Abstract
Arabidopsis is believed to be mostly self-pollinated, although several lines of genetic and morphological evidence indicate that insect-mediated outcrossing occurs with at least a low frequency in wild populations. Here, we show that Arabidopsis flowers emit both monoterpenes and sesquiterpenes, potential olfactory cues for pollinating insects. Of the 32 terpene synthase genes in the Arabidopsis genome, 20 were found to be expressed in flowers, 6 of these exclusively or almost exclusively so. Two terpene synthase genes expressed exclusively in the flowers and one terpene synthase gene expressed almost exclusively in the flowers were characterized and found to encode proteins that catalyze the formation of major floral volatiles. A β-glucuronidase fusion construct with a promoter of one of these genes demonstrated that gene expression was restricted to the sepals, stigmas, anther filaments, and receptacles, reaching a peak when the stigma was receptive to cross pollen. The observation that Arabidopsis flowers synthesize and emit volatiles raises intriguing questions about the reproductive behavior of Arabidopsis in the wild and allows detailed investigations of floral volatile biosynthesis and its regulation to be performed with this model plant system.
INTRODUCTION
One of the principal reasons for the success of Arabidopsis as a model system in plant biology is the ability of this species to set copious numbers of seeds under greenhouse and growth-room conditions that arise nearly exclusively from self-pollination (Meyerowitz and Somerville, 1994). This high degree of self-pollination, along with its small flower size (2 to 3 mm long), inconspicuous white petals, and lack of strong scent, suggest that Arabidopsis has not been selected to attract floral visitors for cross-pollination. However, several lines of evidence indicate that insect-mediated cross-pollination may occur in wild populations. First, in the ontogeny of the flower, the stigma becomes receptive and protrudes out of the petals before the stamens mature, providing a brief window for cross-pollination (Jones, 1971). Second, at the base of the stamens, Arabidopsis has floral nectaries that secrete a mixture of sugars that could serve as a reward for floral visitors (Davis et al., 1998). Third, small insects, including hover flies, also have been observed to visit Arabidopsis flowers (Jones, 1971; Snape and Lawrence, 1971). These insects might act as vectors for pollen transfer, although no direct evidence for their role in pollination has been presented. Finally, the frequency of polymorphic loci in Arabidopsis populations is high enough to exclude simple mutation as the source of variation (Loridon et al., 1998). Cross-pollination has been estimated to be as high as 2%, based on segregation for some morphological traits in the progeny of Arabidopsis inbred lines grown outdoors (Snape and Lawrence, 1971), although lower estimates have been obtained from analyses of allozyme loci in wild populations (Abbott and Gomes, 1989).
If Arabidopsis flowers are visited by insect pollinators, this species is likely to possess visual or olfactory cues for pollinator attraction. The emission of volatiles is a major feature of many insect-pollinated flowers (Knudsen et al., 1993; Dudareva and Pichersky, 2000). Mixtures of volatile compounds drawn from several classes of plant metabolites, including terpenes, phenylpropanoids, fatty acid derivatives, and nitrogen- or sulfur-containing compounds (Dudareva and Pichersky, 2000), have been shown to attract a large variety of insects (as well as some mammals) that vector pollen. However, there are no previous reports regarding the detection of volatiles emanating from Arabidopsis flowers either by chemical methods or the human nose.
The detection of floral volatile emission in this species would allow the vast genetic and genomic resources available for this model plant to be applied to studying fundamental questions about the regulation and evolution of volatile production in pollinator attraction. However, it should be borne in mind that, given the low levels of cross-pollination in Arabidopsis, the emission of floral volatiles may serve functions unrelated to attracting pollinators or simply may be a vestigial trait.
An indication of the metabolic potential of Arabidopsis to produce volatile compounds is afforded by examination of its genomic sequence. Of the major groups of plant volatiles, terpenes appear to be produced in great variety, based on the large number of possible terpene synthase (TPS) genes present. More than 30 genes with sequence homology with known TPS genes from other species are predicted to be present in the Arabidopsis genome (Aubourg et al., 2002). The enzymes of this family catalyze the conversion of allylic prenyl diphosphate intermediates of the terpene pathway, including dimethylallyl diphosphate (C5), geranyl diphosphate (GPP; C10), farnesyl diphosphate (FPP; C15), and geranylgeranyl diphosphate (C20), to hemiterpenes (C5), monoterpenes (C10), sesquiterpenes (C15), and diterpenes (C20), respectively (Cane, 1999; MacMillan and Beale, 1999; Wise and Croteau, 1999). Nearly all of the products of hemiterpene, monoterpene, and sesquiterpene synthases are volatile under ambient temperature and atmospheric pressure, and monoterpenes and sesquiterpenes are very typical floral volatiles. However, to date only one characterized terpene synthase, (S)-linalool synthase from Clarkia breweri (Dudareva et al., 1996), has been shown to be involved in the production of floral volatiles. Work on the TPS genes of Arabidopsis also has just begun. Of the 32 predicted functional sequences, only 4 have been characterized. Two monoterpene synthases have been described, a myrcene/(E)-β-ocimene synthase (Bohlmann et al., 2000) and an (E)-β-ocimene synthase (Fäldt et al., 2002). Two diterpene synthases also have been reported that are involved in gibberellin formation (Sun and Kamiya, 1994; Yamaguchi et al., 1998).
In this investigation, we searched for volatiles released from flowers and vegetative parts of Arabidopsis using headspace sampling methods. We found a blend of volatile terpenes to be emitted specifically from inflorescences and characterized three TPS genes involved in their biosynthesis.
RESULTS
Arabidopsis Flowers Emit Monoterpenes and Sesquiterpenes
The volatile constituents of Arabidopsis flowers were sampled by passing air over flowering plants and then through a charcoal trap to adsorb organic constituents. A semiopen headspace sampling method was used as well as a sensitive closed-loop stripping method (Donath and Boland, 1995). After desorption of the trap with solvent and analysis by gas chromatography–mass spectrometry (GC-MS), a range of monoterpene and sesquiterpene constituents was detected.
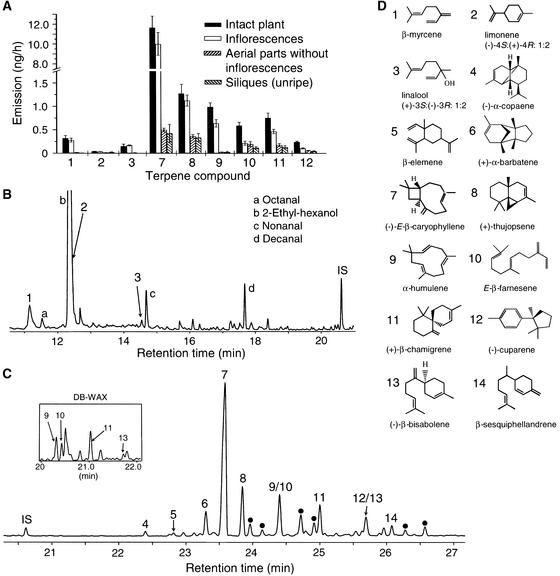
To determine whether these substances were emitted by the flowers, fruits, or vegetative parts, we performed a series of headspace collections in which inflorescences or siliques (fruits) were removed from plants (Figure 1A). By comparing the major volatiles detected, it was determined that the three most abundant monoterpenes (β-myrcene, limonene, and linalool) and most of the major sesquiterpenes, including (−)-(E)-β-caryophyllene, (+)-thujopsene, α-humulene, (E)-β-farnesene, (+)-β-chamigrene, and (−)-cuparene, were emitted almost exclusively from flowers. Together, monoterpenes and sesquiterpenes represented >60% of the total amount of volatiles collected from the flowers (estimated by relative GC-MS peak area; Figures 1B and 1C). The remainder consisted primarily of aliphatic aldehydes or alcohols, including octanal, 2-ethyl-hexanol, nonanal, and decanal. These non-terpenoids also were present in headspace collections from siliques and vegetative parts of flowering plants (data not shown).
Figure 1.
Volatile Terpenes Emitted from Arabidopsis (Columbia Ecotype).
