] and [D]) at 48 h after inoculation (hpi; [A] and [B]) or 6 days after inoculation (dpi; [C] and [D]).
(E) and (F) Leaves from wild-type (Ws-4) and hlm1 plants after inoculation with the avirulent strain of Xanthomonas (Xcc 147) at 48 h (E) and 6 days (F) after inoculation.
Dots on the leaves indicate inoculated leaves. Three days after inoculation with strains 147, DC3000/avrRps4, and DC3000/avrRpt2, 10, 11, and 27% of the inoculated leaves, respectively, responded when inocula of 5 × 107 cfu/mL were used, whereas 100% of leaves responded in the case of wild-type control plants. Six days after inoculation, 80, 80, and 71% of leaves responded to the same strains, respectively, showing either chlorotic symptoms (avrRps4 and Xanthomonas strain 147) or HR-like symptoms (avrRpt2).
(G) Evaluation of cell death in wild-type (open bars) and hlm1 (closed bars) leaves after inoculation with avirulent strain 147 of Xanthomonas. Uptake of Evans blue by leaves was quantified by spectrophotometry. Data are expressed as OD units and result from four independent experiments.
To confirm the absence of hypersensitive cell death in response to avirulent pathogens in the mutant, cell death was assessed using the Evans blue leaf disc assay (Baker and Mock, 1994). When challenged with the avirulent strain of Xanthomonas, cells of the wild-type line located at the inoculation site showed an increase of Evans blue uptake at 24 to 48 h after inoculation, indicating cell death (Figure 3G). Because this dye is not retained after tissue collapse, we observed a decrease in cell death rate at 72 h after inoculation. By contrast, a very low rate of cell death occurred when leaves of the mutant line were inoculated with the same avirulent strain, confirming the macroscopic observations.
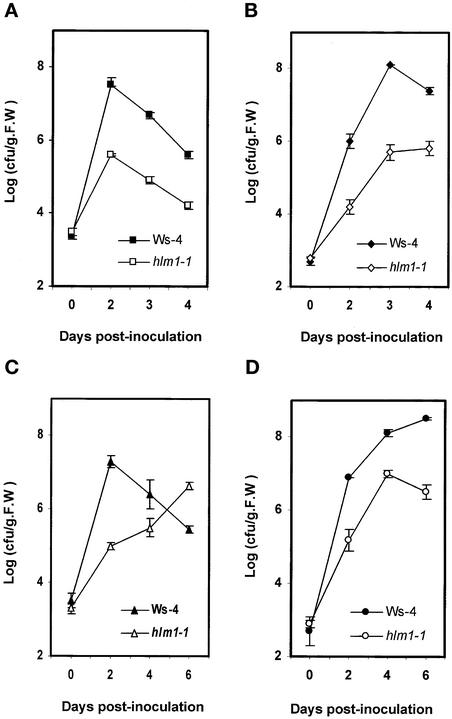
To determine whether the strong decrease of hypersensitive cell death in the hlm1 mutant is associated with altered disease resistance, the growth of four strains of Pseudomonas, one virulent and three avirulent, was measured over time. Growth of the Pseudomonas strains was clearly altered in the mutant line (Figures 4A to 4D). Virulent Pseudomonas bacteria grew less in mutant leaf tissue than in Ws-4 wild-type tissue (Figure 4D), the degree of reduction being similar to that observed for Pseudomonas expressing the avirulence genes avrRpm1, avrRpt2, and avrRps4 (Figures 4A to 4C). In hlm1, avirulent bacterial strains grew even less than in the wild type, at least during the first 2 days of infection. Four and/or 6 days after inoculation, these results were confirmed for the avirulent strains expressing the avrRpm1 and avrRpt2 genes, but surprisingly, the bacterial growth of Pseudomonas containing the avrRps4 gene increased (40-fold between days 2 and 6), whereas it decreased in the wild type (70-fold between days 2 and 6), indicating that the increased resistance observed during the early stages of infection was transient and that the HR in this case seemed to be required directly or indirectly for resistance. These results suggest that the absence, or at least the strong reduction and/or delay of HR cell death, results in alterations of disease resistance depending on the gene-for-gene interaction being studied.
Figure 4.
In Planta Growth of Virulent and Avirulent Strains of Pseudomonas in hlm1.
Growth of avirulent Pseudomonas expressing the avrRpm1 gene (A), the avrRpt2 gene (B), or the avrRps4 gene (C) and of virulent Pseudomonas (DC3000) (D) in wild-type (Ws-4) and hlm1 plants. Inoculation was performed with a bacterial suspension of 105 cfu/mL, and bacterial growth determinations were performed at the times indicated. Mean bacterial densities are shown (three replicates with corresponding standard deviations) for one representative experiment from two to three independent experiments performed with each strain. F.W., fresh weight.
The reduced resistance in response to Pseudomonas (avrRps4) was observed in spite of the increased expression of defense mechanisms in the mutant described in Figure 2. However, the higher level of resistance exhibited by the mutant plants to the other virulent or avirulent bacterial strains might be attributable to the constitutive expression of defense mechanisms. In favor of this hypothesis, the PR1 gene was not only expressed constitutively in the uninoculated mutant plants but also induced over this constitutive level in response to an avirulent strain of Xanthomonas (Figure 5). By contrast, the induction of PDF1-2, another defense marker related to the ethylene/jasmonate signaling pathway, was repressed in the mutant, probably because of the constitutively high levels of SA in the mutant. Athsr3, a molecular marker of the HR (Lacomme and Roby, 1999), showed an expression profile similar to that of PR1 and thus may be more related to defense than to cell death occurring during incompatible interactions. In summary, the HR is affected in hlm1, and certain defense markers are expressed constitutively, whereas other broad-spectrum disease resistance mechanisms function normally.
Figure 5.
RNA Gel Blot Analysis of the Expression of Defense Genes (PR1, PDF1-2, and Athsr3) and CNGC2 in Wild-Type (Ws-4) and hlm1 Plants at Different Times (Hours) after Inoculation with the Avirulent Strain of Xanthomonas (Xcc 147) or with Water.
Similar results (not shown) were obtained in a second independent experiment.
Genetic Analysis of hlm1, Cloning of HLM1, and Identification of Two Other Mutant Alleles
The hlm1 mutation (called hlm1-1 because of the subsequent identification of other mutant alleles) was isolated in the homozygous state. The original homozygous hlm1-1/hlm1-1 was backcrossed to the Ws-4 wild type. The HLM1/hlm1-1 F1 plants were allowed to self-pollinate, and the segregation of the mutant phenotype was monitored in the F2 population. The mutation segregated as a single recessive allele (data not shown), and this analysis indicated that the hlm1-1 mutation probably was T-DNA tagged. The T-DNA contained genes that conferred dominant traits for resistance to kanamycin and the herbicide ammonium glufosinate (Bouchez et al., 1993), which segregated together in a 3:1 ratio of resistant-to-sensitive plants in a population of ∼400 individuals in the F2 generation (data not shown), indicating a single integration locus in the mutant line. Because the T-DNA cosegregated with the mutant phenotype, the mutation probably was tagged. DNA gel blot analysis confirmed the presence of a single copy of the T-DNA (data not shown).
