Abstract
Plants respond to environmental stress by activating “stress genes.” The plant hormone abscisic acid (ABA) plays an important role in stress-responsive gene expression. Although Ca2+ serves as a common second messenger in signaling stress and ABA, little is known about the molecular basis of Ca2+ action in these pathways. Here, we show that CIPK3, a Ser/Thr protein kinase that associates with a calcineurin B–like calcium sensor, regulates ABA response during seed germination and ABA- and stress-induced gene expression in Arabidopsis. The expression of the CIPK3 gene itself is responsive to ABA and stress conditions, including cold, high salt, wounding, and drought. Disruption of CIPK3 altered the expression pattern of a number of stress gene markers in response to ABA, cold, and high salt. However, drought-induced gene expression was not altered in the cipk3 mutant plants, suggesting that CIPK3 regulates select pathways in response to abiotic stress and ABA. These results identify CIPK3 as a molecular link between stress- and ABA-induced calcium signal and gene expression in plant cells. Because the cold signaling pathway is largely independent of endogenous ABA production, CIPK3 represents a cross-talk “node” between the ABA-dependent and ABA-independent pathways in stress responses.
INTRODUCTION
To survive, plants have evolved complex molecular mechanisms by which they tolerate and adapt to adverse growth conditions. When they encounter stress conditions, plant cells reprogram their cellular processes by triggering a network of signaling events that start with stress perception and end with a cellular response, such as gene expression in the nucleus. Genes that normally are silent and activated under stress conditions often are referred as “stress genes.” Abscisic acid (ABA), a phytohormone, has been shown to play a critical role in plant stress responses, because many stress signals increase the level of ABA. Furthermore, the application of ABA to plants in many ways mimics the effect of stress conditions. For example, there is much overlap in the expression pattern of stress genes after cold, drought, high salt, or ABA application, suggesting that these stress signals and ABA share common elements in the signaling pathways (Leung and Giraudat, 1998; Thomashow, 1999; Rock, 2000; Shinozaki and Yamaguchi-Shinozaki, 2000; Finkelstein et al., 2002).
Recent studies suggest that osmotic stress imposed by high salt or drought is transmitted through at least two pathways, one ABA dependent and the other ABA independent. Cold, on the other hand, exerts its effect on gene expression largely through an ABA-independent pathway (Thomashow, 1999; Shinozaki and Yamaguchi-Shinozaki, 2000). This paradigm has been established largely by molecular studies that identify the cis- and trans-acting elements responsible for gene induction by stress signals and ABA (Thomashow, 1999; Shinozaki and Yamaguchi-Shinozaki, 2000). For example, some cold- and drought-inducible (COR/RD/KIN) genes contain a cis-acting element called DRE/CRT (for drought/cold-responsive element) that is critical for drought/cold-induced gene expression but not for ABA induction. Conversely, ABA-induced expression often relies on the presence of a different cis-acting element called ABRE (for ABA-responsive element) (Thomashow, 1999; Shinozaki and Yamaguchi-Shinozaki, 2000; Uno et al., 2000; Finkelstein et al., 2002). Although salt and drought trigger a sustained increase in endogenous ABA levels and are sufficient to induce the expression of ABRE-containing genes, cold induces only a transient increase in ABA level and often is insufficient to induce ABRE-containing genes. This fact suggests that salt and drought, but not cold, have an ABA-dependent branch in the signaling network. More evidence regarding the ABA-independent pathway comes from gene marker analysis in ABA biosynthesis mutants (Thomashow, 1999; Shinozaki and Yamaguchi-Shinozaki, 2000). Because cold- and drought-induced expression of genes with DREs is largely normal in ABA-deficient mutants (aba), the pathway leading to the activation of the DRE is independent of endogenous ABA.
On the other hand, genetic analysis indicates that ABA-dependent and ABA-independent pathways may cross-talk (or even converge) through components in the signaling pathways (Knight and Knight, 2001; Xiong and Zhu, 2002). A strong candidate that may mediate such cross-talk is calcium, which serves as a common second messenger for abiotic stress conditions and ABA. A number of studies have demonstrated that ABA, cold, drought, and high-salt conditions result in a rapid increase in Ca2+ levels in plant cells (Sanders et al., 1999, 2002; Knight and Knight, 2000; Rudd and Franklin-Tong, 2001). In addition, studies have shown that cellular Ca2+ changes precede the activation of ABA-, cold-, and osmotic stress–inducible genes (Knight et al., 1996; Sheen, 1996; Tahtiharju et al., 1997; Wu et al., 1997; Thomashow, 1999; Shinozaki and Yamaguchi-Shinozaki, 2000; Knight and Knight, 2001). However, the specific molecular components that mediate Ca2+-associated gene activation remain unclear.
As a second messenger, Ca2+ transmits the primary signal into cellular responses (such as gene expression) most likely through Ca2+-regulated proteins that include Ca2+ sensors and their targets. The major Ca2+ sensors in plants include calmodulin (CaM) (Zielinski, 1998; Snedden and Fromm, 2001; Luan et al., 2002), CaM domain–containing protein kinases (CDPKs) (Harmon et al., 2000; Sanders et al., 2002), and the more recently discovered calcineurin B–like proteins (CBLs) (Luan et al., 2002). Although CDPKs act as both Ca2+ sensors and kinases, CaMs and CBLs are small Ca2+ binding proteins that do not have any enzymatic activity and that function by interacting with their target proteins. CaMs interact with a variety of target proteins, including a number of enzymes and structural proteins, whereas CBLs specifically target a family of protein kinases referred to as CIPKs (CBL-interacting protein kinases) (Luan et al., 2002). Although studies have identified a number of genes that encode CaMs, CDPKs, and CBLs/CIPKs from various plants, much less is understood regarding their role in stress- and ABA-regulated gene expression. One study showed that a constitutively active form of CDPK is sufficient to activate an ABA- and stress-responsive gene promoter in maize leaf cells (Sheen, 1996). Another study showed that overexpression of a CDPK leads to the expression of stress genes under normal conditions and increased stress tolerance in rice (Saijo et al., 2000). A CaM gene has been implicated as a negative regulator of stress- and ABA-induced gene expression, because overexpression of this CaM gene caused a reduced level of gene induction (Townley and Knight, 2002).
Genetic analysis of loss-of-function mutants is lacking regarding the function of Ca2+ sensors or their targets in stress-regulated gene expression. Here, we present evidence that CIPK3 regulates the response to ABA during seed germination and modulates stress gene expression induced by cold, salt, and ABA treatment. Interestingly, gene induction by drought and hyperosmotic stress was not affected by disruption of CIPK3 function, suggesting that CIPK3 regulates specific pathways that lead to stress gene expression. This study also identified CIPK3 as a cross-talk “node” that mediates the interaction between ABA and abiotic stress signal transduction pathways
RESULTS
CIPK3 Is Highly Expressed in Germinating Seeds and Is Inducible in Seedlings by Abiotic Stress Conditions and ABA Treatment
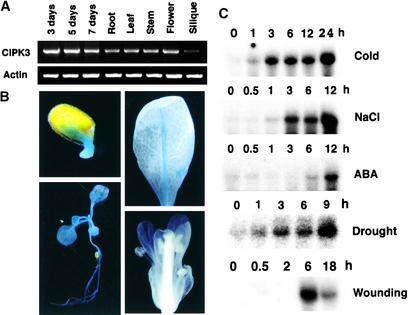
In our previous studies (Shi et al., 1999; Kim et al., 2000), we identified a number of CIPKs that associate with CBL-type calcium sensors. However, little is known about the function of the members of the CIPK gene family (Luan et al., 2002). As a step toward dissecting the function of each CIPK in calcium signaling in plants, the expression patterns of these genes were surveyed by RNA gel blot analyses. We found that the expression of one CIPK family member, CIPK3, was highly regulated. The mRNA of the CIPK3 gene was accumulated at very low levels in adult plants and was barely detectable in most organs by RNA gel blot analysis (data not shown). Therefore, we determined CIPK3 mRNA levels using reverse transcriptase–mediated (RT) PCR (Figure 1A). Germinating seeds and young seedlings (3 to 7 days after sowing) expressed the highest levels of CIPK3 mRNA, whereas levels in all organs of older plants were significantly lower. The siliques accumulated the lowest level of all tissues analyzed. To analyze the expression of the CIPK3 gene in more detail, we fused the putative promoter region of the CIPK3 gene to the β-glucuronidase (GUS) reporter and analyzed GUS activity in transgenic plants by a histochemical procedure. As shown in Figure 1B, CIPK3 promoter was highly active in the germinating seeds (3 days after sowing). GUS activity also was detected in almost all tissues of young seedlings. However, GUS activity was significantly lower in adult plants, consistent with the RT-PCR results shown in Figure 1A. A typical rosette leaf exhibited detectable GUS activity only in the vascular tissues of the main vein. Flower organs showed minimal levels of GUS activity that were detectable only in the sepals (Figure 1B). Siliques and mature seeds did not show any detectable GUS activity (data not shown).