(A) Release rates of major terpenes from 6-week-old flowering plants and parts of these plants determined by dynamic headspace sampling. Inflorescences were the source of emission for the monoterpenes and most of the sesquiterpenes. Data represent the mean of four replications ± se. The emission rate presented is that of a single plant per hour, although collections were made from five plants simultaneously over 8-h periods.
(B) Gas chromatography of monoterpenes collected from 150 cut inflorescences during 12 h of closed-loop stripping (Donath and Boland, 1995). The peak of limonene coeluted with 2-ethyl-hexanol on the (5% phenyl)-polymethylsiloxane column, as shown here, but it was separable from this compound on a polyethylene glycol column.
(C) A later portion of the same chromatogram as in (B), showing the sesquiterpene hydrocarbon region. The inset depicts a portion of the chromatogram of the same sample run on a polyethylene glycol (DB-WAX) column, which shows an improved separation of some components. Compounds marked with dots represent additional sesquiterpene hydrocarbons identified by GC-MS but not yet confirmed by comparison with authentic standards. IS indicates the internal standard, nonyl acetate.
(D) Structures of the compounds identified by numbers in (A), (B), and (C). The number of each compound in (D) corresponds to the numbered columns in (A) and the numbered peaks in (B) and (C). Chirality was determined for all compounds except β-elemene and β-sesquiphellandrene.
Several Arabidopsis TPS Genes Are Expressed Exclusively or Almost Exclusively in the Flowers
To determine which of the predicted functional Arabidopsis TPS genes might be involved in the formation of floral volatiles, the expression of all 32 of these (Aubourg et al., 2002) (Table 1) was determined in various organs of the plant with reverse transcriptase–mediated (RT) PCR. For each gene, specific internal primers designed to span more than one exon were used to discriminate against amplification products from contaminating genomic DNA. No such amplification products were observed, and control PCR experiments omitting reverse transcriptase also resulted in no product. Analysis was performed with RNA isolated from leaves, flowers, siliques, stems, and roots of 6-week-old flowering plants. In these experiments, results for different genes in the same organ are directly comparable, because an identical aliquot of cDNA from the original RT reaction was used in each PCR procedure. To determine whether equal amounts of cDNA were included in the reactions involving different organs, we also performed RT-PCR with primers designed for β-tubulin.
Table 1.
Primers for RT-PCR Analysis
| Gene Name | Sequences (5′ to 3′) |
|---|---|
| At3g25810 | F: TATATTTGATGTAATCATCG |
| R: TTGAACCATAGCAGTGAAGAG | |
| At3g25830 | F: TATTTGATGTGATCATCGACC |
| R: GGAACACTTAAGATATAAAAGGT | |
| At2g24210 | F: CTGGTGGATGGAGACAGGTTT |
| R: CGGTGAGGTTACAAGGTCGTT | |
| At4g16740 | F: GAGACAAAGGATCAAATGGAG |
| R: CCATTGGTCTTTAACATAGAA | |
| At4g16730 | F: TAAAGAAGAGGTGAGGAAGAC |
| R: CTAGAAATAAGTTTAAGTTCT | |
| At1g61680 | F: ATGATCGATGTCATTCAAAGT |
| R: TTAAATGTTTGAGACATTTCTC | |
| At1g70080 | F: CAGGAAATGGATGATCTTTGGA |
| R: GTGGTTAGTGTTTTCAGATCTTG | |
| At5g48110 | F: GAAATTGATAGCCTTGGGAGA |
| R: TTCACGATAGTTTGAAGCTCT | |
| At2g23230 | F: GATATGGATATGCTTACAATAG |
| R: CTTTCGACACAATCAAGAAGC | |
| At4g15870 | F: GAAATGAATGCCCTTGCCCA |
| R: TCAGAAACATTACCGTATCTA | |
| At3g29110 | F: GAAATGGATGTTCTTGAAAGA |
| R: ATTGATGAGGCTTTCGACCTC | |
| At4g20210 | F: GAAATGGATGCGCTTAGGAAA |
| R: CTCTCCAGACTATTGGTGAGG | |
| At3g29190 | F: ATTTCTTTAGGAGATTCAAAGG |
| R: CTCTTCTATTGTGGCACACAC | |
| At1g66020 | F: ATGTATTCAGAAGATTCAAAGGGAG |
| R: TAGAATCGCTAAACAAGTGAAGTAC | |
| At4g20200 | F: GAAATGGATGCCCTTAGAAAA |
| R: CTCTTCAAAAGTATCCAAAAT | |
| At4g20230 | F: GAAATGGATGCACTTAAAGAA |
| R: GTGTATTTCACACTGTAAGAT | |
| At4g13280+At4g13300 | F: GTGTATTCAAGAGATTTACAG |
| R: CTTTCCACACAATCTACGAGC | |
| At5g44630 | F: TGGAGGAAAATATAGTGATAT |
| R: CGGTGCTGAGGTATGTGAAGA | |
| At3g29410 | F: AAACAAATGTCCAGATGGGAT |
| R: TCAAAGAGGTATTGGATGGAG | |
| At3g32030 | F: ATGGCAGTAGCAAGAACGGTT |
| R: CTCTTGGATGTAATGTAAACG | |
| At1g33750 | F: ATGGGTTTTCGAGCTAAAACT |
| R: TTGGTGAGAGTTTTTAGCTCT | |
| At3g14490 | F: TGAAGCCAAAAGTGAGAGACATG |
| R: GCTCTTGGATATAATGTAGCTGGC | |
| At1g31950 | F: GAGTTTGATGAGCTAGAAAGAGAGATTG |
| R: GTGAGCGTTTTGAGTTCTTGGAC | |
| At3g14520+At3g14540 | F: ATGCCTTTGATAGATTCAGAG |
| R: CTCGTAGGTTCCTACGTCATT | |
| At5g23960 | F: GGAACTGAGACGTTCAAAGAG |
| R: CGCTGTGAATAAGATTAGTGC | |
| At1g61120 | F: ATGGGAAGGAGAAGAGCTTAA |
| R: TTAGTAGAAGCATGGTGCGAAT | |
| At1g79460 | F: TCAAAGGGAAGAGAAGCATA |
| R: CGATATGAGGTTTTTGTAACTC | |
| At4g02780 | F: ATACCAAAAGAGATAATGCA |
| R: CTCGCCAGGTAAGTCTTTCAT | |
| At1g48800 | F: GCTTGTGAGTCTTGGTCTCGC |
| R: AGCGAGGTGCAGTTCCACGAT | |
| At1g48820 | F: GCTTGTGAGTCTTGGTCTCGC |
| R: CGCGAAGTGACATTCAACGAG | |
| β-tubulin | F: CTCAAGAGGTTCTCAGCAGTA |
| R: TCACCTTCTTCATCCGCAGTT |
F, forward primer; R, reverse primer.
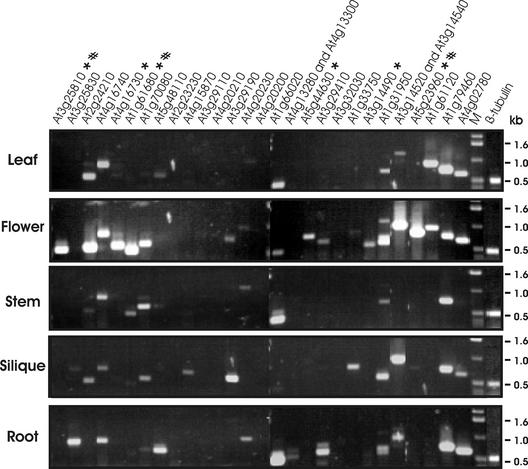
After RT-PCR, amplified fragments from mRNAs of 25 of the 32 TPS genes were obtained from at least one organ (Figure 2). The expression of 11 genes was detected in leaves, 20 in flowers, 11 in siliques, 9 in stems, and 12 in roots. Seven genes (At2g23230, At3g29110, At4g20210, At4g20200, At3g32030, At1g48800, and At1g48820) showed no expression in any of the five organs, whereas eight genes, including the two involved in gibberellin biosynthesis (At1g79460 and At4g02780), were expressed in nearly all of them. Six genes (At3g25810, At4g16730, At1g61680, At5g44630, At3g14490, and At5g23960) showed complete or almost complete flower-specific expression.
Figure 2.
Organ-Specific Expression of Arabidopsis TPS Genes.