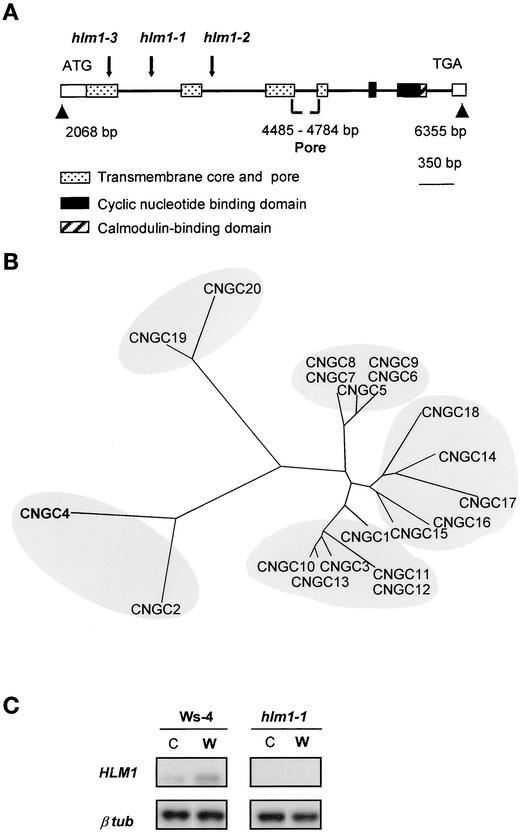
The sequence of the plant DNA flanking the T-DNA borders was determined and used in a Basic Local Alignment Search Tool (BLAST) search of the Arabidopsis genome database, revealing that the T-DNA was inserted into the first intron of an ion channel gene, CNGC4 (Figure 6A). The genomic sequence available through the sequence of the MDK4 BAC clone containing CNGC4 and the cDNA sequence we determined revealed the intron/exon structure of a 2068-bp gene composed of eight exons. The putative HLM1/CNGC4 protein, like other CNGCs, has both cyclic nucleotide and calmodulin domains at the C terminus, a transmembrane core, and a pore domain (Figure 6A) resembling those of the Shaker family, another group of ion transport proteins (Maser et al., 2001). Alignment of the putative pore regions of HLM1/CNGC4 and the other 19 members of the CNGC gene family gave a phylogenetic tree (Figure 6B) showing that CNGC4 and CNGC2, a previously identified CNGC gene (Köhler et al., 1999; Leng et al., 1999; Clough et al., 2000), are closely related and distant from the other CNGCs. As expected, no HLM1 transcript was detected in the hlm1-1 mutant leaves (Figure 6C), and the levels of transcripts found in the corresponding wild-type plants (Ws-4) were barely detectable.
Figure 6.
Genetic and Molecular Identification of the HLM1 Gene.
(A) Genomic organization of hlm1-1, hlm1-2, and hlm1-3. Arrows indicate the insertion site of the T-DNA in each mutant allele within the HLM1 gene sequence. Gene organization in exons (open box) and introns (thick black line) and the localization of the different domains of the deduced protein are presented.
(B) Comparison of the predicted HLM1/CNGC4 protein with the other members of the CNGC family. The phylogenetic tree of Arabidopsis CNGC proteins was drawn using the Treeview program (http://taxonomy.zoology.gla.uk/rod/treeview.html) after alignment of the putative pore region with the CLUSTAL X program (Thompson et al., 1997). The regions delimiting the pore domains were deduced from the available cDNA sequences or predicted by a hydrophobicity profile of the protein (Kyte and Doolittle, 1982) and/or by sequence homology with animal CNGCs. The CNGCs identified are named according to Maser et al. (2001).
(C) Reverse transcriptase–mediated PCR analysis of HLM1/CNGC4 transcript accumulation at 24 h after treatment in wild-type (Ws-4) and hlm1-1 leaves untreated (C) or infiltrated with water (W). HLM1, an 887-bp region of the HLM1 transcript; β tub, a 317-bp region of the β-tubulin4 gene.
Because genetic complementation by transformation was difficult as a result of the very low fertility of the mutant plants, an alternative strategy was chosen to demonstrate that the disruption in CNGC4 was responsible for the mutant phenotype in hlm1-1 mutant plants. A search for other mutant alleles was performed using a flanking insertion site library derived from the original mutant library from which hlm1-1 was isolated (Samson et al., 2002) and by PCR screening of DNA pools prepared from plants of the same library. Two additional mutant alleles were found, hlm1-2 and hlm1-3, that exhibited single T-DNA insertions in the second intron and the first exon of the gene, respectively. These two mutants showed the same lesion-mimic phenotype as hlm1-1 (Figure 1A) and both were altered in the HR (data not shown), indicating that the T-DNA insertion into the CNGC4 gene is responsible for the mutant phenotype of hlm1.
Xenopus Oocytes Expressing CNGC4 (HLM1) Display an Exogenous Ion Channel Gated by Both cGMP and cAMP
Xenopus laevis oocytes were injected with CNGC4 (HLM1) complementary RNA (cRNA) or a combination of CNGC4/KAT1 cRNAs, and control oocytes were injected with KAT1 cRNA alone or water (see Methods; five independent experiments). Membrane patches excised after 3 to 6 days then were voltage clamped in the inside-out configuration. Injection of the K+ channel encoding KAT1 cRNA was used as a control for the oocyte's ability to express foreign transcripts. KAT1 cRNA–injected oocytes always displayed typical KAT1 current in two-electrode voltage-clamp control experiments performed as described previously (Véry et al., 1995) (data not shown). Currents activated specifically by cGMP and cAMP (i.e., never observed in the absence of cyclic nucleotide monophosphate [cNMP] and never observed in control oocytes) were observed in one of these experiments: in three oocytes (among five tested) injected with CNGC4 cRNA and in one oocyte (one tested) injected with CNGC4/KAT1 cRNAs, but not in five (of five tested) water-injected or in four (of four tested) KAT1 cRNA–injected oocytes. This finding suggested a very low expression level of the CNGC4-related conductance in the oocyte plasma membrane. Attempts to increase CNGC4 expression by increasing the amount of injected CNGC4 cRNA usually resulted in high mortality of the oocytes.
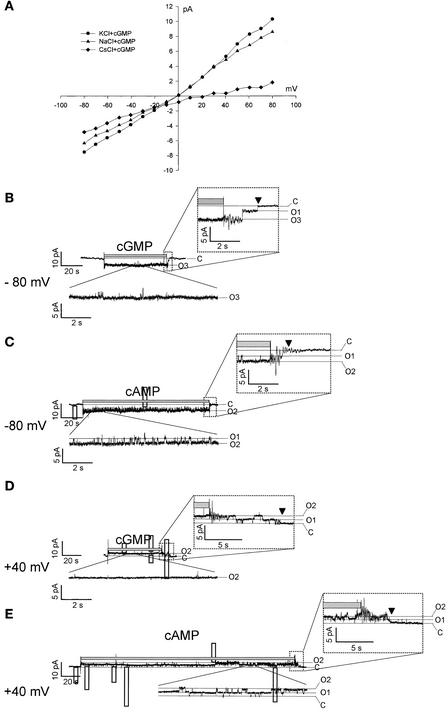
Only single-channel patches were obtained, except in the case illustrated in Figure 7. Perfusion with saturating 0.5 mM cGMP or cAMP caused immediate and reversible increases in current, suggesting that these channels were gated directly by both cGMP and cAMP. Unitary currents that could be resolved in all four positive patches showed cNMP-gated currents flowing through ∼20-pS unitary conductance channels (facing 100 mM K+ plus 0.5 mM cGMP; −80 mV) (Figure 7B). Thus, the inside-out patch shown in Figure 7 harbored at least four channels at the beginning of the recordings (Figure 7A), whereas only two channels could be seen in recordings made 5 to 6 min later (Figures 7D and 7E). Subsequent recordings from this patch showed even less channel activity, suggesting that CNGC4 channels underwent rapid rundown after patch excision (channel activity vanished completely after 11 min).