Figure 1.
Expression Patterns of the CIPK3 Gene.
(A) RT-PCR analysis of CIPK3 transcripts during seed germination and in different organs of Arabidopsis plants. Total RNA was isolated from various tissues (root, leaf, stem, flower, and silique) of 4-week-old wild-type plants grown under long-day conditions or from germinating seeds and young seedlings (3, 5, and 7 days after sowing). RT-PCR was performed with either CIPK3-specific primers (top gel) or Actin2-specific primers (bottom gel).
(B) Histochemical GUS analysis of CIPK3 promoter–GUS transgenic plants. Top left, a germinating seed; bottom left, a 2-week-old seedling; top right, a rosette leaf from a 4-week-old plant; bottom right, a flower showing faint blue color in the sepal only.
(C) Accumulation of CIPK3 gene transcripts in response to cold (4°C), NaCl (300 mM), ABA (100 μM), drought, or wounding treatment. Each lane was loaded with 10 μg of total RNA from 3-week-old Arabidopsis plants. The numbers indicate hours after each treatment.
Interestingly, stress signals (including cold, drought, high salt, and wounding) and ABA strongly induced the expression of the CIPK3 gene (Figure 1C). Cold induction was the strongest, followed by drought, high salt, wounding, and ABA. The mRNA levels appeared to increase with time during the 24-h cold treatment. A number of studies indicate that wounding often dehydrates plants and therefore contains an osmotic stress component (Reymond et al., 2000; Cheong et al., 2002). The stress- and ABA-inducible expression pattern of the CIPK3 gene identified CIPK3 as a stress gene, implicating it in stress and ABA signal transduction.
Isolation and Phenotypic Analysis of a cipk3 Insertional Mutant
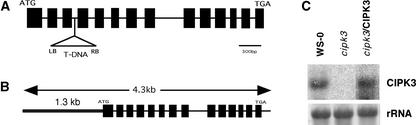
To genetically dissect the in vivo function of the CIPK3 gene, we isolated a T-DNA insertional allele of the CIPK3 gene in Arabidopsis. A single insertion was identified in homozygous plants harboring the CIPK3 gene knockout allele by DNA gel blot analysis (data not shown). Sequencing of the genomic DNA flanking the T-DNA insertion revealed that the insertional site of the T-DNA was located in the third intron in the CIPK3 gene (Figure 2A). RNA gel blot analysis demonstrated that such an insertion event abolished CIPK3 transcript expression in cipk3 knockout mutant plants (Figure 2C). For complementation analysis, we constructed a binary vector containing a 4.3-kb genomic DNA that spans the complete CIPK3 gene for transformation of the cipk3 mutant (Figure 2B). Transgenic plants containing the complementation construct expressed a comparable level of CIPK3 mRNA as the wild-type plants (Figure 2C). No significant phenotypic changes were observed in cipk3 mutant plants compared with wild-type plants under normal growth conditions in the growth chamber or greenhouse, suggesting that CIPK3 may not function in plant growth and development. This finding is consistent with a low level of expression of CIPK3 in adult plants under normal conditions.
Figure 2.
Isolation and Complementation of the cipk3 T-DNA Insertional Mutant.
(A) Scheme of the Arabidopsis CIPK3 gene. Exons (solid boxes) and introns (lines) are indicated. The position and orientation of the T-DNA insertion is shown (not to scale).
(B) Complementation genomic DNA fragment. A 4.3-kb DNA of the CIPK3 gene including 1.3 kb of the 5′ flanking region upstream from the ATG and the complete coding region was amplified by PCR and cloned into the pCAMBIA1300 vector for plant transformation.
(C) RNA gel blot analysis of CIPK3 mRNA in the wild-type (ecotype Wassilewskija [Ws-0]), mutant (cipk3), and complementation transgenic (cipk3/CIPK3) lines. Four independent complementation lines were analyzed with similar results, and results from one typical line are shown. Ten micrograms of total RNA from 3-week-old seedlings was probed with CIPK3 cDNA. rRNA on the membrane was visualized by staining with methylene blue as an equal loading control.
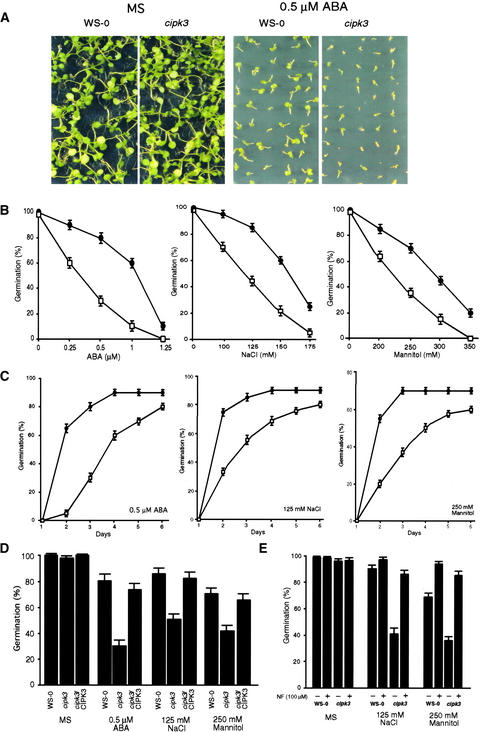
Two lines of evidence suggested that CIPK3 may play a role in stress and/or ABA signaling processes. First, Ca2+ is a common second messenger and CIPK3 is a target protein kinase of CBL-type calcium sensors. Second, CIPK3 is highly responsive to stress conditions and ABA in Arabidopsis plants. We tested whether cipk3 mutant plants were altered in their responses to ABA and abiotic stress conditions including high-salt and high osmotic stress. The seeds of cipk3 mutant and wild-type plants were plated on MS medium (Murashige and Skoog, 1962) containing various concentrations of ABA, NaCl, and mannitol. The early germination rate and growth of the seedlings were monitored. We found that the germination of cipk3 mutant seeds was more sensitive to ABA (Figure 3A). The germination of mutant seeds on ABA was delayed significantly, and the subsequent growth of seedlings also was retarded compared with that in the wild type. On the high-salt and mannitol medium, germination of cipk3 mutant seeds also was inhibited to a greater extent than in the wild-type seeds (Figure 3B).
Figure 3.
Germination of cipk3 Mutant Seeds Is Hypersensitive to ABA and Osmotic Stress Conditions.
(A) ABA inhibition of early seedling growth in cipk3 mutant and wild-type plants. Wild-type (Ws-0) and cipk3 mutant seeds on MS agar medium supplemented with 0 μM ABA (MS) or 0.5 μM ABA were incubated at 4°C for 4 days before transfer to 23°C for germination. The photograph was taken on day 10 after transfer to 23°C.