Leaves, flowers, siliques, stems, and roots were collected from 6-week-old flowering plants. Total RNA was extracted and used for RT-PCR analysis. The expression of the 32 Arabidopsis TPS genes in each organ was examined in 30 RT-PCR procedures, of which the results from 28 are shown in this composite picture. As a result of the high sequence similarity between At4g13280 and At4g13300 and between At3g14520 and At3g14540, PCR products obtained from one or the other member of each pair of genes could not be distinguished. The results for At1g48800 and At1g48820, which showed no expression in any of the tested tissues, are not shown. M indicates DNA markers. The RT-PCR product for β-tubulin is shown at right. To prevent saturation conditions in the reactions with the β-tubulin primers, the cDNA amount was reduced twofold compared with that in the reactions with terpene synthase primers, and the number of cycles was reduced to 26, compared with 30 (see Methods for further details). The six TPS genes that are expressed exclusively or almost exclusively in the flowers are indicated with asterisks. The three TPS genes that were characterized further also are indicated (#).
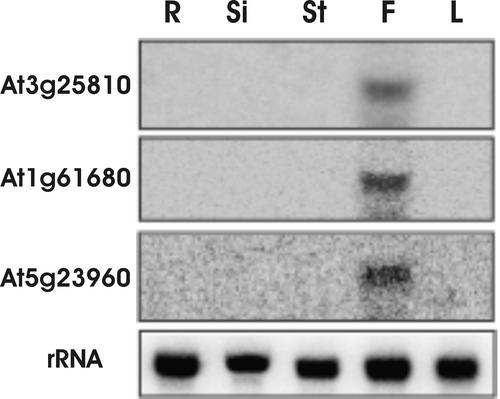
We chose three of these flower-specific genes for further analysis. RNA gel blots (Figure 3) verified that At3g25810 and At5g23960 were expressed exclusively in the flowers. The blots also showed exclusive expression of At1g61680 in the flowers, even though RT-PCR showed that relatively low levels of the transcript of this gene also occurred in the stem, in addition to the much higher levels of transcript in the flowers (Figure 2). This discrepancy likely is attributable to the higher sensitivity of RT-PCR compared with RNA gel blot analysis.
Figure 3.
RNA Gel Blot Analysis of Selected TPS Genes.
Gene-specific probes were amplified by PCR using internal primers as described for RT-PCR analysis, labeled with 32P-dCTP, and hybridized to RNA gel blots. mRNA transcripts of At3g25810, At1g61680, and At5g23960 were detected in flower tissue only. R, root; Si, silique; St, stem; F, flower; L, leaf.
Three Flower-Specific TPS Genes Encode Proteins That Catalyze the Formation of Floral Volatiles
To investigate the catalytic activities of the proteins encoded by At3g25810, At1g61680, and At5g23960, the three flower-specific TPS genes, cDNAs of these obtained by RT-PCR were expressed in Escherichia coli, and the resulting proteins were assayed for terpene synthase activity. Control assays were performed with crude extracts of E. coli (same strain) carrying the same expression vector without any TPS genes inserted.
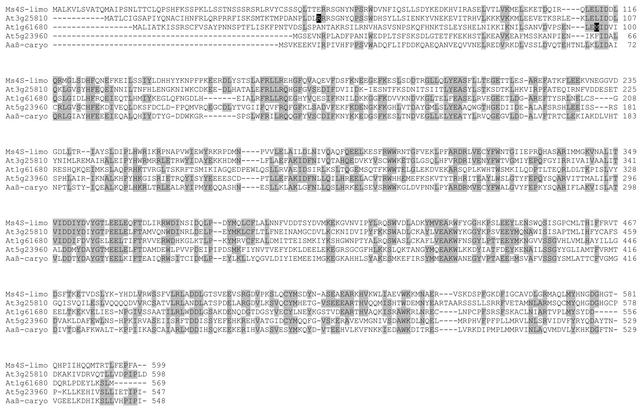
In plants, the biosynthesis of monoterpenes appears to be localized in plastids, the site of formation of the monoterpene precursor GPP, whereas sesquiterpenes are synthesized in the cytosol/endoplasmic reticulum compartment, the site of FPP formation (Lichtenthaler, 1999). Thus, the known monoterpene synthases, all of which are encoded by nuclear genes, possess an N-terminal transit peptide that directs their import into the plastid followed by cleavage of the transit peptide (Turner et al., 1999). Based on sequence analyses, two of the three flower-specific TPS genes, At3g25810 and At1g61680, have transit peptide–like sequences at the N terminus of their proteins (Figure 4) (Aubourg et al., 2002), and they fall into a clade that includes the two previously identified monoterpene synthases of Arabidopsis, At2g24210 and At4g16740 (Bohlmann et al., 2000; Aubourg et al., 2002; Fäldt et al., 2002). Because it has been shown for other monoterpene synthases that the mature (cleaved) protein often has higher specific activity when expressed in E. coli than the full-length preprotein (Williams et al., 1998), we prepared constructs both with and without the N-terminal sequence.
Figure 4.
Derived Amino Acid Sequences of Three Arabidopsis TPS Genes Involved in Floral Volatile Biosynthesis Aligned with Related Sequences.
The sequences of the proteins encoded by At3g25810, At1g61680, and At5g23960 are aligned with the sequences of the monoterpene synthase 4S-limonene cyclase from spearmint (Ms4S-limo) (Colby et al., 1993) and the sesquiterpene synthase β-caryophyllene synthase from A. annua (Aaβ-caryo) (Cai et al., 2002). Amino acid residues conserved in three or more sequences are shaded. The R residue shown by a white letter on a black background in the At3g25810 sequence indicates the position of the first amino acid in the truncated At3g25810 construct described in the text (replaced by a Met). The M residue shown by a white letter on a black background in the At1g61680 sequence indicates the position of the first amino acid in the truncated At1g61680 construct described in the text.
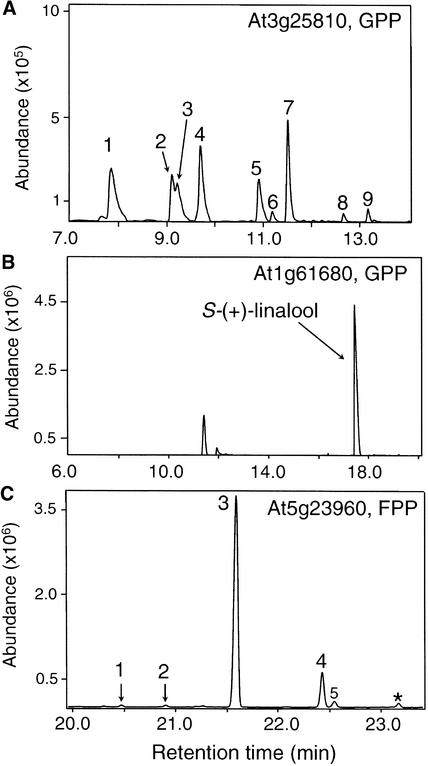
Both the full-length protein encoded by At3g25810 and its mature version lacking the putative transit peptide catalyzed the formation of eight monoterpenes from GPP, including six major products [α-pinene, sabinene, β-pinene, β-myrcene, limonene, and (E)-β-ocimene] (Figure 5A). When assayed with FPP, the At3g25810 protein also catalyzed the formation of some sesquiterpenes, including (E,E)-α-farnesene, (E)-β-farnesene, and trans-α-bergamotene. However, these are not thought to be physiologically relevant products because they are not constituents of the floral volatile blend and the specific activity of the At3g25810 enzyme with GPP was >40-fold higher than that with FPP (1.7 ± 0.1 pkat/mg protein versus 0.04 ± 0.01 pkat/mg protein).
Figure 5.
Gas Chromatographic Separation of Products Formed by Three Terpene Synthases Found in Arabidopsis Flowers.
Extracts of E. coli expressing proteins derived from At3g25810, At1g61680, and At5g23960 were assayed in vitro with GPP or FPP, and products were separated on a (5%-phenyl)-methylpolysiloxane ([A] and [C]) or a β-cyclodextrin ([B]) column. Details of product separation, identification, and enantiomeric determination are described in Methods.