Figure 7.
Single-Channel Currents Recorded (Inside-Out Configuration) through a Membrane Patch Excised from a CNGC4 cRNA–Injected Oocyte.
(A) Current-voltage relationship for a patch facing 100 mM KCl (circles), 100 mM NaCl (triangles), or 100 mM CsCl (diamonds) with saturating 0.5 mM cGMP. The pipette contained 100 mM KCl. The background currents (i.e., in the absence of cGMP) were subtracted for each trace.
(B) to (E) Current traces from the same patch described in (A), with the cytosolic face exposed to 100 mM KCl plus 0.5 mM cGMP ([B] and [D]) or plus 0.5 mM cAMP ([C] and [E]), at the times indicated by the gray bar above the traces and the membrane potentials indicated. A record sample, in the presence of cNMP at steady state, is shown magnified in the inset below each current trace. The time course of the current after the removal of the cNMP is shown magnified in the inset above each current trace. C, the leak-current level (channels closed); 01–03, the current levels corresponding to one to three open channels.
Figure 7A shows current recorded in the presence of cGMP minus current in the absence of cGMP. When 100 mM KCl was used on both sides of the patch, the cGMP-gated current (closed circles) showed a weak outward rectification (i.e., more current at positive voltages than at negative voltages). Changing K+ to Na+ at the cytoplasmic side of the patch affected the cGMP-gated current poorly, indicating a weak K+/Na+ selectivity of CNGC4 channels. Changing K+ to Cs+ reduced the outward current dramatically, suggesting either a much lower mobility of Cs+ ions through the CNGC4 pore or partial blockage of the pore by Cs+. This effect was fully reversible (data not shown).
The same patch then was analyzed in a gap-free mode (i.e., by continuous voltage clamping first at −80 mV [Figures 7B and 7C] and then at +40 mV [Figures 7D and 7E]). At both of these potential values, only a few discrete closing/opening events were resolved during exposure of the cytoplasmic side to cGMP (Figures 7B and 7D, magnified insets below the records), whereas such events were frequent during exposure to cAMP (Figures 7C and 7E, magnified insets below the records). Upon removal of cGMP, in addition to transients caused by the operation of the perfusion apparatus, discrete closing events were resolved that resulted in a tailing of the current (Figures 7B and 7D, magnified insets above the records). Upon removal of cAMP, essentially no discrete closing events were resolved (Figures 7B and 7C, magnified insets above the records), and the time course of tailing (from the end of the gray bar to the arrow) was much shorter than that after cGMP removal at both of the tested membrane potentials. With both cGMP and cAMP, the tailing time course was shorter at −80 mV than at +40 mV. These data suggest that cGMP is bound more efficiently to CNGC4 than cAMP and also that cNMP's affinity for CNGC4 is higher at +40 mV than at −80 mV.
Together, the available data suggest the following: (1) CNGC4 is a CNGC that is activated more efficiently by cGMP than by cAMP; (2) CNGC4 is permeable to both K+ and Na+ and seemingly is blocked by Cs+, a feature that will be useful in future studies; (3) voltage has some effect on CNGC4 gating; and (4) further functional characterization of CNGC4 in oocytes is problematic because of the low expression level and the rapid rundown after patch excision.
HLM1 Gene Expression in Response to Pathogen Infection
Because the hlm1 mutation results in the alteration of plant defenses, we tested whether inoculation with pathogens could affect the expression of HLM1. Inoculation with an avirulent strain of Xanthomonas did not result in any change in the expression of the HLM1 gene in Ws-4, the genetic background of the mutant (data not shown). However, HLM1 expression was induced by the same pathogen in the Columbia (Col-0) background (Figure 8A) between 8 and 24 h after inoculation. For comparison, PR1 expression was observed clearly at 24 h after inoculation in all cases. These results were confirmed by monitoring β-glucuronidase (GUS) activity in transgenic plants (Col-0 background) containing fusions between the HLM1 gene promoter and the GUS coding region (Figure 8B). After inoculation with the avirulent strain of Xanthomonas, transient expression of HLM1 was observed starting at 8 to 10 h after inoculation, with a maximum at ∼24 h and decreasing during the next 24 h. As a comparison, the same analysis was performed using transgenic lines containing a CNGC2 promoter–GUS fusion gene, CNGC2 being a CNGC gene identified previously by a mutation (dnd1) that causes an altered HR in response to bacterial pathogens. The CNGC2 promoter was activated constitutively in leaves, except in the inoculation sites at 24 h after inoculation, where the promoter clearly was repressed (Figure 8B). Interestingly, in the mutant background, CNGC2 seemed to be expressed at higher levels (Figure 5), suggesting that some compensation process might occur in the absence of CNGC4.
Figure 8.
HLM1/CNGC4 Expression after Pathogen Inoculation.
(A) HLM1 and PR1 transcript accumulation in wild-type plants (Col-0 and Ws-4) at different times after inoculation (0, 8, and 24 h) with an avirulent strain of Xanthomonas.
(B) Histochemical localization of GUS activity in leaves from transgenic plants containing a HLM1 promoter–GUS or a CNGC2 promoter–GUS fusion gene, both healthy (H) and after inoculation with the avirulent strain of Xanthomonas (Xcc 147; I) or water (C). Undetached leaves were infiltrated in a small region (1 cm2) with the bacterial strain at 108 cfu/mL, and the leaves were collected at the times (hours) indicated after inoculation (hpi). Arrows indicate the localization of inoculated zones.
Other signals in the activation of defense mechanisms, such as SA, 1-aminocyclopropane-1-carboxylic acid (ACC), and methyl jasmonate (Glazebrook, 2001), were tested as potential inducers of HLM1 gene expression. Although SA and ACC did not affect HLM1 expression significantly, methyl jasmonate, and more particularly ACC and methyl jasmonate used together, efficiently induced HLM1 expression. By contrast, CNGC2 was expressed constitutively regardless of the treatment (data not shown).