(B) Germination rate of wild-type and cipk3 mutant seeds at 3 days after transfer to 23°C in the presence of different concentrations of ABA (left), NaCl (middle), or mannitol (right).
(C) Germination time course (days after incubation at 23°C) on medium containing 0.5 μM ABA (left), 125 mM NaCl (middle), or 250 mM mannitol (right).
(D) Seed germination rate in wild-type (Ws-0), cipk3 mutant (cipk3), and four complementation (cipk3/CIPK3) lines on MS medium (control) or MS medium containing 0.5 μM ABA, 125 mM NaCl, or 250 mM mannitol. Germination was scored at 3 days after incubation at 23°C. Data from one typical complementation line are shown.
(E) Effect of the ABA biosynthesis inhibitor norflurazon (NF) on the germination of cipk3 mutant seeds on medium containing 125 mM NaCl or 250 mM mannitol.
Results in (B) to (E) are presented as average values and standard errors from three experiments. Closed circles, wild type; open squares, cipk3 mutant.
A germination time course is shown in Figure 3C. On medium containing 0.5 μM ABA, 90% of wild-type seeds and 80% of cipk3 seeds germinated in a 6-day period. However, the germination of cipk3 mutant seeds was delayed dramatically and was most pronounced at 2 days after transfer to the growth chamber, when 65% of wild-type seeds and 5% of cipk3 seeds were observed to have germinated. On medium with 125 mM NaCl, 75% of wild-type seeds had germinated after 2 days, but cipk3 seeds reached a comparable level of germination only at day 6. Likewise, in 250 mM mannitol, germination of both wild-type and cipk3 seeds was inhibited significantly. However, cipk3 seeds were much more sensitive to such hyperosmotic conditions than wild-type seeds, with a germination rate in the 6-day period of <60%, whereas wild-type seeds had achieved 70% germination after 3 days (Figure 3C). Figure 3D shows that seeds from complementation transgenic plants exhibited comparable germination levels under ABA and stress conditions, indicating that hypersensitivity to ABA and stress conditions in the cipk3 mutant resulted from disruption of the CIPK3 gene.
Because high-salt and hyperosmotic stress have been shown to increase endogenous ABA levels (Leung and Giraudat, 1998; Seo and Koshiba, 2002), we investigated whether the observed inhibition of seed germination by salt and mannitol was a consequence of ABA accumulation. We tested this possibility by including the ABA biosynthesis inhibitor norflurazon (Zeevaart and Creelman, 1988) in the medium (Figure 3E). On normal MS medium, norflurazon did not affect the germination of wild-type and cipk3 seeds. However, on the salt- or mannitol-containing medium, norflurazon rescued the hypersensitive phenotype of cipk3 mutant seeds, suggesting that salt and mannitol exerted their effect on cipk3 germination through ABA.
Based on hypersensitivity to ABA in the germination assay, we tested whether ABA-sensitive stomatal movements in adult plants was altered in the cipk3 mutant. Our results indicated that ABA-induced stomatal closure was not affected in the mutant plants (data not shown), suggesting that CIPK3 plays a role in ABA response specifically in seed germination but not in stomatal movement. This finding is consistent with the high-level expression of CIPK3 in seeds but not in guard cells, as indicated by GUS activity assays (Figure 1B and data not shown). Moreover, cipk3 mutant and wild-type plants exhibited similar growth under a variety of stress conditions. First, 7-day-old seedlings grown on normal medium were transferred to stress media such as those used in germination assays, and their growth (both roots and leaves) was monitored. Second, we assayed the drought and salt tolerance of wild-type and cipk3 mutant plants by withdrawing watering or watering with salt solution, as described previously (Liu et al., 1998). Third, we grew plants in the dark condition on media containing high concentrations of salt and mannitol and assayed hypocotyl elongation in wild-type and cipk3 mutant plants. In all of these assays, no significant difference was observed between cipk3 and wild-type plants (data not shown), suggesting that stress tolerance in cipk3 is not altered significantly during later developmental stages beyond germination.
CIPK3 Regulates Stress Gene Expression in Arabidopsis Plants
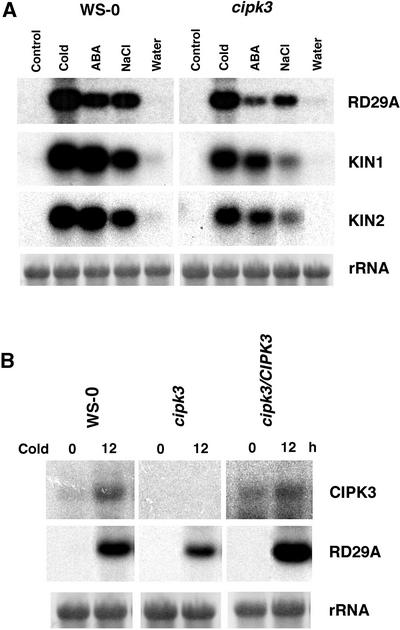
We have shown that CIPK3 is activated strongly by stress conditions and ABA, implicating CIPK3 in the regulation of stress and ABA responses in plants. Although the stress tolerance at the whole-plant level did not appear to change in the cipk3 mutant in several assays, such whole-plant stress tolerance assays often lack sufficient sensitivity to resolve changes in a particular signaling pathway in plants. Therefore, it is possible that certain signaling pathways in the stress response are altered but may not result in dramatic changes in stress tolerance at the whole-plant level. We decided to examine the specific signaling pathways at the molecular level. One of the best characterized signaling pathways for ABA and stress responses is the induction of stress gene expression upon stress or ABA treatment. A number of different stress genes were used as markers to ascertain which stress signaling pathway might involve CIPK3 function. Included were RD29A, KIN1, and KIN2 genes, which are induced strongly by cold, drought, high salt, and ABA (Kurkela and Borg-Franck, 1992; Yamaguchi-Shinozaki and Shinozaki, 1994; Tahtiharju et al., 1997; Liu et al., 1998).
As shown in Figure 4A, all three gene markers were induced by ABA, cold, and NaCl in the wild-type plants, consistent with the results from previous studies (Kurkela and Borg-Franck, 1992; Yamaguchi-Shinozaki and Shinozaki, 1994; Tahtiharju et al., 1997; Liu et al., 1998). These genes also were induced in cipk3 mutant plants, although the induction levels in the cipk3 mutant plants were significantly (∼50%) lower compared with those in the wild type. This finding suggests that CIPK3 is involved in the regulation of stress gene expression in Arabidopsis. We also examined RD29A expression in the complemented transgenic plants and found the induction level in these plants to be comparable to that in the wild-type plants (Figure 4B), indicating that the lower level of induction in the cipk3 mutant was the result of CIPK3 disruption.
Figure 4.
Expression of Stress-Responsive Genes in Wild-Type (Ws-0) and cipk3 Mutant Plants.
(A) Expression of stress genes under stress conditions and ABA application. RNA gel blot analysis of RD29A, KIN1, and KIN2 in plants after treatment with cold (4°C for 12 h), ABA (100 μM for 5 h), NaCl (300 mM for 3 h), and water control (3 h).
(B) Transcript levels of CIPK3 and RD29A in Ws-0, cipk3 mutant, and a complementation line (cipk3/CIPK3) after cold treatment for 12 h.
rRNA on the membrane was visualized by staining with methylene blue as an equal loading control.