(A) At3g25810 mature protein with GPP. Peak 1, α-pinene [98% (−)-1S]; peak 2, sabinene [75% (−)-1S, 25% (+)-1R]; peak 3, β-pinene [98% (−)-1S]; peak 4, β-myrcene; peak 5, limonene [92% (−)-4S, 8% (+)-4R]; peak 6, (Z)-β-ocimene; peak 7, (E)-β-ocimene; peak 8, α-terpinolene; peak 9, linalool (racemic, also produced in the assay of control E. coli cultures transformed with vector lacking the terpene synthase insert).
(B) At1g61680 mature protein with GPP. Enantiomerically pure (+)-3S-linalool is the sole product. Unlabeled peaks also occurred in the control assay with an extract of E. coli transformed with the same expression vector lacking an insert.
(C) At5g23960 with FPP. Peak 1, (−)-α-copaene; peak 2, β-elemene (chirality not determined); peak 3, (−)-(E)-β-caryophyllene; peak 4, α-humulene; peak 5, (E)-β-farnesene (also produced by the control). The asterisk indicates an unidentified product.
The complete protein encoded by At1g61680 and its mature version without the putative transit peptide both catalyzed the conversion of GPP to only one product, (+)-3S-linalool (Figure 5B). Incubation of the expressed protein with FPP led to the formation of very low amounts of nerolidol, but this sesquiterpene alcohol is not present in Arabidopsis floral volatiles, and the rate of this reaction was >50-fold lower than the rate with GPP.
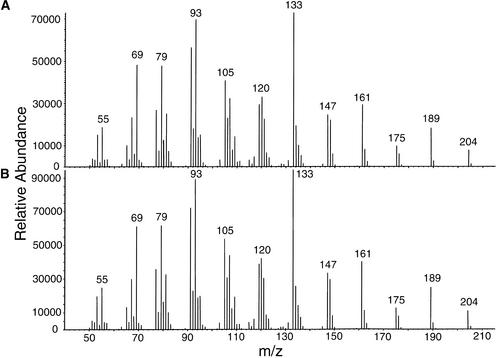
The open reading frame of At5g23960 appears to encode a protein with no transit peptide (Figure 3) (Aubourg et al., 2002). When synthesized in E. coli, the At5g23960 protein catalyzed the formation of (−)-E-β-caryophyllene and α-humulene as well as trace amounts of (−)-α-copaene and β-elemene from FPP (Figures 5C and 6). The enzyme did not convert GPP to any monoterpenes.
Figure 6.
Identification of (E)-β-Caryophyllene as the Major Enzyme Product of At5g23960 Catalysis.
(A) Mass spectrum of (E)-β-caryophyllene produced by the incubation of FPP with a cell-free extract of E. coli BL21 Codon Plus expressing the At5g23960 protein.
(B) Mass spectrum of authentic (E)-β-caryophyllene standard obtained under the same conditions as in (A).
m/z, mass-to-charge ratio.
The Promoter of At3g25810 Is Active in Specific Floral Parts
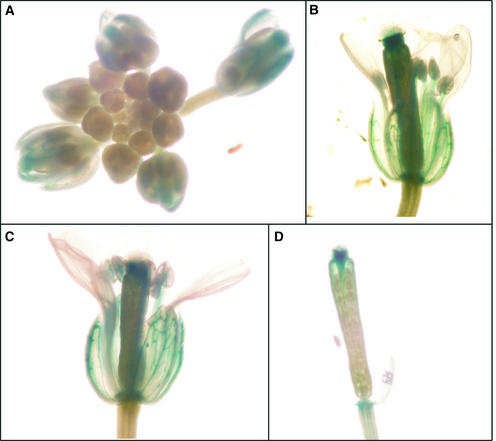
RT-PCR and RNA gel blot analyses showed flower-exclusive or nearly exclusive expression of At3g25810, At1g61680, and At5g23960 (Figures 2 and 3) but could not identify the specific part(s) of the flower where gene activity occurs. To investigate the tissue localization of At3g25810 expression, the promoter region (1.4 kb) of At3g25810 was fused to β-glucuronidase (GUS), and the resulting construct was used to transform Arabidopsis of the Columbia ecotype. Flowers and leaves of transgenic plants then were stained for GUS expression. GUS staining was observed only in flowers, confirming the results of the RT-PCR and the RNA gel blot experiments. Among the floral parts, staining was evident in the sepals, stigmas, anther filaments, and receptacles and was visible in mature, but not immature, buds (Figure 7A), in newly opened flowers in which the stigma protrudes above the anthers and petals (Figure 7B), and at a more mature stage of floral development at which the stamens have elongated and the pollen shed (Figures 7C and 7D).
Figure 7.
Expression Patterns of the At3g25810 Promoter–GUS Fusion Gene during Flower Development.
GUS staining was observed in sepals, stigmas, anther filaments, and receptacles of mature, but not immature, buds (A), newly opened flower (B), and older flower (C). In (D), petals and sepals were removed to show clearly the staining in the stigma of the older flower. The newly opened flower (B) is in the protogynous stage, in which the receptive stigma, protruding above the petals and the immature anthers, is accessible to cross-pollination. The older flower (C) is in the autogamous stage, in which the stamens have elongated to the level of the stigma and dehisced. The proximity of the stamens to the stigma at this stage facilitates self-pollination.
DISCUSSION
Arabidopsis Flowers Release a Mixture of Volatiles Dominated by Terpenes
Arabidopsis inflorescences emit a complex blend of monoterpenes, sesquiterpenes, aliphatic aldehydes, and alcohols. Although the aldehydes and alcohols also are emitted from vegetative parts of mature plants and were detected in volatile collections of younger, rosette-stage plants (Van Poecke et al., 2001), the terpenoids are almost completely specific to the flowers and represent >60% of the total amount of volatiles collected from these organs. The individual terpene constituents are nearly all well known from higher plants. For example, the sesquiterpene hydrocarbon (−)-(E)-β-caryophyllene, the most abundant terpene collected, is very widely distributed in the plant kingdom (Joulain and König, 1998; Cane, 1999) and is a floral volatile of many other plant species (Knudsen et al., 1993). However, the tricyclic α-barbatene, which is common in liverworts, has been reported only rarely from vascular plants (König et al., 1996).
The terpene emission level from Arabidopsis flowers is considerably lower than what is observed from the strongly scented C. breweri, a model wild plant for the study of floral scent. A single mature Arabidopsis plant, which has on average 80 open flowers, emits a total of 23.4 ± 1.4 ng of terpenes per hour. By contrast, a single mature C. breweri plant, which has on average only two flowers in the wild, emits a total of 3.3 to 5.9 μg of volatiles (terpenes, phenylpropanoids, and benzenoids) per hour (calculated from Raguso and Pichersky, 1995), a 140- to 250-fold difference. However, emission from flowers of the closely related species C. concinna is 30- to 60-fold lower than that from C. breweri (Raguso and Pichersky, 1995). Unfortunately, it is difficult to make further interspecific comparisons because absolute floral emission values for wild species generally are not reported. Nevertheless, Arabidopsis flowers clearly emit a terpene-rich blend of volatiles that, although readily detectable by modern gas chromatography, is scentless to the human nose as a result of its high proportion of sesquiterpene hydrocarbons, the low abundance of monoterpenes, and the absence of pungent phenylpropanoids and benzenoids.
The Formation of Arabidopsis Floral Terpenes Is Catalyzed by Terpene Synthases
The volatile terpenes of Arabidopsis inflorescences are biosynthesized by the action of terpene synthases, enzymes that catalyze the conversion of the ubiquitous isoprenoid pathway intermediates, GPP and FPP, to monoterpene and sesquiterpene products, respectively (Cane, 1999; Wise and Croteau, 1999). Of the 32 intact genes in the Arabidopsis genome that have sequence homology with known TPS genes, we selected three (At3g25810, At1g61680, and At5g23960) for further characterization that were expressed exclusively or almost exclusively in flowers, based on both RT-PCR (Figure 2) and RNA gel blot analysis (Figure 3), and that appeared likely to encode monoterpene and sesquiterpene synthases, based on their sequence similarity to other members of this gene family. When assayed with GPP, At3g25810 catalyzed the formation of multiple monoterpenes. Three of these, limonene, β-myrcene, and (E)-β-ocimene, were detected as Arabidopsis floral volatiles in our headspace collections (Figure 1), whereas others, including α-pinene and sabinene, were detected occasionally in low amounts (data not shown). The remainder may be retained in the cell and not emitted at detectable levels, or the mixture of products produced by this enzyme in vitro may be different from what occurs in vivo (Jia et al., 1999; Crowell et al., 2002). The ability of a single terpene synthase to form multiple products is a rather common feature of this enzyme family, attributed to the generation of carbocationic intermediates during the reaction that can have more than one metabolic fate (Crock et al., 1997; Cane, 1999; Wise and Croteau, 1999).