DISCUSSION
We identified a novel lesion-mimic mutant, hlm1, that was affected in the HR and in resistance to diverse pathogens. Although lesion-mimic mutants could result from mutations that affect plant cell physiology and that may be unrelated to disease defense responses, at least some of them are thought to represent defects in genes that regulate the HR cell death program directly. For example, the lsd and acd mutants are affected in resistance to pathogens (Greenberg et al., 1993; Dietrich et al., 1994). In addition, mutations that result in the constitutive expression of defense mechanisms, such as the cpr mutations cpr1 (Bowling et al., 1994), cpr5 (Bowling et al., 1997), cpr20 and cpr21 (Silva et al., 1999), and cpr22 (Yoshioka et al., 2001), cause spontaneous lesion phenotypes. Resistance genes or genes that act downstream of the R gene, such as Prf (Oldroyd and Staskawicz, 1998), ssi1 (Shah et al., 1999), Pto (Tang et al., 1999), mpk4 (Petersen et al., 2000), and ssi2-1 (Shah et al., 2001), also can cause such phenotypes when they are overexpressed. These mutants provide the opportunity to dissect the defense signaling pathways and the communication between multiple pathways that might not be available by studying the responses of wild-type plants to pathogens. Thus, among the different lesion-mimic mutants identified to date, some of them present a normal HR (all of the lsd mutants except lsd1, acd5, cpr22, and dll1) (Dietrich et al., 1994; Greenberg et al., 2000; Yoshioka et al., 2001; Pilloff et al., 2002), one of them exhibits an accelerated HR (cpn1) (Jambunathan et al., 2001), some of them show runaway cell death (lsd1 and acd2) (Dietrich et al., 1994; Greenberg et al., 1994), and some others show a suppressed HR phenotype (dnd1, acd6, agd2, and hrl1) (Yu et al., 1998; Rate et al., 1999; Rate and Greenberg, 2001; Devadas and Raina, 2002). Among these mutants, lesion formation can be either nahG and/or NPR dependent or nahG and/or NPR independent (lsd2, lsd4, cpr5, and ssi2) (Bowling et al., 1997; Hunt et al., 1997; Shah et al., 2001). Such studies highlight the complexity involved in the regulation of the defense responses in plants, about which relatively little is known. The hlm1 mutant, which is affected strongly in its ability to mount an HR, provides a useful tool with which to study the mechanisms that control programmed cell death, gene-for-gene resistance, and broad-spectrum disease resistance.
The HLM1 gene encodes a CNGC, CNGC4. The CNGCs possess cyclic nucleotide and calmodulin binding domains together at the C-terminal region, and their transmembrane domains resemble those of the Shaker channel family. Functional heterologous expression of CNGCs has been difficult to achieve; some data suggest that some of them might form K+-permeable channels (Leng et al., 1999), whereas other studies show that they may be permeable to divalent cations (Sunkar et al., 2000; Leng et al., 2002). In mammalian systems, CNGC isoforms may be components of functional tetrameric channels, some of them being competent as channels, others being required for the function of such channels. CNGC4 has not been characterized functionally. To this end, CNGC4 cRNA was injected into Xenopus oocytes, and the inside-out configuration of the patch-clamp technique was used as in most reports on animal CNGCs (Biel et al., 1996; Zagotta and Siegelbaum, 1996). Only a few recordings were obtained, clearly indicating that functional characterization of CNGC4 is problematic in this system. This effect may be caused by its very low level of expression and the fact that the few channels that were expressed showed quick inactivation in the inside-out configuration. However, these preliminary data suggest that CNGC4 expression results in the formation of poorly selective (permeable to both K+ and Na+) channels that are open in the presence of the second messengers cGMP and cAMP. We have not yet determined whether these channels are permeable to (or controlled by) Ca2+, a property that also could support a role in HR signaling. Clearly, additional studies aimed at determining the functional features of CNGC4 should be performed, preferably in another heterologous context such as HEK cells.
Mutants that show a similar phenotype (spontaneous lesions and HR suppression) have been isolated (Yu et al., 1998; Rate et al., 1999; Rate and Greenberg, 2001), including the dnd mutants. One of their corresponding genes, DND1, has been cloned and encodes a CNGC, CNGC2. The CNGC gene family of Arabidopsis comprises 20 members with overall sequence similarities ranging between 55 and 83%, and according to Maser et al. (2001), they can be divided in four groups, group IV genes being more distantly related to the other CNGCs. CNGC4 and CNGC2 are very closely related and belong to the subgroup IVB, which contains only these two genes (Maser et al., 2001; this study). These two CNGCs, closely related on the basis of both their structures and their possible function in plants, might participate in signaling pathways that lead to the HR. However, they are not redundant, because mutations in either gene confer an altered HR phenotype. Thus, the physiological functions of these two channels in the plant are of particular interest for future research.
A number of differences in the phenotypes conferred by the corresponding mutations and in the expression of both genes have been identified in this study, addressing the question of their respective roles in plant resistance to pathogens. Both mutants show spontaneous lesions and constitutive activation of defense mechanisms. These effects could be explained by perturbation of cell homeostasis; these channels might have functions in the absence of pathogen attack and, additionally, might be recruited for signal transduction, leading to defense establishment upon infection. This hypothesis would explain the dual phenotype of spontaneous lesions and activation of defense and suppression of the HR that is observed in both mutants. Alternatively, the increased defense responses observed in hlm1 could play a role in HR suppression (a form of desensitization), as suggested recently for the hrl1 mutant (Devadas and Raina, 2002). However, this model seems less convincing for two reasons: (1) several reports suggest that the HR and defense gene expression result from distinct pathways (Jakobek and Lindgren, 1993; Morel and Dangl, 1997; Zhou et al., 1997), and (2) hlm1 does not seem to be desensitized, because it is still able to respond to pathogen attack, as shown by the increased expression of PR1 and Athsr3 (8 to 72 h after inoculation), compared with noninoculated mutant plants.
Several genes are known to be important for both defense regulation and growth and development in plants and animals. In animals, certain genes important for the innate immune response also are involved in dorsoventral patterning (Anderson, 2000), and NFκB is important for controlling cell death and growth in addition to defense responses (Hatada et al., 2000). In plants, AGD2 represents one example: the agd2 mutant not only shows increased resistance to pathogens, increased SA levels, and spontaneous lesions but also an alteration in cell growth (Rate and Greenberg, 2001). ACD2, the accelerated cell death gene, encodes red chlorophyll catabolite reductase, which suppresses the spread of disease symptoms (Mach et al., 2001). Similarly, acd6 is affected in both cell death and cell enlargement (Rate et al., 1999). Such pleiotropic effects are not related specifically to lesion-mimic mutants, because the mutants defective in jasmonate biosynthesis or perception not only are deficient in defense responses but also are male sterile (Vijayan et al., 1998; Turner et al., 2002), and recent reports show that protein degradation may be crucial for pathogen responses and resistance signaling (Dodds and Schwechheimer, 2002). Thus, in addition to those proteins involved specifically in defense responses, such as the R proteins, some proteins may function in defense signaling and be involved in plant development or other physiological responses.