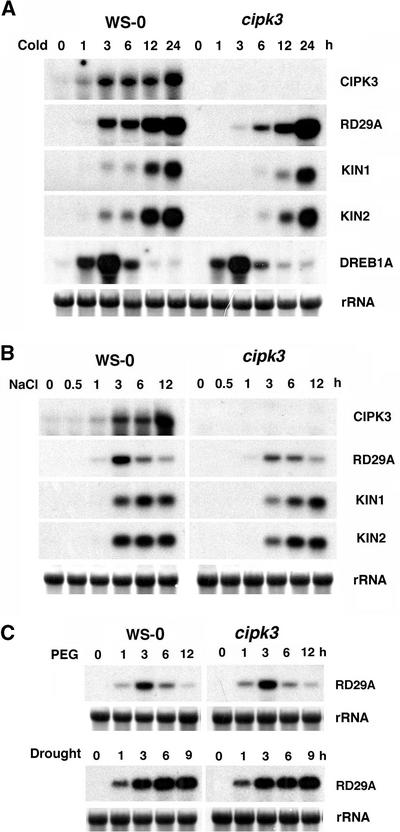
CIPK3 Modulates Cold- and Salt-Induced Gene Expression but Not Drought-Induced Gene Expression
Although the survey shown in Figure 4 revealed a difference in the induction level of stress genes in cipk3 mutant versus wild-type plants, a more detailed analysis of the induction process may provide insight into how stress gene expression is changed. For example, induction of a stress gene can be changed in terms of maximal mRNA accumulation level or in the kinetic pattern or other parameters. We compared the gene induction time course in the cipk3 mutant and wild-type plants under various stress conditions (Figure 5).
Figure 5.
Expression of Stress Genes in the Wild Type (Ws-0) and the cipk3 Mutant Induced under Various Conditions.
Conditions included 4°C cold (A), 300 mM NaCl (B), and 30% polyethylene glycol (PEG) or drought (C). rRNA on the membrane was visualized by staining with methylene blue as an equal loading control.
Under cold conditions (Figure 5A), the RD29A gene was induced rapidly in the wild-type plants and the transcript level was still increasing >24 h after the beginning of cold treatment. This pattern of cold-inducible expression for RD29A is consistent with the results reported in earlier studies (Yamaguchi-Shinozaki and Shinozaki, 1994; Liu et al., 1998). Interestingly, the cold-induced expression of RD29A was delayed significantly in the cipk3 mutant plants. The RD29A transcript level was much lower in the early phase of induction (first 12 h), although maximal induction comparable to that in the wild type was achieved at 24 h. At 3 h of cold treatment, the RD29A mRNA level in the wild-type plants was nine times higher than the level in the cipk3 mutant. At the 6-h treatment, RD29A induction in the wild type was twice as much as in mutant plants. Induction of the KIN1 and KIN2 genes was affected similarly in the cipk3 mutant (i.e., the induction was delayed significantly, but the maximal level of induction at 24 h was not altered).
As reported previously, RD29A and KIN1/KIN2 gene expression is under the control of CBFs/DREBs, transcription factors that bind to the cis-acting elements (CRE/DRE) in the promoter regions of RD29A and KIN1/KIN2 (Liu et al., 1998; Thomashow, 1999). Genes that encode CBFs/DREBs also are stress genes themselves, because they are induced by stress conditions and ABA. Therefore, a signal transduction cascade has been proposed whereby the stress signal is first transmitted to factors that activate the expression of CBFs/DREBs, which in turn bind to CRE/DREs in the RD29A and KIN1/KIN2 genes, thus activating their transcription (Thomashow, 1999). The results shown in Figure 5A demonstrate that CIPK3 may lie between the cold stress signal and the expression of CBF/DREB target genes. To further elucidate the role of CIPK3 in such a signaling cascade, we examined the induction pattern of the DREB1A/CBF3 gene, which is highly induced by cold stress (Gilmour et al., 1998, 2000; Liu et al., 1998). In both wild-type and cipk3 mutant plants, DREB1A/CBF3 was induced by cold treatment with similar kinetics and induction levels. This finding indicates that CIPK3 regulates the expression of CBF/DREB target genes but not the transcription of the CBF/DREB genes themselves.
Although cold and hyperosmotic stress caused by salt and drought share common elements in the regulation of stress gene expression, significant differences remain in the signaling processes (Knight and Knight, 2001). Figure 5B shows a comparison of stress gene induction by high salt in the wild-type and cipk3 mutant plants. The maximal level of RD29A induction was reduced by ∼50% in the cipk3 mutant. The induction of KIN1/KIN2 genes appeared to be delayed in the cipk3 mutant. Compared with cold induction, changes in salt-induced gene expression were less dramatic. Nevertheless, these changes were significant and reproducible in three independent experiments using new plant material each time. Typical patterns are shown in Figure 5B. In particular, the early induction at the 3-h time point for all three gene markers was significantly lower (each ∼50%) in the cipk3 mutant compared with wild-type plants.
Both salt and drought cause hyperosmotic stress and therefore share common elements in their signaling pathways. For example, most drought-induced genes also are induced by high salt and vice versa (Hasegawa et al., 2000; Zhu, 2002). Surprisingly, however, RD29A expression was not altered in the cipk3 mutant when treated with polyethylene glycol or under drought conditions, although polyethylene glycol–induced RD29A gene expression exhibited a very different pattern from drought-regulated induction (Figure 5C). The drought stress was imposed by either removing plants from the soil, as described previously (Yamaguchi-Shinozaki and Shinozaki, 1994; Kudla et al., 1999), or by exposing seedlings to dehydration in a laminar flow hood (data not shown). The expression of KIN1/KIN2 was not altered in cipk3 mutant plants (data not shown). These studies indicate that the drought-induced expression of RD29A and KIN1/KIN2 is not regulated by CIPK3.
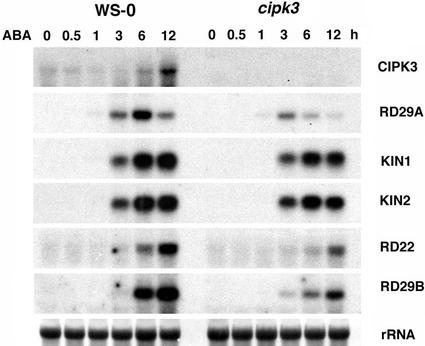
ABA-Responsive Gene Expression Is Reduced in the cipk3 Mutant
Although drought and salt increase ABA levels and thus have an ABA-dependent branch in their signaling pathways, cold has been shown to signal through an ABA-independent pathway (Shinozaki and Yamaguchi-Shinozaki, 2000; Knight and Knight, 2001). CIPK3 regulates cold- and salt-induced gene expression but not drought-induced gene expression, implicating CIPK3 in a distinct pathway(s) that apparently is independent of ABA. However, our results indicate that CIPK3 plays a role in ABA-responsive gene expression (Figures 4 and 6). Unlike cold induction that increases with time, the ABA-induced RD29A gene transcript reached its maximal level at ∼6 h after ABA application. In cipk3 mutant plants, RD29A gene induction by ABA was diminished significantly. At the 6-h time point, the RD29A transcript level in the cipk3 mutant was only ∼25% of the wild-type level. The induction of KIN1/KIN2 genes also was reduced in the cipk3 mutant, although the changes were not as dramatic as with RD29A induction.
Figure 6.
Expression of ABA-Responsive Genes in the Wild Type (Ws-0) and the cipk3 Mutant after Application of 100 μM ABA.
rRNA on the membrane was visualized by staining with methylene blue as an equal loading control.
Two other ABA-induced genes, RD22 and RD29B, were examined to further address the function of CIPK3 in ABA-responsive gene expression. The expression of RD29B is controlled by ABRE binding factors, whereas RD22 expression is activated by a different mechanism (Shinozaki and Yamaguchi-Shinozaki, 2000). Nevertheless, the expression of both RD22 and RD29B is regulated by CIPK3, as shown by the altered induction pattern in the cipk3 mutant. In all of the genes examined, the cipk3 mutant showed a significantly lower level of induction but retained similar kinetics of induction. These results clearly show that CIPK3 affects the cold-, salt-, and ABA-induced expression of RD29A and KIN1/KIN2 genes, but the mechanisms underlying the regulation are different. In both cold and salt induction, CIPK3 appears to be required for the early phase but not the late phase of the gene induction. In ABA-induced expression, CIPK3 is essential for maintaining the maximal level of induction.
