Abstract
Aquaporins are ubiquitous channel proteins that facilitate the transport of water across cell membranes. Aquaporins show a typically high isoform multiplicity in plants, with 35 homologs in Arabidopsis. The integrated function of plant aquaporins and the function of each individual isoform remain poorly understood. Matrix-assisted laser desorption/ionization time-of-flight analyses suggested that Plasma Membrane Intrinsic Protein2;2 (PIP2;2) is one of the abundantly expressed aquaporin isoforms in Arabidopsis root plasma membranes. Two independent Arabidopsis knockout mutants of PIP2;2 were isolated using a PCR-based strategy from a library of plant lines mutagenized by the insertion of Agrobacterium tumefaciens T-DNA. Expression in transgenic Arabidopsis of a PIP2;2 promoter–β-glucuronidase gene fusion indicated that PIP2;2 is expressed predominantly in roots, with a strong expression in the cortex, endodermis, and stele. The hydraulic conductivity of root cortex cells, as measured with a cell pressure probe, was reduced by 25 to 30% in the two allelic PIP2;2 mutants compared with the wild type. In addition, free exudation measurements revealed a 14% decrease, with respect to wild-type values, in the osmotic hydraulic conductivity of roots excised from the two PIP2;2 mutants. Together, our data provide evidence for the contribution of a single aquaporin gene to root water uptake and identify PIP2;2 as an aquaporin specialized in osmotic fluid transport. PIP2;2 has a close homolog, PIP2;3, showing 96.8% amino acid identity. The phenotype of PIP2;2 mutants demonstrates that, despite their high homology and isoform multiplicity, plant aquaporins have evolved with nonredundant functions.
INTRODUCTION
Aquaporins form a large family of membrane channels that facilitate the diffusion of water and small neutral solutes across the cellular membranes of most living organisms (Agre et al., 1998). The subcellular localization, primary transport, and regulation properties of some plant aquaporins have been described in detail (for recent reviews, see Maurel et al., 2002; Tyerman and Niemietz, 2002). Yet, the full significance of aquaporins in plant water relations remains unclear.
The uptake of water from the soil and its delivery to the xylem requires that water move radially across living root tissues. The limiting role of cellular membranes in this process has long remained conjectural, because this so-called transcellular path might be bypassed by transport along cell walls (apoplastic path) or cytoplasmic continuities (symplastic path) (Steudle and Peterson, 1998; Javot and Maurel, 2002). Because of their ability to block the pores of most aquaporins, mercury ions have been used in recent years to investigate aquaporin function in planta. A mercury-sensitive water channel path was revealed in the roots of a large variety of plant species that contributes to >50% of water uptake (reviewed by Javot and Maurel, 2002). It also was proposed that this path can mediate the regulation of root hydraulic conductivity in response to environmental stimuli, such as stresses, day and night cycles, or changes in nutrient availability.
Because of the overall toxicity of mercury compounds and their effects on other physiological functions besides water transport (Zhang and Tyerman, 1999; Javot and Maurel, 2002), reverse genetics provides a more specific approach to address aquaporin function in planta. Compared with other living organisms, plants appear to have a particularly large number of aquaporin homologs. The genome of Arabidopsis, for instance, encodes 35 full-length aquaporin genes with 13 homologs in the Plasma Membrane Intrinsic Protein (PIP) subgroup (Johanson et al., 2001; Quigley et al., 2001). In view of such high isoform multiplicity, gene silencing by antisense suppression first appeared as an efficient reverse genetics approach, because concomitant downregulation of several aquaporin homologs and consequently more pronounced phenotypes are expected (Kaldenhoff et al., 1998; Barrieu et al., 2000). Arabidopsis plants that express a PIP1;2 antisense transgene have been instrumental in establishing the contribution of aquaporins of the PIP1 subgroup, with five members in Arabidopsis, to water transport in isolated protoplasts (Kaldenhoff et al., 1998). The same plants also exhibited a fivefold increase in root growth, which was interpreted as an increase in root absorption surface to compensate for reduced hydraulic conductivity (Kaldenhoff et al., 1998). Antisense expression of a PIP1 homolog (NtAQP1) in tobacco recently demonstrated that the PIP1 subgroup of aquaporins indeed contributes to at least 55% of root hydraulic conductivity (Siefritz et al., 2002). Surprisingly, no morphological alteration was exhibited by the tobacco antisense plants. By contrast, a critical role for PIP1 aquaporins in the mobilization of water into leaf tissues and in water stress avoidance in leaves was revealed in the antisense plants (Siefritz et al., 2002).