The consequences of the absence of either CNGC protein in the two mutants seem to be different, at least in one important respect: in dnd1, the loss-of-HR phenotype did not affect the gene-for-gene-mediated restriction of pathogen growth (Yu et al., 1998), whereas in hlm1, after a transient stage of increased broad-spectrum resistance, gene-for-gene resistance was affected in response to certain pathogens (DC3000/avrRps4) but not in response to others (DC3000 harboring avrRpm1 or avrRpt2) (this work). Although this observation is difficult to interpret at this time, especially if we consider the constitutive induction of defense mechanisms in the uninoculated mutant plants, it suggests that these CNGCs have different functions in defense signaling. The differential patterns of expression of the two genes also favor this hypothesis. CNGC2 is expressed constitutively at rather high levels and is repressed upon infection, whereas CNGC4 is barely detectable in the absence of pathogen attack and is induced upon infection, at least in a Col-0 genetic background. Interestingly, CNGC2 appeared to be expressed at a higher level in hlm1/hlm1 plants, suggesting the existence of coordinated regulation for the expression of the two genes.
In mammals, CNGCs are essential components of signaling pathways involved in the olfactory and visual systems (Firestein, 2001; Hardie and Raghu, 2001). They constitute the final targets of a cascade, downstream from the receptors (odor or light receptors), and participate in the regulation of the entire response through a feedback pathway that involves other ion channels. Because the HR is suppressed in hlm1/hlm1 plants in response to different pathogens whose signal perception acts through different pathways (Aarts et al., 1998), CNGC4, like animal CNGCs, might be a common downstream component of signaling pathways leading to the HR. Although the suppression or strong decrease and/or delay of HR in the mutant lines have different consequences on plant resistance depending on the resistance gene–avirulence gene interaction in question, the importance of the HR in plant disease resistance cannot be addressed directly using hlm1 mutant plants, because spontaneous lesions and broad-spectrum resistance also are induced as a direct or indirect consequence of the mutation. It should be possible to gain further insights into the roles of CNGC4 and CNGC2 within the complex interplay of plant defense signaling networks by examination of the mutant phenotypes in response to other pathogens whose specificities are mediated by the EDS1 signaling pathway (Aarts et al., 1998), by exploration of the roles of SGT1, RAR1, and NPR1, other downstream signaling components, and through expression analysis of a much wider array of genes. The rapid induction of the transcription of HLM1 by ethylene/jasmonate and by Xanthomonas, similar to that observed for PAD4 (Jirage et al., 1999), EDS1 (Falk et al., 1999), NDR1 (Century et al., 1997), and EDS5 (Nawrath et al., 2002) in response to pathogens, would favor such an approach.
Although the mechanisms by which HLM1 activates its associated responses remain to be defined, we have shown that this mutation affects the HR, broad-spectrum resistance, and certain specificities of gene-for-gene resistance. Future analysis of the hlm1 mutant should help to clarify the defense signal transduction network involving ion fluxes, some of which play essential roles in HR cell death (Scheel, 1998; Grant et al., 2000). Finally, this work opens exciting perspectives for addressing the biological function of at least one of the CNGC channels, about which almost nothing is known in plants.
METHODS
Plants and Growth Conditions
Arabidopsis thaliana plants, accessions Columbia and Wassilewskija (Ws-4), were used in these experiments. The mutant hlm1-1 line was isolated from a Ws-4 ecotype population mutagenized with T-DNA (Bechtold et al., 1993). The mutant allele hlm1-2 was identified by comparing the hlm1-1 sequence with the flanking insertion site database (Genoplante, Evry, France), and hlm1-3 was found by PCR screening of ∼40,000 T-DNA insertion mutants (Ws-4 ecotype) (Institut National de la Recherche Agronomique, Versailles, France) with primers corresponding to CNGC4 and the T-DNA left and right borders. Primers used for the identification of the positive line were 5′-ATGGCCACAGAACAAGAATTCACACGC-3′ and 5′-CTACAAATT-GCCTTTTCTTATCGAC-3′ for the CNGC4 gene. The exact position of the T-DNA insertion was determined by sequencing the T-DNA flanking sequences. Plants homozygous for the disruption and hemizygous plants were identified in the progeny of the positive line by PCR.
For all experiments, mutant and wild-type seeds were sterilized in sodium hypochlorite (12%) for 20 min, rinsed five times with sterile distilled water, and sown on Murashige and Skoog (1962) medium. Seeds were incubated for 1 week under a 12-h photoperiod at 22°C to allow germination. Seedlings then were picked out of Jiffy pots and grown in a culture chamber with a 9-h photoperiod at 22°C and 70% RH. Four-week-old plants were used for all inoculation experiments.
Bacterial Strains and Inoculation Procedures
Xanthomonas campestris pv campestris and Pseudomonas syringae pv tomato were grown as described previously (Godard et al., 2000). Plant inoculations and bacterial growth measurements were performed as described by Lummerzheim et al. (1993).
DNA Constructs and Plant Transformation
The CNGC4 promoter region was isolated by PCR with proofreading Pfu polymerase (Promega, Charbonnières, France) on genomic DNA with a forward primer containing a BamHI site (5′-TTTTTGGAT-CCTCTTTTAGTTTCGTTTTGTCTTAATATACCC-3′) and a reverse primer introducing an NcoI site just upstream of the ATG codon (5′-TTTTTCCATGGAAAGTACTAACGCATGCAACACAGAGAG-3′). The amplified fragment (2068 bp) was sequenced and introduced in the pBI320.X vector carrying the β-glucuronidase (GUS) coding sequence. The resulting translational fusion then was cloned into the pBIB-Hygro binary vector. This final vector was used to transform Agrobacterium tumefaciens MP90. Arabidopsis plants (ecotype Columbia) were transformed by the floral dip method (Clough and Bent, 1998). Transformants were selected on Murashige and Skoog (1962) medium supplemented with 1% Suc, 0.7% agar, and 30 μg/mL hygromycin B (H7772; Sigma).
GUS Assay
Plant tissue was ground in liquid nitrogen, homogenized in 1× GUS buffer (50 mM NaPO4, pH 7; 10 mM β-mercaptoethanol; 10 mM NaEDTA; 0.1% Sarcosyl; 0.1% Triton X-100), and centrifuged for 5 min at 10,000g, and the supernatant was assayed for GUS activity as described previously (Roby et al., 1990).
Histochemistry and Microscopy
Cell death was quantified by monitoring the uptake of Evans blue (0.25%) by leaf discs from hlm1-1 and Ws-4 inoculated or healthy plants (Baker and Mock, 1994). The assay was performed with eight leaf discs (5 mm diameter) from the inoculated zones of four to five plants. Three replicates were tested per point. Localization of cell death was observed after vacuum infiltrating 4-week-old plants in a solution of Evans blue (0.25%). Plants were destained in water, and leaves were observed by light microscopy (Axiophot; Zeiss, Jena, Germany). Autofluorescence examinations were performed with a Zeiss Axiophot microscope using an FT510 filter. The whole leaf was observed with a Leica MZ FLIII fluorescence stereomicroscope using a GFP3 filter (Wetzlar, Germany). For histochemical GUS assays, inoculated leaves were collected at different times after inoculation and infiltrated under vacuum with GUS staining buffer (Jefferson et al., 1987). Samples were incubated overnight at 37°C in the staining buffer, and leaves were fixed and cleared in 70% ethanol.
RNA Isolation and Hybridization
Total RNA was isolated from frozen leaf material using the Extract-All solution according to the recommendations of the manufacturer (Eurobio, Les Ulis, France). RNA gel blot analysis was performed as described previously (Godard et al., 2000).