DISCUSSION
We have shown that CIPK3, a calcium sensor–associated kinase, serves multiple functions in the early development and stress responses of Arabidopsis. In germinating seeds, CIPK3 is highly expressed and plays an important role in regulating ABA sensitivity. Germination of cipk3 mutant seeds was not affected under normal conditions. However, cipk3 was hypersensitive to exogenous ABA and showed a significant delay in germination on ABA-containing medium. After germination, CIPK3 expression declined dramatically under normal conditions but was highly inducible upon stress or ABA treatment. Such an expression pattern suggests that CIPK3 is not required for the normal development of the plants after germination but may play a role in stress responses. Indeed, results in this study show that signaling pathways that connect stress and ABA to gene expression are altered by the disruption of CIPK3 function. These findings identify CIPK3 as a signaling component that mediates the action of Ca2+ signals and serves as a cross-talk node in stress and ABA signal transduction.
CIPK3, a Protein Kinase That Mediates Ca2+-Regulated Stress Gene Expression
Stress acclimation in plants depends on changes in the molecular processes, especially gene expression, that initiate readjustment of physiology and metabolism. A critical issue in understanding stress acclimation is the mechanism by which plants perceive and translate the stress signal into changes in gene expression. In the past decade, significant progress has been made in the identification of stress genes and cis- and trans-acting factors that control stress-responsive expression. For example, RD29A/COR78 has been shown to be responsive to a variety of stress signals and ABA and has served as a model system for the dissection of promoter regions responsive to stress- and ABA-induced expression in plants (Liu et al., 1998; Shinozaki and Yamaguchi-Shinozaki, 2000; Seki et al., 2001). The RD29A gene promoter contains separate cis-acting elements for stress and ABA responses: the DRE/CRT for drought and cold responses, and the ABRE for ABA response. The transcription factors that bind to DRE/CRT and ABRE have been identified and shown to function in stress- and ABA-responsive gene activation (Liu et al., 1998; Choi et al., 2000; Uno et al., 2000). In addition, genes that encode these transcription factors, including CBFs/DREBs, AREBs, and ABFs, also are induced by stress and ABA, respectively. Therefore, it is thought that these transcription factors are early stress genes that must be activated before the downstream target genes, including RD29A/COR78, can be activated (Thomashow, 1999). The initial transcription factors that activate the expression of CBFs/DREBs and AREBs remain to be identified.
Although the nuclear events for stress gene activation have begun to be identified, the early signaling steps before the transcription factors are largely unknown. These include the initial receptors/sensors for the stress signals and ABA and the signal transducers in the cytoplasm (Thomashow, 1999; Shinozaki and Yamaguchi-Shinozaki, 2000; Knight and Knight, 2001). Increasing evidence suggests that Ca2+ serves as a second messenger in stress and ABA responses. Both stress conditions (e.g., cold, high salt, and drought) and ABA elicit cellular Ca2+ changes (Trewavas, 1999; Knight and Knight, 2000). Inhibition of such changes results in the inhibition of stress or ABA responses. Regarding stress- and ABA-induced gene expression, several studies have linked Ca2+ changes in the cell to the induction level of a particular target gene. For instance, Sheen (1996) used a transient expression system to show that Ca2+ is required and sufficient for the activation of a stress- and ABA-inducible promoter. Others have shown that the manipulation of Ca2+ fluxes leads to changes in stress gene expression and stress tolerance (reviewed by Thomashow, 1999). One important issue is to identify the signaling components that sense the Ca2+ signal and translate it into gene expression in the nucleus.
Several Ca2+ sensors, including CaMs and CDPKs, have long been implicated in plant stress responses. The available evidence has been built largely on the stress-inducible expression of some CaM/CDPK genes and gain-of-function analysis using overexpression and transient assays (Sheen, 1996; Saijo et al., 2000; Sanders et al., 2002; Townley and Knight, 2002). For example, transient expression of a constitutively active form of a CDPK member activates the promoter of a stress- and ABA-responsive gene in maize protoplasts under nonstress conditions (Sheen, 1996), implicating CDPK in the signal transduction pathway connecting stress/ABA to gene expression. Overexpression of a CDPK leads to the expression of stress genes under normal conditions and increased stress tolerance in rice (Saijo et al., 2000). Loss-of-function analysis using a genetics approach has yet to be conducted to determine which CaM/CDPK members are involved in which signaling pathways.
Recent studies have discovered a new type of Ca2+ sensors unique to higher plants, the so-called CBLs for their similarity to yeast and animal calcineurin B (Kudla et al., 1999; Luan et al., 2002). Unlike calcineurin B, which interacts and regulates the activity of a protein phosphatase, plant CBLs interact with and regulate the activity of a group of Ser/Thr protein kinases, referred to as CIPKs (Shi et al., 1999; Luan et al., 2002). To date, at least 10 CBL genes and 25 CIPK genes have been identified from Arabidopsis. Among these genes, SOS3/SOS2 (CBL4/CIPK24) were shown to play a role in ionic homeostasis in Arabidopsis (Xiong and Zhu, 2002). Loss of function in either SOS3 (CBL4) or its interacting kinase SOS2 (CIPK24) renders plants hypersensitive to high-salt conditions as a result of impaired ionic homeostasis. None of the CBLs and CIPKs has yet been shown to regulate stress- or ABA-regulated gene expression in plants. Our results here show that one of the CIPK genes, CIPK3, plays a role in the regulation of stress gene expression in Arabidopsis. The gene that encodes CIPK3 is highly inducible by stress and ABA, implicating CIPK3 in stress and ABA responses. This hypothesis is supported by the finding that disruption of CIPK3 function altered the pattern of stress gene induction by cold, high salt, and ABA. Interestingly, CIPK3 does not regulate the gene expression induced by drought stress. Because both salt and drought have a common ABA-dependent pathway and CIPK3 functions only in salt-induced expression, CIPK3 appears to regulate an ABA-independent pathway of salt stress but is involved in the pathway initiated by exogenous ABA application.
It is noteworthy that CIPK3 function appears to be most important in the cold induction of gene expression. Of all the marker genes examined (RD29A and KIN1/KIN2), induction was delayed most dramatically under cold conditions in the cipk3 mutant plants, although the maximal level of gene induction was not altered. This finding suggests that the cold-induced expression of RD29A and KIN1/KIN2 genes may consist of two components, the early phase and the late phase. Only the early-phase component may involve CIPK3 function. These two phases could involve two different (sets of) transcription factors or the same factors under different regulations (e.g., transcriptional versus post-translational). Transcription factors for RD29A activation, such as DREBs/CBFs, are activated at the transcriptional level by cold stress. Because the overexpression of DREB1A/CBF3 is not sufficient for a maximal induction of its target genes under normal conditions (Liu et al., 1998; Kasuga et al., 1999), it is likely that DREB1A/CBF3 also is subject to post-translational control by stress conditions. Because it takes time from the transcriptional induction of DREB1A/CBF3 to the production of proteins that in turn activate the RD29A gene, it is tempting to suggest that DREB1A/CBF3 produced from transcriptional activation by cold stress may function in the “late phase” of RD29A induction and is not controlled by CIPK3. Indeed, CIPK3 does not appear to affect the transcription of DREB1A/CBF3. The preexisting DREB1A or other transcription factors may function in the “early phase” of the induction and are regulated by post-translational modification such as phosphorylation. Such post-translational modification may involve CIPK3, consistent with the fact that the early-phase induction of RD29A and KIN1/KIN2 genes is affected in the cipk3 mutant. This hypothesis also is consistent with the general observation that Ca2+ changes often occur very rapidly after abiotic stress (Trewavas, 1999; Knight and Knight, 2000). Identification of the physiological substrates for CIPK3 will be helpful to test this hypothesis.