Genetic studies in humans and mice have established that, with the exception of aquaporin-2 (AQP2), the dysfunction of which is a major cause of genetically transmitted nephrogenic diabetes insipidus (Deen et al., 1994), patients or transgenic animals with individually inactive aquaporins were phenotypically normal in standard living conditions (Preston et al., 1994; Agre, 1998). A closer inspection of fluid transport together with physiological challenge in extreme conditions was necessary to reveal a function for AQP1 and AQP3 in renal filtration (Schnermann et al., 1998; Ma et al., 2000), for AQP5 in fluid secretion in secretory epithelia and glands (Ma et al., 1999; Song and Verkman, 2001; Nejsum et al., 2002), and for AQP4 in water balance in the brain (Manley et al., 2000).
In plants, understanding the function of specific aquaporin isoforms is a crucial objective for implementing our molecular understanding of plant water relations. Cellular function for specific aquaporin homologs can be inferred on the basis of distinct subcellular localizations or tissue-specific expression patterns (Santoni et al., 2000a). Furthermore, functional properties specific to certain isoforms, such as mixed selectivity or regulation by phosphorylation (reviewed by Johansson et al., 2000; Tyerman and Niemietz, 2002), have been reported, but these properties have not been evaluated at the whole gene family level.
Single aquaporin knockouts should be a tool of choice for addressing the function of specific isoforms. However, it is unclear whether the high isoform multiplicity and the potential functional redundancy of plant aquaporins would preclude such an approach. To date, mutation of a single PIP1 homolog has been linked to a loss of pollen self-incompatibility in Brassica (Ikeda et al., 1997), but the mechanistic interpretation of this phenotype remains unclear.
In the present work, we address the function of PIP aquaporins in Arabidopsis and identify PIP2;2 as one of the abundantly expressed PIP isoforms in roots. Using two independently isolated knockout mutants, we determine the contribution of this aquaporin to the water permeability (hydraulic conductivity) of root cortex cells and demonstrate its role in osmotic water transport in the whole root. These studies establish in plants the function of a single aquaporin gene at the cell and tissue level. The existence of PIP2;3 (which forms a tandem duplicated aquaporin cluster with PIP2;2) suggests that despite their high isoform multiplicity, plant aquaporins have evolved with nonredundant functions.
RESULTS
PIP2;2 Is One of the Abundantly Expressed PIP Isoforms in Roots
To make a rapid inventory of aquaporins that are expressed abundantly in the Arabidopsis root, proteins of purified root plasma membranes were separated by SDS-PAGE and analyzed by mass spectrometry. Proteins that comigrated with a 28-kD band immunodetected with both anti-PIP1 and anti-PIP2 antibodies were digested by trypsin and examined using matrix-assisted laser desorption/ionization time-of-flight analysis. A complex fingerprint with >40 peptidic fragments was revealed, in agreement with the expected presence of numerous proteins in this crude 28-kD fraction. To determine which of these fragments might correspond to PIPs, theoretical fingerprints of each of the 13 Arabidopsis PIPs were compared with the observed fingerprint. Matching peptides were identified on the basis of a <50-ppm difference between the observed (mass-to-charge ratio) and predicted masses. Six, four, and four matching peptides were detected for PIP2;1, PIP2;2, and PIP2;4, respectively (Table 1). All other PIP homologs yielded three (PIP1;1) or fewer matching peptides (Table 1) (V. Santoni, J. Vinh, D. Pflieger, N. Sommerer, and C. Maurel, unpublished data). Noticeably, no peptide corresponding to the PIP2;3 sequence was observed (Table 1). As a result of the high sequence homology between PIPs, certain peptides can be allocated to several isoforms (Table 1). However, we observed that three of the four PIP2;2 peptides were specific for this isoform. The presence of PIP2;1 and PIP2;4 also can be predicted on the basis of three and two specific peptides, respectively.