Reverse Transcriptase–Mediated PCR
First-strand cDNA synthesis was performed using 8 μg of total RNA and Superscript II RNase H− reverse transcriptase (Invitrogen, Cergy Pontoise, France). cDNA synthesized from the total RNA of hlm1-1 and Ws-4 plants was tested for CNGC4 expression by PCR using primers 5′-GACCCGAGATCCAAATGGGTTCG-3′ (forward) and 5′-CCTATCTTTTGGGGTCTCATGACT-3′ (reverse) (annealing at 65°C). As a control, PCR was performed to amplify the cDNA of a constitutively expressed β-tubulin gene using primers 5′-GTCCAGTGTCTG-TGATATTGCACC-3′ (forward) and 5′-GGCTCCCTCGGATTCGTA-AGC-3′ (reverse) (annealing 65°C).
Measurement of Salicylic Acid
Total salicylic acid (SA) (free SA plus SA conjugate) concentration was measured as described by Baillieul et al. (1995).
Identification of the HLM1 Gene and Isolation of CNGC4 cDNA
The full-length CNGC4 open reading frame was amplified with Pfu polymerase (Promega) by reverse transcriptase–mediated PCR on poly(A+) RNA from leaves using primers 5′-ACTAGTATGGCCACAGAACAAGAATTCACACGC-3′ and 5′-GGTACCTCAATAATCATC-AAAATCGTCGGGATTGGG-3′, introducing an SpeI site just upstream of the ATG and a KpnI site just downstream of the stop codon. The amplified fragment was cloned in the pBluescript II KS (+/−) vector and sequenced.
Patch Clamp in CNGC4-Expressing Oocytes
Oocytes were obtained surgically from cold-anesthetized Xenopus laevis (Véry et al., 1995). CNGC4 and KAT1 complementary RNA (cRNA) solutions (typical concentration of 0.5 to 1 ng/nL) were obtained after in vitro transcription of the corresponding cDNA. Stage V and VI oocytes were injected with 50 nL of water or KAT1 cRNA solution (controls) or with 50 nL of CNGC4 cRNA solution or a mixture (3:1) of the CNGC4/KAT1 cRNAs. After 3 to 6 days of incubation at 19°C, the vitelline membrane was removed and oocytes were placed in a bath solution containing 100 mM KCl, 2.5 mM EGTA, and 10 mM Hepes-KOH, pH 7.2. Pipettes with a tip resistance of 2 to 5 MΩ were filled with the same solution. Inside-out patches were excised and perfused with different solutions (as indicated in the legend to Figure 7) with the help of a Rapid Solution Changer (RSC-160; Bio-Logic SA, Claix, France). Currents were recorded using a patch-clamp amplifier (Axopatch 200-A; Axon Instruments, Foster City, CA). Data were filtered at 2 kHz and digitized at 10 kHz through a 1200 Digidata AC/DC converter using pClamp6 software (Axon Instruments). The single-channel data then were analyzed with Clampfit8.1 software (Axon Instruments) and filtered digitally (emulating an eight-pole Bessel filter) at 150 Hz for display. All potentials mentioned in the text and figures are given with respect to the physiological polarity (i.e., for inside-out patch-clamp configuration, the opposite of the imposed voltage).
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes.
Accession Number
The GenBank accession number for CNGC4 is AB010695.
Supplementary Material
Acknowledgments
We thank Patrick Saindrenan for SA level determinations, Nigel Grimsley for critical reading of the manuscript, and Maurice Tronchet and Marie Christine Auriac for technical assistance. We also are grateful to Hervé Sentenac and Jean Baptiste Thibaud for helpful discussions. S.M. was supported by a Marie Curie Fellowship of the European Community program Improving Human Research Potential and the Socio-Economic Knowledge Base under contract HPMFCT 2000-00634.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.006999.
Footnotes
Online version contains Web-only data.
References
- Aarts, N., Metz, M., Holub, E., Staskawicz, B.J., Daniels, M.J., and Parker, J.E. (1998). Different requirements for EDS1 and NDR1 by disease resistance genes define at least two R gene-mediated signaling pathways in Arabidopsis. Proc. Natl. Acad. Sci. USA 95, 10306–10311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, K.V. (2000). Toll signaling pathways in the innate immune response. Curr. Opin. Immunol. 12, 13–19. [DOI] [PubMed] [Google Scholar]
- Atkinson, M.M., Keppler, L.D., Orlandi, E.W., Baker, C.J., and Mischke, C.F. (1990). Involvement of plasma membrane calcium influx in bacterial induction of the K+/H+ and hypersensitive responses in tobacco. Plant Physiol. 92, 215–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baillieul, F., Genetet, I., Kopp, M., Saindrenan, P., Fritig, B., and Kauffman, S. (1995). A new elicitor of the hypersensitive response in tobacco: A fungal glycoprotein elicits cell death, expression of defense genes, production of salicylic acid, and induction of systemic acquired resistance. Plant J. 8, 551–560. [DOI] [PubMed] [Google Scholar]
- Baker, C.J., and Mock, N.M. (1994). An improved method for monitoring cell death in cell suspension and leaf disc assays using Evans blue. Plant Cell Tissue Organ Cult. 39, 7–12. [Google Scholar]
- Bechtold, N., Ellis, J., and Pelletier, G. (1993). In planta Agrobacterium-mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C. R. Acad. Sci. Paris 316, 1194–1199. [Google Scholar]
- Biel, M., Zong, X., and Hofman, F. (1996). Cyclic nucleotide-gated cation channels: Molecular diversity, structure and cellular functions. Trends Cardiovasc. Med. 6, 274–280. [DOI] [PubMed] [Google Scholar]
- Bouchez, D., Camilleri, C., and Caboche, M. (1993). A binary vector based on Basta resistance for in planta transformation of Arabidopsis thaliana. C. R. Acad. Sci. Paris 316, 1188–1193. [Google Scholar]
- Bowling, S.A., Clarke, J.D., Liu, Y., Klessig, D.F., and Dong, X. (1997). The cpr5 mutant of Arabidopsis expressed both NPR1-dependent and NPR1-independent resistance. Plant Cell 9, 1573–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowling, S.A., Guo, A., Cao, H., Gordon, A.S., Klessig, D.F., and Dong, X. (1994). A mutation in Arabidopsis that leads to constitutive expression of systemic acquired resistance. Plant Cell 6, 1845–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Century, K.S., Shapiro, A.D., Repetti, P.P., Dahlbeck, D., Holub, E., and Staskawicz, B.J. (1997). NDR1, a pathogen-induced component required for Arabidopsis disease resistance. Science 278, 1963–1965. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., Fengler, K.A., Yu, I., Lippok, B., Smith, R.K., Jr., and Bent, A.F. (2000). The Arabidopsis dnd1 “defense, no death” gene encodes a mutated cyclic nucleotide-gated ion channel. Proc. Natl. Acad. Sci. USA 97, 9323–9328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devadas, S.K., and Raina, R. (2002). Preexisting systemic acquired resistance suppresses hypersensitive response-associated cell death in Arabidopsis hrl1 mutant. Plant Physiol. 128, 1234–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich, R.A., Delaney, T.P., Uknes, S.J., Ward, E.R., Ryals, J.A., and Dangl, J.L. (1994). Arabidopsis mutants simulating disease resistance response. Cell 77, 565–577. [DOI] [PubMed] [Google Scholar]