CIPK3 Represents a Cross-Talk Node for Cold and ABA Signal Transduction
Although cold, drought, salt, and ABA often activate the expression of the same genes, the distinct signaling pathways that mediate this response have been shown to differ significantly. Although drought and salt induce ABA synthesis and therefore have an ABA-dependent branch in signal transduction, cold-induced gene expression has been shown to be independent from ABA production. Nevertheless, cross- talk between cold and ABA signaling pathways may exist, based on some genetic studies (Knight and Knight, 2001; Xiong and Zhu, 2002). A study of sfr (sensitivity to freezing) mutants in Arabidopsis (Knight et al., 1999) effectively illustrates such cross-talk. The sfr6 mutant exhibits not only reduced gene induction by cold, but ABA-induced gene expression is affected as well. Because ABA and cold induce gene expression through different pathways, SFR6 clearly is involved in both the cold and ABA signaling pathways and is considered a cross-talk node for the interaction between ABA and cold signal transduction. Identification of the SFR6 gene will provide more information regarding the molecular nature of the SFR6 protein.
Ca2+ may serve as a common second messenger for stress and ABA; therefore, some calcium sensors and their targets may function as cross-talk nodes (Knight and Knight, 2001). Our study identifies a Ca2+-regulated protein kinase, CIPK3, as a cross-talk node for ABA, cold, and high-salt signal transduction, because disruption of CIPK3 function simultaneously altered gene induction patterns by cold, salt, and ABA. In particular, the cold-induced expression of RD29A has been shown to be independent of ABA production (Thomashow, 1999; Shinozaki and Yamaguchi-Shinozaki, 2000). Finding a single gene involved in two separate pathways clearly indicates that CIPK3 plays a role in both pathways that can interact with each other through CIPK3. Based on the available data on stress gene induction by cold, drought, and ABA, cold and drought both work through a DRE present in the stress gene promoter, although the transcription factor(s) responsible for the regulation may differ. The drought signal may be transmitted through DREB2, whereas the cold signal may be transmitted through DREB1/ CBF3 (Liu et al., 1998). Therefore, we propose that drought and cold signaling pathways also may differ upstream of trans-acting factors (e.g., DREBs/CBFs). Such a difference is exemplified by the results presented here that CIPK3 regulates only cold-induced but not drought-induced gene expression. We propose that CIPK3 is located upstream of transcription factors and downstream of the Ca2+ signal.
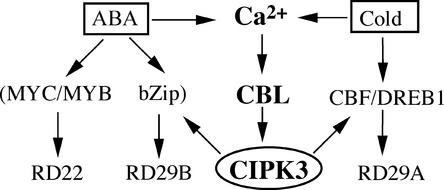
This putative location for CIPK3 in the signaling pathways is further supported by the changes in the ABA induction of RD29A and other target genes in the cipk3 mutant. ABA induction of RD29A and RD29B is mediated by the cis-acting element ABRE (Yamaguchi-Shinozaki and Shinozaki, 1994; Shinozaki and Yamaguchi-Shinozaki, 2000; Uno et al., 2000; Seki et al., 2001). Although RD29A has one copy of ABRE, RD29B has two copies of ABRE in the promoter region. Induction of both RD29A and RD29B was inhibited significantly in the cipk3 mutant, suggesting that transcription factors that bind to ABREs may be controlled by CIPK3. In addition, RD22 with a different cis-acting element also was affected in the cipk3 mutant. The transcription factors for the RD22 activation may include MYC/MYB-type proteins (Abe et al., 1997; Shinozaki and Yamaguchi-Shinozaki, 2000). The fact that CIPK3 regulates both RD22 and RD29A/RD29B expression again suggests that CIPK3 is located upstream of transcription factors in ABA-induced gene expression. Figure 7 presents a model of how CIPK3 may function in the cold and ABA signaling network.
Figure 7.
Hypothetical Model of CIPK3 Function.
CIPK3 positively regulates the ABA- and cold-induced expression of stress genes. It may mediate the cross-talk between ABA-dependent and ABA-independent pathways by modulating different transcription factors. bZip, basic domain/Leu zipper.
Because ABA, cold, and salt signaling pathways may include Ca2+ as a common messenger, CIPK3 serves as a cross-talk point for these pathways to interact with each other. Identification of such cross-talk nodes may help explain many puzzling phenomena in stress and ABA signal transduction. For example, cold does not produce a significant level of ABA in plants and thus is considered ABA independent. However, ABA appears to be required for a maximal cold response in gene induction, because some ABA-deficient mutants appear to be impaired in cold induction of the RD29A gene (Xiong et al., 2001). Such cold–ABA pathway interaction could be transmitted through the activation of CIPK3 and other cross-talk nodes. In the case of CIPK3 as a cross-talk node, cold and ABA together may trigger a higher magnitude of Ca2+ increase so that the CIPK3 kinase is activated at a higher level, leading to a more robust response in gene expression. Such a synergistic effect would be impaired in the ABA-deficient mutant despite an intact pathway for cold response. Another example is the cold-induced weak expression of RAB18, which appears to contain only the ABRE and is highly inducible by ABA (Lang and Palva, 1992; Nordin et al., 1993; Ishitani et al., 1998). The cold signaling pathway may not directly and effectively activate ABRE-containing genes. Rather, the weak activation could be a result of the cold activation of a cross-talk node (such as CIPK3) that can weakly activate the ABRE binding factors even in the absence of ABA.
In summary, this study has identified not only a signaling component downstream of Ca2+ in the stress and ABA signal transduction pathways but also a cross-talk node for the interaction between ABA and cold stress. Because CIPK3 is a member of a large family of CBL-associated kinases, additional studies on the CIPK family may identify other CIPK members in the signal transduction pathways for stress responses and other stimulus-response coupling processes in plants.
METHODS
Plant Materials, Stress Treatments, and RNA Analysis
Arabidopsis thaliana plants (ecotype Wassilewskija [Ws-0]) were grown in the greenhouse under long-day conditions (16-h-light/8-h-dark cycle) to the flowering stage for plant transformation and RNA analysis. For treatment under different stress conditions, 3-week-old seedlings grown on MS medium (Murashige and Skoog, 1962) were used. For aseptic growth, seeds were treated with isopropanol for 2 min and with 50% household bleach for 5 min, washed three times with sterile water, and plated on MS medium solidified with 0.8% agar.
For abscisic acid (ABA) treatment, 100 μM (±)-cis,trans-ABA was sprayed on the seedlings to ensure total coverage of the foliage area. The plants were incubated at room temperature under white light. For NaCl, mannitol, and polyethylene glycol treatment, 300 mM NaCl, 300 mM mannitol, or 30% polyethylene glycol 6000 was added to the seedlings on MS plates, and the seedlings were incubated at room temperature under white light. For cold treatment, seedlings were transferred to the 4°C cold room under white light. For drought and wounding treatment, 4-week-old Arabidopsis (ecotype Ws-0) seedlings grown under short-day conditions (8 h of light at 500 μmol·m−2·s−1 and 21 to 23°C, with 75% humidity for both day and night) were used. For drought treatments, plants grown in potted soil were carefully removed and dehydrated on the filter paper as described by Yamaguchi-Shinozaki and Shinozaki (1994). Wounding was performed by puncturing leaves with a hemostat as described by Kudla et al. (1999).