Table 1.
Mass Spectrometry Analysis of Peptides Isolated from a 28-kD Root Plasma Membrane Fraction and Matching with the Sequence of PIP2 Homologs
| Observed Massa(m/z) | Predicted Mass (D) |
Difference in Mass (ppm) |
Predicted Peptide | Isoformb | Position in Sequence |
|---|---|---|---|---|---|
| 886.50 | 886.457 | −48.504 | KWSFYR | PIP2;1/PIP2;6 | 34 to 39/33 to 38 |
| 1069.56 | 1069.568 | 7.106 | SFGAAVIYNK | PIP2;1/PIP2;2 | 232 to 241/230 to 239 |
| 1122.55 | 1122.521 | −25.477 | AFQSSYYTR | PIP2;1/PIP2;4 | 145 to 153/145 to 153 |
| 1136.55 | 1136.501 | −43.376 | AFQSSYYDR | PIP2;2 | 143 to 151 |
| 1234.63 | 1234.570 | −48.759 | DVEGPEGFQTR | PIP2;2 | 4 to 14 |
| 1312.65 | 1312.653 | 2.438 | SFGAAVIYNNEK | PIP2;4/PIP2;7/PIP2;8 | 232 to 243/225 to 236/223 to 234 |
| 1340.68 | 1340.644 | −26.851 | DLDVNESGPPAAR | PIP2;4 | 4 to 16 |
| 1404.72 | 1404.675 | −31.820 | DVEAVPGEGFQTR | PIP2;1 | 4 to 16 |
| 1872.90 | 1872.901 | 0.748 | DYQDPPPAPFIDGAELK | PIP2;1 | 17 to 33 |
| 2000.97 | 2000.996 | 13.144 | DYQDPPPAPFIDGAELKK | PIP2;1 | 17 to 34 |
| 2067.00 | 2066.953 | −22.882 | DYKDPPPAPFFDMEELR | PIP2;4 | 17 to 33 |
| 2096.960 | 2096.933 | −12.684 | DYEDPPPTPFFDADELTK | PIP2;2 | 15 to 32 |
Peptide masses (mass-to-charge ratio [m/z]) were measured by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Representative data from one of five independent root plasma membrane protein extractions and mass spectrometry analyses are shown.
Note that certain peptides can be allocated to several PIP2 isoforms; however, all PIP2;2 peptides contain residues that discriminate between the PIP2;2 and PIP2;3 sequences.
The putative PIP2;2 peptide with a mass of 1136.55 was analyzed by tandem mass spectrometry, and a peptidic sequence (AFQSSYYDR) was established that corresponds to the PIP2;2 sequence at positions 143 to 151. Note that this peptide is specific for PIP2;2 with respect to the overlapping peptides expected from tryptic digestion of PIP2;1 (AFQSSYYTR) and PIP2;3 (no cleavage at position 151), the closest homologs of PIP2;2. Similar analyses indicated that the sequence of another of the putative PIP2;2-specific peptides, with a mass of 2096.96, perfectly matched that of the predicted peptide (V. Santoni, J. Vinh, D. Pflieger, N. Sommerer, and C. Maurel, unpublished data). Thus, mass spectrometry and tandem mass spectrometry results unambiguously establish the presence of PIP2;2 in root plasma membranes and suggest that PIP2;1, PIP2;2, and PIP2;4 are among the most abundantly expressed PIP isoforms in these membranes. Because of the very high structural homology between PIP2;2 and PIP2;3 (96.8% identity at the amino acid level), it is unlikely that the two aquaporins show significantly different behavior during membrane preparation and matrix-assisted laser desorption/ionization. Thus, our failure to detect any PIP2;3 peptide shows that PIP2;3 is expressed at a much lower level than PIP2;2. This is remarkable because PIP2;2 and PIP2;3 share a common 3-kb promoter region and form an inverted tandem repeat on chromosome II, suggesting that they have evolved through a recent gene duplication (Figure 1).
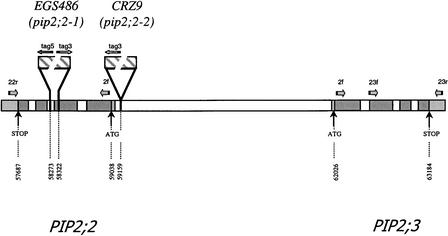
Figure 1.