- Dietrich, R.A., Richberg, M.H., Shmidt, R., Dean, C., and Dangl, J.L. (1997). A novel zinc finger protein is encoded by the Arabidopsis LSD1 gene and functions as a negative regulator of plant cell death. Cell 88, 685–694. [DOI] [PubMed] [Google Scholar]
- Dodds, P.N., and Schwechheimer, C. (2002). A breakdown in defense signaling. Plant Cell 14 (suppl.), S5.–S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk, A., Feys, B.J., Frost, L.N., Jones, J.D.G., Daniels, M.J., and Parker, J.E. (1999). EDS1, an essential component of R gene-mediated disease resistance in Arabidopsis, has homology to eukaryotic lipases. Proc. Natl. Acad. Sci. USA 96, 3292–3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firestein, S. (2001). How the olfactory system makes sense of scents. Nature 413, 211–218. [DOI] [PubMed] [Google Scholar]
- Glazebrook, J. (2001). Genes controlling expression of defense responses in Arabidopsis: 2001 status. Curr. Opin. Plant Biol. 4, 301–308. [DOI] [PubMed] [Google Scholar]
- Godard, F., Lummerzheim, M., Saindrenan, P., Balagué, C., and Roby, D. (2000). Hxc-2, an Arabidopsis mutant with altered hypersensitive response to Xanthomonas campestris pv. campestris. Plant J. 24, 749–762. [DOI] [PubMed] [Google Scholar]
- Grant, J.J., Yun, B.-W., and Loake, G.J. (2000). Oxidative burst and cognate redox signalling reported by luciferase imaging: Identification of a signal network that functions independently of ethylene, SA, and Me-JA but is dependent on MAPKK activity. Plant J. 24, 569–582. [DOI] [PubMed] [Google Scholar]
- Greenberg, J.T. (1997). Programmed cell death in plant pathogen interactions. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48, 525–545. [DOI] [PubMed] [Google Scholar]
- Greenberg, J.T., Guo, A., Klessig, D.F., and Ausubel, F.M. (1994). Programmed cell death in plants: A pathogen-triggered response activated coordinately with multiple defense functions. Cell 77, 551–563. [DOI] [PubMed] [Google Scholar]
- Greenberg, J.T., Klessig, D.F., and Ausubel, F.M. (1993). Arabidopsis thaliana mutants compromised for the control of cellular damage during pathogenesis and aging. Plant J. 4, 327–341. [DOI] [PubMed] [Google Scholar]
- Greenberg, J.T., Silverman, F.P., and Liang, H. (2000). Uncoupling salicylic acid-dependent cell death and defense-related responses from disease resistance in the Arabidopsis mutant acd5. Genetics 156, 341–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardie, R.C., and Raghu, P. (2001). Visual transduction in Drosophila. Nature 413, 186–193. [DOI] [PubMed] [Google Scholar]
- Hatada, E.N., Krappman, D., and Scheidereit, C. (2000). NF-κB and the innate immune response. Curr. Opin. Immunol. 12, 52–58. [DOI] [PubMed] [Google Scholar]
- He, S.Y., Huang, H.C., and Collmer, A. (1993). Pseudomonas syringae pv. syringae harpinPss: A protein that is secreted via the Hrp pathway and elicits the hypersensitive response in plants. Cell 73, 1255–1266. [DOI] [PubMed] [Google Scholar]
- Hunt, M.D., Delaney, T.P., Dietrich, R.A., Weymann, K.B., Dangl, J.L., and Ryals, J.A. (1997). Salicylate-independent lesion formation in Arabidopsis lsd mutants. Mol. Plant-Microbe Interact. 10, 531–536. [DOI] [PubMed] [Google Scholar]
- Jabs, T., Tschope, M., Colling, C., Hahlbrock, K., and Scheel, D. (1997). Elicitor-stimulated ion fluxes and O2− from the oxidative burst are essential components in triggering defense gene activation and phytoalexin synthesis in parsley. Proc. Natl. Acad. Sci. USA 94, 4800–4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobek, J.L., and Lindgren, P.B. (1993). Generalized induction of defense responses in bean is not correlated with the induction of the hypersensitive reaction. Plant Cell 5, 49–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jambunathan, N., Siani, J.M., and McNellis, T.W. (2001). A humidity-sensitive Arabidopsis copine mutant exhibits precocious cell death and increased disease resistance. Plant Cell 13, 2225–2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson, R.A., Kavanagh, T.A., and Bevan, M.W. (1987). GUS gene fusions: β-Glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6, 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jirage, D., Tootle, T.L., Reuber, T.L., Frost, L.N., Feys, B.J., Parker, J.E., Ausubel, F.M., and Glazebrook, J. (1999). Arabidopsis thaliana PAD4 encodes a lipase-like gene that is important for salicylic acid signaling. Proc. Natl. Acad. Sci. USA 96, 13583–13588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keen, N.T., Ersek, T., Long, M., Bruegger, B., and Holliday, M. (1981). Inhibition of the hypersensitive response of soybean leaves to incompatible Pseudomonas spp. by blastocidin S, streptomycin or elevated temperature. Physiol. Plant Pathol. 18, 325–337. [Google Scholar]
- Köhler, C., Merkle, T., and Neuhaus, G. (1999). Characterisation of a novel gene family of putative cyclic nucleotide- and calmodulin-regulated ion channels in Arabidopsis thaliana. Plant J. 18, 97–104. [DOI] [PubMed] [Google Scholar]
- Kyte, J., and Doolittle, R.F. (1982). A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157, 105–132. [DOI] [PubMed] [Google Scholar]
- Lacomme, C., and Roby, D. (1999). Identification of new early markers of the hypersensitive response in Arabidopsis thaliana. FEBS Lett. 459, 149–153. [DOI] [PubMed] [Google Scholar]
- Leng, Q., Mercier, R.W., Hua, B.G., Fromm, H., and Berkowitz, G.A. (2002). Electrophysiological analysis of cloned cyclic nucleotide-gated ion channels. Plant Physiol. 128, 400–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leng, Q., Mercier, R.W., Yao, W., and Berkowitz, G.A. (1999). Cloning and first functional characterization of a plant cyclic nucleotide-gated cation channel. Plant Physiol. 121, 753–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine, A., Pennell, R.I., Alvarez, M.E., Palmer, R., and Lamb, C. (1996). Calcium-mediated apoptosis in a plant hypersensitive disease resistance response. Curr. Biol. 6, 427–437. [DOI] [PubMed] [Google Scholar]
- Lummerzheim, M., de Olivera, D., Castresana, C., Miguens, F.C., Louzada, E., Roby, D., Van Montagu, M., and Timmerman, B. (1993). Identification of compatible and incompatible interactions between Arabidopsis thaliana and Xanthomonas campestris pv. campestris and characterization of the hypersensitive response. Mol. Plant-Microbe Interact. 6, 532–544. [Google Scholar]