Not all terpene synthases make multiple products. At1g61680 catalyzed the formation from GPP of only (+)-S-linalool, which also was detected in Arabidopsis floral scent (Figure 1). When assayed with FPP, both the At3g25810 and At1g61680 proteins catalyzed the formation of some sesquiterpenes. However, these are likely not to be significant reactions in planta because (1) they occur at significantly lower rates in vitro compared with reactions that form monoterpenes from GPP, (2) the sesquiterpene products of such reactions were not found in the blend of floral volatiles, and (3) the proteins themselves may never contact free FPP because they contain transit peptide–like sequences at the N terminus and so are likely targeted to the plastids (Turner et al., 1999), where free FPP is not present (Lichtenthaler, 1999).
The third TPS gene, At5g23960, encodes a protein that catalyzed the formation of five sesquiterpenes. Of these, (−)-(E)-β-caryophyllene and α-humulene were two of the major sesquiterpenes detected in the collected floral volatiles, and (−)-α-copaene and β-elemene were found in low amounts (Figure 1). Although (−)-(E)-β-caryophyllene is a common component of floral scent (Knudsen et al., 1993) and one of the most common sesquiterpenes in the plant kingdom (Joulain and König, 1998), a gene encoding a terpene synthase capable of producing this substance has been reported previously only from Artemisia annua (Cai et al., 2002). The proteins encoded by At5g23960 and the A. annua β-caryophyllene synthase show 38% sequence identity (Figure 4), a level of conservation that is not particularly high among terpene synthases (Bohlmann et al., 1998).
Sesquiterpenes account for >95% of the total terpene volatiles emitted from Arabidopsis flowers, and the volatiles produced by the enzyme encoded by At5g23960, mostly (−)-(E)-β-caryophyllene and α-humulene, constitute ∼45% of the emitted sesquiterpenes. The analysis of the AtTPS family by Aubourg et al. (2002) indicated that only three AtTPS genes (not including At5g23960) encode proteins lacking a transit peptide. Adding gene At5g23960 characterized in this study, Arabidopsis is likely to have only four sesquiterpene synthases. Two of these, At4g13280 and At4g13300, are not expressed in any organ under normal conditions (Figure 2). We have preliminary data (D. Tholl, F. Chen, J. Gershenzon, and E. Pichersky, unpublished data) showing that a third sesquiterpene synthase, At5g44630, does not catalyze the formation of (−)-(E)-β-caryophyllene and α-humulene but instead is responsible for the synthesis of most or all of the other sesquiterpenes emitted from the flowers. Thus, gene At5g23960, characterized here, is by itself responsible for the synthesis of (−)-(E)-β-caryophyllene and α-humulene, which together account for 43% of the total terpene volatiles emitted from the flowers.
Aubourg et al. (2002) also suggested that there probably are only six monoterpene synthases in Arabidopsis. Five of these are expressed in flowers (At3g25810, At2g24210, At4g16740, At4g16730, and At1g61680), but not always exclusively so, and the functions of four of them have now been determined: At2g24210 and At4g16740 were characterized previously to encode enzymes capable of producing β-myrcene plus (E)-β-ocimene and (E)-β-ocimene, respectively (Bohlmann et al., 2000; Fäldt et al., 2002), and we have shown here that At3g25810 and At1g61680 encode enzymes that synthesize β-myrcene plus several other monoterpenes and S-linalool, respectively. The function of At4g16730 remains undetermined. The emission data indicate that β-myrcene and S-linalool are the two main monoterpenes emitted from Arabidopsis (ecotype Columbia) flowers. Thus, the formation of β-myrcene in flowers may be a consequence of the action of at least two different enzymes, whereas At1g61680 may be solely responsible for the emission of S-linalool. However, a better quantitative assessment of the contribution of each monoterpene synthase to the total floral monoterpene output will require completing the functional analysis of At4g16730 as well as making separate measurements of the activities of each enzyme in floral tissue—a difficult task.
The Members of the Arabidopsis TPS Gene Family Have Different Patterns of Organ Expression
Although many plant terpenes, such as sterols, carotenoids, and gibberellins, have well-known functions in plant growth and development, the roles of the vast majority of this large class of plant natural products are unknown. The presence of a large set of terpene synthase genes in the model species Arabidopsis affords an excellent opportunity to identify the functions of plant terpenes. Based on the RT-PCR analysis presented here (Figure 2), the Arabidopsis TPS genes have dramatically different organ expression patterns. Some are expressed almost exclusively in flowers, roots, or siliques, whereas others are expressed to a significant degree in two or more organs. These results suggest that some TPS genes, such as those demonstrated to be involved in floral volatile production in this study, may produce specialized products, whereas others may have a more general function. Further studies to characterize additional Arabidopsis TPS genes, manipulate terpene production, and assess phenotypic changes should significantly increase our understanding of the biological raison d'être of plant terpenoid metabolites.
Arabidopsis Floral Volatiles May Function in Pollinator Attraction and in Other Roles
It is tempting to hypothesize that the biological role of the terpenes emitted from Arabidopsis inflorescences is to attract insects for cross-pollination. Flowers of many species emit terpenes and other volatiles for pollinator attraction. Although Arabidopsis readily self-pollinates when grown indoors, the presence of floral nectaries (Davis et al., 1998), the protogynous mode of floral development (early maturation of the stigma while the anthers are still immature) (Jones, 1971), and several experimental studies (Snape and Lawrence, 1971; Abbott and Gomes, 1989; Loridon et al., 1998) all suggest that at least a low degree of cross-pollination exists in wild populations. Thus, floral volatiles could provide cues for pollinator attraction. Indeed, anecdotal observations of various insects visiting Arabidopsis flowers have been reported (Jones, 1971; Snape and Lawrence, 1971), and we have observed syrphid flies (Diptera: Syrphidae) visiting Arabidopsis flowers near Jena, Germany. However, no information is available regarding whether these insects facilitate cross-pollination and whether they can perceive olfactory cues from Arabidopsis. It has been shown that the antennae of the moth Hyles lineata can detect the monoterpene linalool and some other C. breweri floral volatiles at concentrations similar to those produced by emission from Arabidopsis flowers (Raguso et al., 1996). Nevertheless, similar experiments need to be conducted with potential insect visitors of Arabidopsis to determine if the compounds emitted from its flowers function as attractants and if the insects so attracted can effect cross-pollination.
The floral volatiles of Arabidopsis also could play a variety of other roles instead of or in addition to pollinator attraction. For example, many terpenes, including β-myrcene, (E)-β-ocimene, linalool, and (E)-β-caryophyllene, react readily with ozone and other reactive oxygen species (Calogirou et al., 1999; Loreto and Velikova, 2001). Thus, Arabidopsis floral volatiles could function to protect the reproductive organs, with their valuable germ line cells, from oxidative damage. A variety of monoterpenes and sesquiterpenes also are reported to have antimicrobial activity (Deans and Waterman, 1993). Hence, Arabidopsis floral terpenes also could help defend floral organs from bacterial or fungal infestation. The moist surface of the stigma may be an ideal environment for fungal growth. Finally, it is conceivable that, although Arabidopsis floral terpenes once had a function in the evolutionary past, they no longer do today. Arabidopsis thaliana diverged only recently (∼5 million years ago) from a clade made up of A. halleri, A. lyrata, and A. petraea (Koch et al., 2000), a lineage of self-incompatible perennials that presumably relied on insect pollination. Thus, floral terpenes may have been present in a recent ancestor of A. thaliana and simply have not yet been eliminated by genetic drift or natural selection.