For RNA gel blot analysis, total RNA (10 μg) isolated with Tripure isolation reagent (Roche Diagnostics, Indianapolis, IN) was separated by electrophoresis on a 1.2% agarose gel, transferred to a GeneScreen Plus nylon membrane, and hybridized with 32P-labeled specific probe as described in Results. The membranes were autoradiographed with Kodak XAR film. Membrane-bound 23S rRNA was stained with methylene blue and used as a loading control. The photographs were scanned by a high-resolution scanner and processed using Adobe Photoshop software (Mountain View, CA). For the quantification of gene expression, we used the NIH Image program (http://rsb.info.nih.gov/nih-image). All RNA gel blot experiments were repeated at least three times, and results from one representative experiment are shown in Figures 4 to 6.
Reverse Transcriptase–Mediated PCR for the Analysis of CIPK3 Gene Expression
To analyze the expression of CIPK3 by reverse transcriptase–mediated PCR, total RNA was extracted from Arabidopsis tissues with Tripure reagent (Roche Diagnostics). Total RNA (1 μg) was heated to 65°C for 7 min and then subjected to reverse transcription reaction using Moloney murine leukemia virus reverse transcriptase (200 units per reaction; Invitrogen, San Diego, CA) and AmpliTaq (2 units per reaction; Perkin-Elmer) for 25 min at 48°C. PCR amplification was performed with initial denaturation at 94°C for 2 min followed by 25 cycles of incubations at 94°C for 20 s, 55°C for 40 s, and 72°C for 1.5 min, and a final extension at 72°C for 10 min using the CIPK3-specific forward (5′-GGAGTGATATTTGTTTGTGGTGTGGTTAG-3′) and reverse (5′-GTCCCAAAAGAAAACTCTCATACATCAC-3′) primers. Actin gene expression level was used as a quantitative control. Aliquots of individual PCR products were resolved by agarose gel electrophoresis and visualized with ethidium bromide under UV light.
Isolation and Complementation of the cipk3 T-DNA Insertional Mutant
The CIPK3 insertion allele was isolated by screening 60,480 T-DNA–tagged Arabidopsis lines (ecotype Ws-0) at the University of Wisconsin Arabidopsis Knockout Facility. The JL-202 primer (5′-CATTTT-ATAATAACGCTGCGGACATCTAC-3′) annealing to the T-DNA left border was used together with CIPK3-specific primers (forward, 5′-CTCTCTCTTTCTCACTCAATCTCTCTGTA-3′; and reverse, 5′-ATCCCACTCCCTTTCTCATCATCCACACA-3′) for the identification of putative mutant lines. The T-DNA insertion in the mutant (cipk3) was confirmed by DNA gel blot analysis, and its exact position was determined by sequencing. Plants homozygous for the cipk3 mutant were used for further analysis.
For complementation of the cipk3 mutant, a 4.3-kb fragment including the CIPK3 coding region and 1306 bp of the 5′ flanking DNA upstream of the ATG codon was amplified by PCR from Arabidopsis genomic DNA (ecotype Ws-0) with forward (5′-AATGTCGACGCA-GCAACTAGATAGTAATT-3′) and reverse (5′-GGCAAGCTTCAAAAG-AAAACTCTCATACA) primers. The PCR product was cloned into the binary vector pCAMBIA1300 (CAMBIA, Canberra, Australia) using SalI and HindIII restriction sites (underlined in the primer sequences). The constructs were transformed into Agrobacterium tumefaciens strain GV3101 and introduced into Arabidopsis plants by the floral dip method (Clough and Bent, 1998). Transgenic seeds were plated on half-strength MS medium containing 0.8% (w/v) agarose, 112 mg/L Gamborg's B5 vitamin mixture, and 50 μg/mL kanamycin or 15 μg/mL hygromycin. The resistant seedlings were transplanted to soil and grown in the greenhouse to produce seeds. Homozygous complemented lines (cipk3/CIPK3) were used for germination assay or RNA gel blot analysis. All of the PCR procedures were performed using Pfu DNA polymerase (Stratagene, La Jolla, CA) to enhance fidelity. All constructs were verified by DNA sequencing.
Germination Assay
Approximately 100 seeds each from the wild type (Ws-0), the cipk3 mutant, and the cipk3/CIPK3 complemented line were planted in triplicate on MS medium with different concentrations of ABA, NaCl, or mannitol and incubated at 4°C for 4 days before being placed at 23°C under long-day conditions. Germination (emergence of radicals) was scored daily for 6 days.
Analysis of CIPK3 Promoter–β-Glucuronidase Expression
To generate the CIPK3 promoter–β-glucuronidase construct, the 5′ flanking DNA of CIPK3 was amplified with forward (5′-AATGTCGACGCAGCAACTAGATAGTAATT-3′) and reverse (5′-AATGGATCC-TTCTCTAACCACACCACAAA-3′) primers. The 1,320-bp PCR fragment was cloned into SalI-BamHI sites (underlined in the primer sequences) in the pBI101.1 vector (Clontech, Palo Alto, CA). The construct was transformed into wild-type (Ws-0) plants, and transformants were selected as described above. T1 seedlings were stained with 5-bromo-4-chloro-3-indolyl-β-d-glucuronide for 12 h followed by incubation in 80% ethanol to remove chlorophyll (Jefferson et al., 1987).
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes.
Acknowledgments
We are grateful to the ABRC (Ohio State University, Columbus, OH) for Arabidopsis seeds and DNA clones and the Arabidopsis Gene Knockout Facility (University of Wisconsin, Madison) for screening the cipk3 mutant. This work was supported by the National Science Foundation and Syngenta Research and Technology.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.006858.
References
- Abe, H., Yamaguchi-Shinozaki, K., Urao, T., Iwasaki, T., Hosokawa, D., and Shinozaki, K. (1997). Role of Arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. Plant Cell 9, 1859–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong, Y.H., Chang, H.-S., Gupta, R., Wang, X., Zhu, T., and Luan, S. (2002). Transcriptional profiling reveals novel interactions between wounding, pathogen, abiotic stress, and hormonal responses in Arabidopsis. Plant Physiol. 129, 661–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, H., Hong, J., Ha, J., Kang, J., and Kim, S. (2000). ABFs, a family of ABA-responsive element binding factors. J. Biol. Chem. 275, 1723–1730. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Finkelstein, R.R., Gampala, S.S.L., and Rock, C.D. (2002). Ab-scisic acid signaling in seeds and seedlings. Plant Cell 14 (suppl.), S15.–S45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour, S.J., Sebolt, A.M., Salazar, M.P., Everard, J.D., and Thomashow, M.F. (2000). Overexpression of the Arabidopsis CBF3 transcriptional activator mimics multiple biochemical changes associated with cold acclimation. Plant Physiol. 124, 1854–1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour, S.J., Zarka, D.G., Stockinger, E.J., Salazar, M.P., Houghton, J.M., and Thomashow, M.F. (1998). Low temperature regulation of the Arabidopsis CBF family of AP2 transcriptional activators as an early step in cold-induced COR gene expression. Plant J. 16, 433–442. [DOI] [PubMed] [Google Scholar]
- Harmon, A.C., Gribskov, M., and Harper, J.F. (2000). CDPKs: A kinase for every Ca2+ signal? Trends Plant Sci. 5, 154–159. [DOI] [PubMed] [Google Scholar]
- Hasegawa, P.M., Bressan, R.A., Zhu, J.-K., and Bohnert, H.J. (2000). Plant cellular and molecular responses to high salinity. Annu. Rev. Plant Physiol. Plant Mol. Biol. 51, 463–499. [DOI] [PubMed] [Google Scholar]
- Ishitani, M., Xiong, L., Lee, H., Stevenson, B., and Zhu, J.-K. (1998). HOS1, a genetic locus involved in cold-responsive gene expression in Arabidopsis. Plant Cell 10, 1151–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson, R.A., Kavanagh, T.A., and Bevan, M.W. (1987). GUS fusions: β-Glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6, 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasuga, M., Liu, Q., Miura, S., Yamaguchi-Shinozaki, K., and Shinozaki, K. (1999). Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat. Biotechnol. 17, 287–291. [DOI] [PubMed] [Google Scholar]