- Mach, J.M., Castillo, A.R., Hoogstraten, R., and Greenberg, J.T. (2001). Arabidopsis-accelerated cell death gene ACD2 encodes red chlorophyll catabolite reductase and suppresses the spread of disease symptoms. Proc. Natl. Acad. Sci. USA 98, 771–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maser, P., et al. (2001). Phylogenetic relationships within cation transporter families of Arabidopsis. Plant Physiol. 126, 1646–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel, J.-B., and Dangl, J. (1997). The hypersensitive response and the induction of cell death in plants. Cell Death Differ. 4, 671–683. [DOI] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioessays with tobacco tissue cultures. Physiol. Plant. 15, 473–497. [Google Scholar]
- Nawrath, C., Heck, S., Parinthawong, N., and Metraux, J.P. (2002). EDS5, an essential component of salicylic acid–dependent signaling for disease resistance in Arabidopsis, is a member of the MATE transporter family. Plant Cell 14, 275–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldroyd, G.E.D., and Staskawicz, B.J. (1998). Genetically engineered broad-spectrum disease resistance in tomato. Proc. Natl. Acad. Sci. USA 95, 10300–10305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen, M., et al. (2000). Arabidopsis map kinase 4 negatively regulates systemic acquired resistance. Cell 103, 1111–1120. [DOI] [PubMed] [Google Scholar]
- Pilloff, R.K., Devadas, S.K., Enyedi, A., and Raina, R. (2002). The Arabidopsis gain-of-function mutant dll1 spontaneously develops lesions mimicking cell death associated with disease. Plant J. 30, 61–70. [DOI] [PubMed] [Google Scholar]
- Rate, D.N., Cuenca, J.V., Bowman, G.R., Guttman, D.S., and Greenberg, J.T. (1999). The gain-of-function Arabidopsis acd6 mutant reveals novel regulation and function of the salicylic acid signaling pathway in controlling cell death, defenses, and cell growth. Plant Cell 11, 1695–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rate, D.N., and Greenberg, J.T. (2001). The Arabidopsis aberrant growth and death2 mutant shows resistance to Pseudomonas syringae and reveals a role for NPR1 in suppressing hypersensitive cell death. Plant J. 27, 203–211. [DOI] [PubMed] [Google Scholar]
- Richberg, M.H., Aviv, D.H., and Dangl, J.L. (1998). Dead cells do tell tales. Curr. Opin. Plant Biol. 1, 480–485. [DOI] [PubMed] [Google Scholar]
- Roby, D., Broglie, K., Cressman, R., Biddle, P., Chet, I., and Broglie, R. (1990). Activation of a bean chitinase promoter in transgenic tobacco plants by phytopathogenic fungi. Plant Cell 2, 999–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson, F., Brunaud, V., Balzergue, S., Dubreucq, B., Lepiniec, L., Pelletier, G., Caboche, M., and Lecharny, A. (2002). FLAGdb/FST: A database of mapped flanking insertion sites (FSTs) of Arabidopsis thaliana T-DNA transformants. Nucleic Acids Res. 30, 94–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasabe, M., Takeuchi, K., Kamoun, S., Ichinose, Y., Govers, F., Toyoda, K., Shiraishi, T., and Yamada, T. (2000). Independent pathways leading to apoptotic cell death, oxidative burst and defense gene expression in response to elicitin in tobacco cell suspension culture. Eur. J. Biochem. 267, 5005–5013. [DOI] [PubMed] [Google Scholar]
- Scheel, D. (1998). Resistance response physiology and signal transduction. Curr. Opin. Plant Biol. 1, 305–310. [DOI] [PubMed] [Google Scholar]
- Schuurink, R.C., Shartzer, S.F., Fath, A., and Jones, R.L. (1998). Characterization of a calmodulin-binding transporter from the plasma membrane of barley aleurone. Proc. Natl. Acad. Sci. USA 95, 1944–1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah, J., Kachroo, P., and Klessig, D.F. (1999). The Arabidopsis ssi1 mutation restores pathogenesis-related gene expression in npr1 plants and renders defensin gene expression salicylic acid dependent. Plant Cell 11, 191–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah, J., Kachroo, P., Nandi, A., and Klessig, D.F. (2001). A recessive mutation in the Arabidopsis SSI2 gene confers SA- and NPR1-independent expression of PR genes and resistance against bacterial and oomycete pathogens. Plant J. 25, 563–574. [DOI] [PubMed] [Google Scholar]
- Silva, H., Yoshioka, K., Dooner, H.K., and Klessig, D.F. (1999). Characterization of a new Arabidopsis mutant exhibiting enhanced disease resistance. Mol. Plant-Microbe Interact. 12, 1053–1063. [DOI] [PubMed] [Google Scholar]
- Sunkar, R., Kaplan, B., Bouche, N., Arazi, T., Dolev, D., Talke, I.N., Maathuis, F.J., Sanders, D., Bouchez, D., and Fromm, H. (2000). Expression of a truncated tobacco NtCBP4 channel in transgenic plants and disruption of the homologous Arabidopsis CNGC1 gene confer Pb2+ tolerance. Plant J. 24, 533–542. [DOI] [PubMed] [Google Scholar]
- Tang, X., Xie, M., Kim, Y.J., Zhou, J., Klessig, D.F., and Martin, G.B. (1999). Overexpression of Pto activates defense responses and confers broad resistance. Plant Cell 11, 15–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J.D., Gibson, T.J., Plewniak, F., Jeanmougin, F., and Higgins, D.G. (1997). The CLUSTALX Windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876.–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner, J.G., Ellis, C., and Devoto, A. (2002). The jasmonate signal pathway. Plant Cell 14 (suppl.), S153.–S164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Véry, A.A., Gaymard, F., Bosseux, C., Sentenac, H., and Thibault, J.-B. (1995). Expression of a cloned plant K+ channel in Xenopus oocytes: Analysis of macroscopic currents. Plant J. 7, 321–332. [DOI] [PubMed] [Google Scholar]
- Vijayan, P., Shockey, J., Levesque, C.A., Cook, R.J., and Browse, J. (1998). A role for jasmonate in pathogen defense of Arabidopsis. Proc. Natl. Acad. Sci. USA 95, 7209–7214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioka, K., Kachroo, P., Tsui, F., Sharma, S.B., Shah, J., and Klessig, D.F. (2001). Environmentally sensitive, SA-dependent defense responses in the cpr22 mutant of Arabidopsis. Plant J. 26, 447–459. [DOI] [PubMed] [Google Scholar]
- Yu, I.C., Parker, J., and Bent, A.F. (1998). Gene-for-gene disease resistance without the hypersensitive response in Arabidopsis dnd1 mutant. Proc. Natl. Acad. Sci. USA 95, 7819–7824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zagotta, W.N., and Siegelbaum, S.A. (1996). Structure and function of cyclic nucleotide-gated channels. Annu. Rev. Neurosci. 19, 235–263. [DOI] [PubMed] [Google Scholar]
- Zhou, J., Tang, X., and Martin, J.B. (1997). The Pto kinase conferring resistance to tomato bacterial speck disease interacts with proteins that bind a cis-element of pathogenesis-related genes. EMBO J. 16, 3207–3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.