Our observations of transgenic plants containing the promoter-GUS fusion construct for the At3g25810 gene also have implications for the function of floral volatiles in Arabidopsis, but these results need to be considered with caution until they are confirmed by additional methods, such as in situ hybridization. GUS activity was observed in four floral parts: sepals, stigmas, anther filaments, and receptacles (Figure 7). Activity in the stigmas is consistent with the idea that terpene volatiles function to protect the stigma from pathogen attack or oxidative damage, whereas the occurrence of GUS staining in the sepals, filaments, and receptacles suggests a function involving the entire flower, such as pollinator attraction. Interestingly, several genes involved in nonterpenoid floral scent biosynthesis in C. breweri and Antirrhinum majus have been reported to be expressed exclusively in petals, rather than in sepals or other parts of the flower (Wang et al., 1997; Dudareva et al., 1998, 2000). However, the linalool synthase gene of C. breweri also is expressed in the stigma, in addition to showing lower activity in the petals (Dudareva et al., 1996).
The Floral Volatiles of Arabidopsis May Have Implications for the Evolution of This Species and Its Population Genetic Structure
The emission of volatiles from flowers of Arabidopsis may have evolutionary as well as ecological significance. If floral volatiles increase visitation by potential insect pollinators, this will raise the degree of outcrossing and so alter the genetic structure of Arabidopsis populations. Even if the resulting degree of outcrossing is low, as observed by Abbott and Gomes (1989), progeny arising from outcrossing may have greater reproductive fitness because of the genetic polymorphisms they contain (Agren and Schemske, 1993). This may have led to the retention of traits that promote outcrossing even in species that are largely self-fertilizing. Thus, floral volatiles and other traits involved in the attraction of floral visitors may have been critical factors in the evolutionary history of Arabidopsis. From this perspective, the TPS genes involved in the formation of the major floral volatiles may represent valuable markers for the study of the phylogenies of this model species and its nearest relatives.
METHODS
Plant Material
Arabidopsis thaliana of the Columbia ecotype was grown from seeds in a controlled-climate room (22°C, 55% RH, and 100 μmol·m−2·s−1 PAR) under long-day conditions (16-h-light/8-h-dark photoperiod) for up to 6 weeks. At this time, plants had developed shoots and flowers (both primary and secondary bolts).
Volatile Analysis
Volatiles from 6-week-old flowering plants were collected in a dynamic headspace sampling system that was installed in a controlled-climate chamber (23°C, 70% RH, and 150 μmol·m−2·s−1 PAR). Five plants with intact root balls wrapped in aluminum foil were placed in a 4-L glass chamber that consisted of a flat flange reaction vessel and a flat flange lid with four necks, closed with Teflon stoppers. Charcoal-purified air entered the chamber at a flow rate of 1 L/min from the top through a Teflon hose (whose outlet was positioned in the lower part of the glass vessel). Volatiles were collected for 8 h by pumping air from the chamber through activated charcoal traps (1.5 mg) at a rate of 0.9 L/min. The remaining air was vented through the top of the chamber. For volatile collections from flowers or siliques, these organs were cut from five plants and transferred to a small beaker containing tap water or moistened filter paper.
To collect less abundant volatiles from inflorescences, a high-sensitivity closed-loop stripping method was applied (Donath and Boland, 1995). Ninety or 150 inflorescences were transferred to small glass beakers filled with tap water and were placed in an 1-L bell jar. Under continuous air circulation, emitted volatiles were collected for 12 h on activated charcoal traps that had been fitted into a steel column connected to the circulation pump. Volatiles were eluted with 40 μL of CH2Cl2, and 120 ng of nonyl acetate was added as an internal standard.
Samples from volatile collections and from solid-phase microextraction of terpene synthase assays (see below) were analyzed on a Hewlett-Packard 6890 gas chromatograph coupled to a Hewlett-Packard 5973 quadrupole mass selective detector. Separation was performed on (5%-phenyl)-methylpolysiloxane or polyethylene glycol columns (J&W Scientific, Folsom, CA) of 30 m × 0.25 mm i.d. × 0.25 m thickness. Helium was the carrier gas (flow rate of 2 mL/min), a splitless injection (injection volume of 2 μL) was used, and a temperature gradient of 5°C/min from 40°C (3-min hold) to 240°C was applied. For determination of the enantiomeric composition, a heptakis(2,3-di-O-methyl-6-O-t-butyldimethyl-silyl)-β-cyclodextrin column was used for all of the monoterpenes and the sesquiterpenes (E)-β-caryophyllene, cuparene, and β-bisabolene, whereas a heptakis(2,6-di-O-methyl-3-O-pentyl)-β-cyclodextrin column was used for α-barbatene, α-copaene, thujopsene, and β-chamigrene. The temperature program for the monoterpenes was as follows: 40°C (3-min hold) followed by a ramp of 5°C/min to 125°C. The program for the sesquiterpenes was as follows: 40°C (1-min hold) with a ramp of 30°C/min to 90°C (30-min hold). Mass spectrometry was performed with a transfer line temperature of 230°C, source temperature of 230°C, quadrupole temperature of 150°C, ionization potential of 70 eV, and scan range of 50 to 400 atomic mass units.
The identities of all compounds were determined by comparison of retention times and mass spectra with those of authentic standards and with mass spectra in the National Institute of Standards and Technology and Wiley libraries (Agilent Technologies, Palo Alto, CA). The racemic standards of α-barbatene, (E)-β-caryophyllene, and cuparene were provided by Wilfried A. König (University of Hamburg, Germany). Chirality was determined by direct comparison with standards of both enantiomers for all compounds except β-chamigrene and thujopsene, for which only the (−)-enantiomers were available. For quantification, representative single-ion peaks of each compound were integrated and compared with the equivalent response of the internal standard (single-ion method).
Determination of Gene Expression by Reverse Transcriptase–Mediated PCR
Total RNA was isolated with an RNeasy Plant Mini Kit (Qiagen, Valencia, CA), and DNA contamination was removed with DNase (Qiagen) treatment for 15 min at room temperature. Five micrograms of RNA was reverse transcribed into cDNA in a 33-μL reaction with poly(dT) priming using a First-Strand cDNA Synthesis Kit (Amersham Pharmacia Biotech). The PCR volume was 25 μL, containing 100 ng of each primer, 2 mM of each deoxynucleoside triphosphate, 1 μL of cDNA, and 0.75 units of Taq DNA polymerase (Fisher, Pittsburgh, PA). The primers used in the reverse transcriptase–mediated (RT) PCR experiments are shown in Table 1.
A PTC-100 Programmable Thermal Controller (MJ Research, Watertown, MA) was used with an initial denaturation step of 96°C for 1 min, followed by 30 cycles of 94°C for 30 s, 54°C for 30 s, 72°C for 60 s, and a final elongation step of 72°C for 10 min. RT-PCR analysis was performed in duplicate using RNA from two different sets of plants. Results of the two replicates were similar, and only one replicate is depicted in Figure 2. The second RT-PCR was performed using the same reagents as those used for the first PCR except that the RNA was isolated from different plants.
RNA Gel Blot Analysis
Total RNA (16 μg per sample) was separated on a 1% agarose gel containing 17% formamide and transferred onto a Hybond-XL nylon membrane (Amersham Pharmacia Biotech). DNA probes were generated by PCR and labeled with 32P-dCTP using a Rediprime II Kit (Amersham Pharmacia Biotech). Standard procedures were followed for prehybridization, hybridization, and washing.
cDNA Cloning by RT-PCR and Protein Expression in Escherichia coli
Full-length cDNAs were obtained by RT-PCR as described above. The primers used were 5′-AATGGCTACTTTGTGTATAGGT-3′ (or 5′-ATCCATATGCGACGCTCGGGCAACTATCAA-3′, containing a NdeI site) and 5′-ATCGGATCCTTAATCTAATGGGATTGGGTC-3′ (containing a BamHI site) for At3g25810, 5′-AATGGCCTTAATAGCTACCAAAATAAG-3′ (or 5′-AATGATCGATGTCATTCAAAGT-3′) and 5′-ATTACATTAGAGACTTGAGATAT-3′ for At1g61680, and 5′-AAT-GGGGAGTGAAGTCAACC-3′ and 5′-ATCAAATGGGTATAGTTTCAA-TG-3′ for At5g23960. The second forward primers (listed in parentheses) for At3g25810 and At1g61680 were chosen to eliminate the region coding for the transit peptide. The resulting fragments were cloned into the vector pCRT7/CT-TOPO (Invitrogen, Carlsbad, CA), and the truncated cDNA of AtTPS1 was subcloned further into the BamHI and NdeI sites of pET11a. An E. coli BL21 Codon Plus strain, transformed with the appropriate expression construct, was used for protein expression. Induction was performed at 18°C overnight with 1 mM isopropyl-1-thio-β-d-galactopyranoside.