- Kim, K.-N., Cheong, Y.H., Gupta, R., and Luan, S. (2000). Interaction specificity of Arabidopsis calcineurin B-like calcium sensors and their target kinases. Plant Physiol. 124, 1844–1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight, H., and Knight, M.R. (2000). Imaging spatial and cellular characteristics of low temperature calcium signature after cold acclimation in Arabidopsis. J. Exp. Bot. 51, 1679–1686. [DOI] [PubMed] [Google Scholar]
- Knight, H., and Knight, M.R. (2001). Abiotic stress signaling pathways: Specificity and cross-talk. Trends Plant Sci. 6, 262–267. [DOI] [PubMed] [Google Scholar]
- Knight, H., Trewavas, A.J., and Knight, M.R. (1996). Cold calcium signaling in Arabidopsis involves two cellular pools and a change in calcium signature after acclimation. Plant Cell 8, 489–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight, H., Veale, E.L., Warren, G.J., and Knight, M.R. (1999). The sfr6 mutation in Arabidopsis suppresses low-temperature induction of genes dependent on the CRT/DRE sequence motif. Plant Cell 11, 875–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudla, J., Xu, Q., Harter, K., Gruissem, W., and Luan, S. (1999). Genes for calcineurin B-like proteins in Arabidopsis are differentially regulated by stress signals. Proc. Natl. Acad. Sci. USA 96, 4718–4723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurkela, S., and Borg-Franck, M. (1992). Structure and expression of kin2, one of two cold- and ABA-induced genes of Arabidopsis thaliana. Plant Mol. Biol. 19, 689–692. [DOI] [PubMed] [Google Scholar]
- Lang, V., and Palva, E.T. (1992). The expression of a rab-related gene, rab18, is induced by abscisic acid during the cold acclimation process of Arabidopsis thaliana (L.) Heynh. Plant Mol. Biol. 20, 951–962. [DOI] [PubMed] [Google Scholar]
- Leung, J., and Giraudat, J. (1998). Abscisic acid signal transduction. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 199–222. [DOI] [PubMed] [Google Scholar]
- Liu, Q., Kasuga, M., Sakuma, Y., Abe, H., Miura, S., Yamaguchi-Shinozaki, K., and Shinozaki, K. (1998). Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10, 1391–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan, S., Kudla, J., Rodriguez-Concepcion, M., Yalovsky, S., and Gruissem, W. (2002). Calmodulins and calcineurin B-like proteins: Calcium sensors for specific signal response coupling in plants. Plant Cell 14 (suppl.), S389.–S400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473–497. [Google Scholar]
- Nordin, K., Vahala, T., and Palva, E.T. (1993). Differential expression of two related, low-temperature-induced genes in Arabidopsis thaliana (L.) Heynh. Plant Mol. Biol. 21, 641–653. [DOI] [PubMed] [Google Scholar]
- Reymond, P., Weber, H., Damond, M., and Farmer, E.E. (2000). Differential gene expression in response to mechanical wounding and insect feeding in Arabidopsis. Plant Cell 12, 707–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock, C.D. (2000). Pathways to abscisic acid-regulated gene expression. New Phytol. 148, 357–396. [DOI] [PubMed] [Google Scholar]
- Rudd, J.J., and Franklin-Tong, V.E. (2001). Unraveling response-specificity in Ca2+ signaling pathways in plant cells. New Phytol. 151, 7–33. [DOI] [PubMed] [Google Scholar]
- Saijo, Y., Hata, S., Kyozuka, J., Shimamoto, K., and Izui, K. (2000). Over-expression of a single Ca2+-dependent protein kinase confers both cold and salt/drought tolerance on rice plants. Plant J. 23, 319–327. [DOI] [PubMed] [Google Scholar]
- Sanders, D., Brownlee, C., and Harper, J.F. (1999). Communicating with calcium. Plant Cell 11, 691–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders, D., Pelloux, J., Brownlee, C., and Harper, J.F. (2002). Calcium at the crossroads of signaling. Plant Cell 14 (suppl.), S401.–S417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki, M., Narusaka, M., Abe, H., Kasuga, M., Yamaguchi-Shinozaki, K., Carninci, P., Hayashizaki, Y., and Shinozaki, K. (2001). Monitoring the expression pattern of 1300 Arabidopsis genes under drought and cold stresses by using a full-length cDNA microarray. Plant Cell 13, 61–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo, M., and Koshiba, T. (2002). Complex regulation of ABA biosynthesis in plants. Trends Plant Sci. 7, 41–48. [DOI] [PubMed] [Google Scholar]
- Sheen, J. (1996). Ca2+-dependent protein kinases and stress signal transduction in plants. Science 274, 1900–1902. [DOI] [PubMed] [Google Scholar]
- Shi, J., Kim, K.-N., Ritz, O., Albrecht, V., Gupta, R., Harter, K., Luan, S., and Kudla, J. (1999). Novel protein kinases associated with calcineurin B-like calcium sensors in Arabidopsis. Plant Cell 11, 2393–2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki, K., and Yamaguchi-Shinozaki, K. (2000). Molecular responses to dehydration and low temperature: Differences and cross-talk between two stress signaling pathways. Curr. Opin. Plant Biol. 3, 217–223. [PubMed] [Google Scholar]
- Snedden, W.A., and Fromm, H. (2001). Calmodulin as a versatile calcium signal transducer in plants. New Phytol. 151, 35–66. [DOI] [PubMed] [Google Scholar]
- Tahtiharju, S., Sangwan, V., Monroy, A.F., Dhindsa, R.S., and Borg, M. (1997). The induction of kin genes in cold-acclimating Arabidopsis thaliana: Evidence of a role for calcium. Planta 203, 442–447. [DOI] [PubMed] [Google Scholar]
- Thomashow, M.F. (1999). Plant cold acclimation: Freezing tolerance genes and regulatory mechanisms. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 571–599. [DOI] [PubMed] [Google Scholar]
- Townley, H.E., and Knight, M.R. (2002). Calmodulin as a potential negative regulator of Arabidopsis COR gene expression. Plant Physiol. 128, 1169–1172. [DOI] [PubMed] [Google Scholar]
- Trewavas, A.J. (1999). How plants learn. Proc. Natl. Acad. Sci. USA 96, 4216–4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uno, Y., Furihata, T., Abe, H., Yoshida, R., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2000). Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc. Natl. Acad. Sci. USA 97, 11632–11637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, Y., Kuzma, J., Marechal, E., Graeff, R., Lee, H.C., Foster, R., and Chua, N.-H. (1997). Abscisic acid signaling through cyclic ADP-ribose in plants. Science 278, 2126–2130. [DOI] [PubMed] [Google Scholar]
- Xiong, L., Ishitani, M., Lee, H., and Zhu, J.-K. (2001). The Arabidopsis LOS5/ABA3 locus encodes a molybdenum cofactor sulfurase and modulates cold stress- and osmotic stress-responsive gene expression. Plant Cell 13, 2063–2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong, L., and Zhu, J.-K. (2002). Molecular and genetic aspects of plant responses to osmotic stress. Plant Cell Environ. 25, 131–139. [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki, K., and Shinozaki, K. (1994). A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell 6, 251–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeevaart, J.A.D., and Creelman, R.A. (1988). Metabolism and physiology of abscisic acid. Annu. Rev. Plant Physiol. Plant Mol. Biol. 39, 439–473. [Google Scholar]
- Zhu, J.-K. (2002). Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 53, 247–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielinski, R.E. (1998). Calmodulin and calmodulin-binding proteins in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 697–725. [DOI] [PubMed] [Google Scholar]