Terpene Synthase Enzyme Assays
Cells from a 100-mL induced culture were harvested at 4°C, washed with 20 mL of washing buffer (20 mM Tris-HCl, pH 7.0, and 50 mM KCl), and resuspended in 5 mL of extraction buffer (50 mM 3-(N-morpholino)-2-hydroxypropanesulfonic acid, pH 7.0, 10% [v/v] glycerol, 5 mM MgCl2, 5 mM DTT, 5 mM sodium ascorbate, and 0.5 mM phenylmethylsulfonyl fluoride). Cells were disrupted by sonication (2 × 4-min treatment at 50% power with a Bandelin model UW2070 instrument [Berlin, Germany]) and then centrifuged at 20,000g at 4°C for 30 min. The supernatant was desalted with a Bio-Rad Econo column, and the resulting 4-mL eluate (in assay buffer, containing 10 mM 3-(N-morpholino)-2-hydroxypropanesulfonic acid, pH 7.0, 10% [v/v] glycerol, and 1 mM DTT) was used for the enzyme assay. The volume of each assay was 1 mL, containing 960 μL of enzyme extract, 20 mM MgCl2, 0.2 mM MnCl2, 0.2 mM NaWO4, 0.1 mM NaF, and 60 μM geranyl diphosphate or farnesyl diphosphate (Echelon Research Laboratories, Salt Lake City, UT). The assay was performed in an 8-mL DuPont autosampler vial with a white solid-top polypropylene cap (Alltech, Deerfield, IL), and a solid-phase microextraction PDMS-100 (polydimethylsiloxane) fiber (Supelco, Bellefonte, PA) was inserted into the tube to collect volatiles. The assay was incubated at 30°C for 1 h and then at 42°C for 15 min. After incubation, the solid-phase microextraction fiber was injected into a gas chromatography–mass spectrometry system for analysis. Alternatively, enzyme products were extracted three times with 1 mL of pentane, and the organic extract was concentrated to ∼100 μL before gas chromatography–mass spectrometry analysis. Controls included assays with crude extracts of induced E. coli (same strain) carrying the expression vector without any insert.
Microscale assays to compare the relative activity of different substrates were performed in a final volume of 500 μL with 450 μL of crude extract and 10 or 30 μM 3H-geranyl diphosphate (2 or 9 MBq/μmol) or 3H-farnesyl diphosphate (9 MBq/μmol). Buffer, salt, and incubation conditions were as described above, but 0.4 mM NaWO4 and 0.2 mM NaF were used in assays with At1g61680 enzyme. After a 10-min incubation, the reaction products were extracted two times with 1 mL of hexane. Total radioactivity of the reaction products was determined by scintillation counting. In assays with the At3g25810 enzyme, the hexane extract was treated with silica gel to remove phosphatase products before counting.
Construction of the At3g25810 Promoter–β-Glucuronidase Reporter Gene Fusion Construct, Arabidopsis Transformation, and Histochemical Localization of β-Glucuronidase Activity
The At3g25810 promoter region (1.4 kb) was isolated via PCR from Arabidopsis genomic DNA using the primers 5′-CTCCGATTATTG-AATTCTAGGGCGGATTG-3′ and 5′-GAATCTTGGATCCATGGCAAT-TATCGTAC-3′, which contain an EcoRI and an NcoI site, respectively. The resulting PCR product was cloned into the pCRT7/CT-TOPO vector and sequenced. Next, the promoter region of At3g25810 was cut out of the pCRT7/CT-TOPO vector with EcoRI and NcoI digestion and inserted into the binary vector pCAMBIA1303 (Hajdukiewicz et al., 1994), replacing the 35S promoter of Cauliflower mosaic virus so that the At3g25810 promoter directs the expression of the uidA (β-glucuronidase [GUS]) gene. The construct was introduced into Agrobacterium tumefaciens strain GV3101, which was used to transform Arabidopsis by floral vacuum infiltration (Bechtold et al., 1993). Transgenic lines transformed with the construct were selected using hygromycin and confirmed by PCR. Enzymatic assays with 5-bromo-4-chloro-3-indolyl-β-d-glucuronide were performed to determine the localization of the enzyme activity of the GUS enzyme (Jefferson et al., 1987). Tissue samples were incubated at 37°C in GUS staining buffer (50 mM sodium phosphate buffer, pH 7.0, 0.1% Triton X-100, and 0.5 mg/mL 5-bromo-4-chloro-3-indolyl-β-d-glucuronide) overnight. After detection of the blue color, chlorophyll was extracted with 70% ethanol for 24 h.
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes.
Acknowledgments
We thank W. König for some of the sesquiterpene standards used, Bettina Raguschke and Katrin Heisse for technical assistance, and J. Bohlmann for sharing results before publication. This research was supported by National Science Foundation Grants MCB-9974463 and IBN-0211697 (E.P.), the Max Planck Society (J.G.), and Alexander von Humboldt Foundation Fellowships to A.F. and E.P.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.007989.
References
- Abbott, R.J., and Gomes, M.F. (1989). Population genetic structure and outcrossing rate of Arabidopsis thaliana (L.) Heynh. Heredity 62, 411–418. [Google Scholar]
- Agren, J., and Schemske, D.W. (1993). Outcrossing rate and inbreeding depression in 2 annual monoecious herbs, Begonia hirsuta and B. semiovata. Evolution 47, 125–135. [DOI] [PubMed] [Google Scholar]
- Aubourg, S., Lecharny, A., and Bohlmann, J. (2002). Genomic analysis of the terpenoid synthase (AtTPS) gene family of Arabidopsis thaliana. Mol. Genet. Genomics 267, 730–745. [DOI] [PubMed] [Google Scholar]
- Bechtold, N., Ellis, J., and Pelletier, G. (1993). In planta Agrobacterium-mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C. R. Acad. Sci. Paris Life Sci. 316, 1194–1199. [Google Scholar]
- Bohlmann, J., Martin, D., Oldham, N.J., and Gershenzon, J. (2000). Terpenoid secondary metabolism in Arabidopsis thaliana: cDNA cloning, characterization, and functional expression of a myrcene/(E)-β-ocimene synthase. Arch. Biochem. Biophys. 375, 262–269. [DOI] [PubMed] [Google Scholar]
- Bohlmann, J., Meyer-Gauen, G., and Croteau, R. (1998). Plant terpenoid synthases: Molecular biology and phylogenetic analysis. Proc. Natl. Acad. Sci. USA 95, 4126–4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai, Y., Jia, J.-W., Crock, J., Lin, Z.-X., Chen, X.-Y., and Croteau, R. (2002). A cDNA clone for β-caryophyllene synthase from Artemisia annua. Phytochemistry 61, 523–529. [DOI] [PubMed] [Google Scholar]
- Calogirou, A., Larsen, B.R., and Kotzias, D. (1999). Gas-phase terpene oxidation products: A review. Atmos. Environ. 33, 1423–1439. [Google Scholar]
- Cane, D.E. (1999). Sesquiterpene biosynthesis: Cyclization mechanisms. In Comprehensive Natural Products Chemistry, Vol. 2, Isoprenoids Including Carotenoids and Steroids, D.D. Cane, ed (Amsterdam: Elsevier), pp. 155–200.
- Colby, S.M., Alonso, W.R., Katahira, E.J., McGarvey, D.J., and Croteau, R. (1993). 4S-Limonene synthase from the oil glands of spearmint (Mentha spicata): cDNA isolation, characterization and bacterial expression of the catalytically active monoterpene cyclase J. Biol. Chem. 268, 23016–23024. [PubMed] [Google Scholar]
- Crock, J., Wildung, M., and Croteau, R. (1997). Isolation and bacterial expression of a sesquiterpene synthase cDNA clone from peppermint (Mentha × piperita, L.) that produces the aphid alarm pheromone (E)-β-farnesene. Proc. Natl. Acad. Sci. USA 94, 12833–12838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowell, A.L., Williams, D.C., Davis, E.M., Wildung, M.R., and Croteau, R. (2002). Molecular cloning and characterization of a new linalool synthase. Arch. Biochem. Biophys. 405, 112–121. [DOI] [PubMed] [Google Scholar]
- Davis, A.R., Pylatuik, J.D., Paradis, J.C., and Low, N.H. (1998). Nectar-carbohydrate production and composition vary in relation to nectary anatomy and location within individual flowers of several species of Brassicaceae. Planta 205, 305–318. [DOI] [PubMed] [Google Scholar]
- Deans, S.G., and Waterman, P.G. (1993). Biological activity of volatile oils. In Volatile Oil Crops: Their Biology, Biochemistry and Production, R.K.M. Hay and P.G. Waterman, eds (Essex, UK: Longman Scientific and Technical), pp. 97–111.
- Donath, J., and Boland, W. (1995). Biosynthesis of acyclic homoterpenes: Enzyme selectivity and absolute configuration of the nerolidol precursor. Phytochemistry 39, 785–790. [Google Scholar]
- Dudareva, N., Cseke, L., Blanc, V.M., and Pichersky, E. (1996). Evolution of floral scent in Clarkia: Novel patterns of S-linalool synthase gene expression in the C. breweri flower. Plant Cell 8, 1137–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudareva, N., D'Auria, J.C., Nam, K.H., Raguso, R.A., and Pichersky, E. (1998). Acetyl-CoA:benzylalcohol acetyltransferase: An enzyme involved in floral scent production in Clarkia breweri. Plant J. 14, 297–304. [DOI] [PubMed] [Google Scholar]
- Dudareva, N., Murfitt, L.M., Mann, C.J., Gorenstein, N., Kolosova, N., Kish, C.M., Bonham, C., and Wood, K. (2000). Developmental regulation of methyl benzoate biosynthesis and emission in snapdragon flowers. Plant Cell 12, 949–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudareva, N., and Pichersky, E. (2000). Biochemical and molecular genetic aspects of floral scents. Plant Physiol. 122, 627–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fäldt, J., Arimura, G.I., Gershenzon, J., Takabayashi, J., and Bohlmann, J. (2002). Functional identification of AtTPS03 as (E)-β-ocimene synthase: A monoterpene synthase catalyzing jasmonate- and wound-induced volatile formation in Arabidopsis thaliana. Planta, in press. [DOI] [PubMed]
- Hajdukiewicz, P., Svab, Z., and Maliga, P. (1994). The small, versatile pZIP family of Agrobacterium binary vectors for plant transformation. Plant Mol. Biol. 25, 989–994. [DOI] [PubMed] [Google Scholar]
- Jefferson, R.A., Kavanagh, T.A., and Bevan, M.W. (1987). GUS fusions: β-Glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6, 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia, J.W., Crock, J., Lu, S., Croteau, R., and Chen, X.Y. (1999). (3R)-Linalool synthase from Artemisia annua L.: cDNA isolation, characterization, and wound induction. Arch. Biochem. Biophys. 372, 143–149. [DOI] [PubMed] [Google Scholar]
- Jones, M.E. (1971). Population genetics of Arabidopsis thaliana. 1. Breeding system. Heredity 27, 39–50. [Google Scholar]
- Joulain, D., and König, W.A. (1998). The Atlas of Spectral Data of Sesquiterpene Hydrocarbons. (Hamburg, Germany: E.B.-Verlag).
- Knudsen, J.T., Tollsten, L., and Bergstrom, L.G. (1993). Floral scents: A checklist of volatile compounds isolated by headspace techniques. Phytochemistry 33, 253–280. [Google Scholar]
- Koch, M.A., Haubold, B., and Mitchell-Olds, T. (2000). Comparative evolutionary analysis of chalcone synthase and alcohol dehydrogenase loci in Arabidopsis, Arabis, and related genera (Brassicaceae). Mol. Biol. Evol. 17, 1483–1498. [DOI] [PubMed] [Google Scholar]
- König, W.A., Rieck, A., Saritas, Y., Hardt, I.H., and Kubeczka, K.-H. (1996). Sesquiterpene hydrocarbons in the essential oil of Meum athamanticum. Phytochemistry 42, 461–464. [Google Scholar]
- Lichtenthaler, H.K. (1999). The 1-deoxy-d-xylulose-5-phosphate pathway of isoprenoid biosynthesis in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 47–65. [DOI] [PubMed] [Google Scholar]
- Loreto, F., and Velikova, V. (2001). Isoprene produced by leaves protects the photosynthetic apparatus against ozone damage, quenches ozone products, and reduces lipid peroxidation of cellular membranes. Plant Physiol. 127, 1781–1787. [PMC free article] [PubMed] [Google Scholar]
- Loridon, K., Cournoyer, B., Goubely, C., Depeiges, A., and Picard, G. (1998). Length polymorphism and allele structure of trinucleotide microsatellites in natural accessions of Arabidopsis thaliana. Theor. Appl. Genet. 97, 591–604. [Google Scholar]
- MacMillan, J., and Beale, M.H. (1999). Diterpene biosynthesis. In Comprehensive Natural Products Chemistry, Vol. 2, Isoprenoids Including Carotenoids and Steroids, D.D. Cane, ed (Amsterdam: Elsevier), pp. 217–243.
- Meyerowitz, E., and Somerville, C.R. (1994). Arabidopsis. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Raguso, R.A., Light, D.M., and Pichersky, E. (1996). Electroantennogram responses of Hyles lineata (Sphingidae: Lepidoptera) to volatile compounds from Clarkia breweri (Onagraceae) and other moth-pollinated flowers. J. Chem. Ecol. 22, 1735–1766. [DOI] [PubMed] [Google Scholar]
- Raguso, R.A., and Pichersky, E. (1995). Floral volatiles from Clarkia breweri and C. concinna (Onagraceae): Recent evolution of floral scent and moth pollination. Plant Syst. Evol. 194, 55–67. [Google Scholar]
- Snape, J.W., and Lawrence, M.J. (1971). Breeding system of Arabidopsis thaliana. Heredity 27, 299–301. [Google Scholar]
- Sun, T.P., and Kamiya, Y. (1994). The Arabidopsis GA1 locus encodes the cyclase ent-kaurene synthetase A of gibberellin biosynthesis. Plant Cell 6, 1509–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner, G., Gershenzon, J., Nielson, E.E., Froehlich, J.E., and Croteau, R. (1999). Limonene synthase, the enzyme responsible for monoterpene biosynthesis in peppermint, is localized to leucoplasts of oil gland secretory cells. Plant Physiol. 120, 879–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Poecke, R.M.P., Posthumus, M.A., and Dicke, M. (2001). Herbivore-induced volatile production by Arabidopsis thaliana leads to attraction of the parasitoid Cotesia rubecula: Chemical, behavioral, and gene-expression analysis. J. Chem. Ecol. 27, 1911–1928. [DOI] [PubMed] [Google Scholar]
- Wang, J., Dudareva, N., Bhakta, S., Raguso, R.A., and Pichersky, E. (1997). Floral scent production in Clarkia breweri (Onagraceae). II. Localization and developmental modulation of the enzyme S-adenosyl-l-methionine:(iso)eugenol O-methyltransferase and phe-nylpropanoid emission. Plant Physiol. 114, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, D.C., McGarvey, D.J., Katahira, E.J., and Croteau, R. (1998). Truncation of limonene synthase preprotein provides a fully active ‘pseudomature’ form of this monoterpene cyclase and reveals the function of the amino-terminal arginine pair. Biochemistry 37, 12213–12220. [DOI] [PubMed] [Google Scholar]
- Wise, M., and Croteau, R. (1999). Monoterpene biosynthesis. In Comprehensive Natural Products Chemistry, Vol. 2, Isoprenoids Including Carotenoids and Steroids, D.D. Cane, ed (Amsterdam: Elsevier), pp. 97–153.
- Yamaguchi, S., Sun, T.P., Kawaide, H., and Kamiya, Y. (1998). The GA2 locus of Arabidopsis thaliana encodes ent-kaurene synthase of gibberellin biosynthesis. Plant Physiol. 116, 1271–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]