Physical Map of the PIP2;2 and PIP2;3 Genes Derived from the Genomic Sequence of the Chromosome.
The sizes of the inserted T-DNAs are not presented to scale. Numbers refer to nucleotides in PIP2;2. Horizontal arrows indicate the positions and orientations of primer sequences used for PCR analyses.
Isolation and Molecular Characterization of PIP2;2 Aquaporin Mutants
In a broad reverse genetics approach to PIP aquaporin function, we screened, using a PCR-based strategy, a library of plant lines mutagenized by the insertion of Agrobacterium tumefaciens T-DNA. Several transformed lines, including EGS486 and CRZ9, that carried possibly disrupted PIP2 genes were identified. Genomic DNA fragments were amplified from the transformed lines by PCR, using a combination of primers specific for aquaporins and T-DNA sequences, respectively. These fragments were sequenced to establish the positions of the T-DNA insertions in the plant genome.
In DNA amplified from EGS486, T-DNA left border sequences were detected in the PIP2;2 gene at positions downstream of nucleotide 58,273 (section 202 of Arabidopsis chromosome II) (data not shown), whereas T-DNA right border sequences were detected at a position upstream of nucleotide 58,322 (Figures 1 and 2A). Thus T-DNA insertion resulted in a 49-bp deletion and disrupted the junction between exon 2 and intron 2 of PIP2;2 (Figure 2). In CRZ9, foreign DNA sequences containing a right border signal are found, at the same genomic location, for nucleotides beyond nucleotide 59,158 (Figure 1), indicating that T-DNA insertion occurred in the PIP2;2 promoter at 121 bp from the start codon (Figure 2).
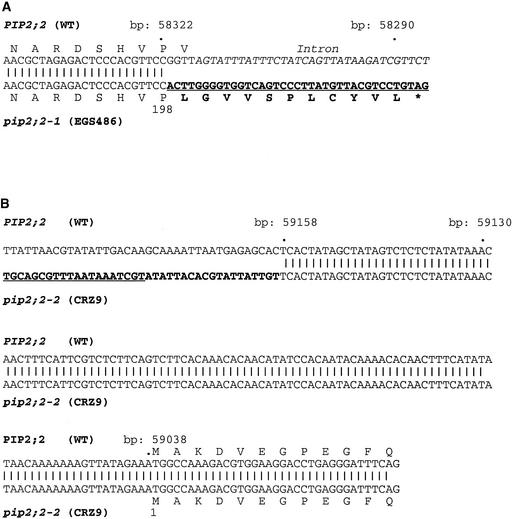
Figure 2.
Sequence Analysis of T-DNA Insertion Sites in Two Arabidopsis PIP2;2 Mutant Lines.
Genomic DNA was amplified by PCR from the wild type and the EGS486 and CRZ9 transformed lines using a pair of primers specific for PIP and T-DNA border sequences, respectively. DNA sequences that are divergent from wild-type genomic DNA are indicated in bold type, and specific T-DNA sequences are underlined. Nucleotide numbers refer to those in PIP2;2. The predicted translation products of the wild-type (WT) and mutated PIP2;2 genes are indicated using single-letter code, with residues being numbered with respect to the initiating Met.
(A) Sequence analysis in EGS486 (pip2;2-1). The T-DNA sequences shown carry part of the right border signal. The sequence of intron 2 in PIP2;2 is indicated in italics. According to the putative membrane topology of PIPs, the amino acid stretch Asn-192 to Val-200 is located in the second intracellular loop of PIP2;2. Note that if translated, the EGS486 mRNA may lead to a truncated protein of 209 amino acids comprising the first 198 residues of PIP2;2 (wild-type PIP2;2 has 285 residues) plus 11 T-DNA–encoded residues.
(B) Sequence analysis in CRZ9 (pip2;2-2). The DNA sequence that reads outside from the T-DNA toward the right border signal is underlined. The remaining 19-bp foreign sequence is of unknown origin and was generated during T-DNA insertion. The insertion occurred at 121 bp upstream of the ATG, downstream of nucleotide 59,158.
For further molecular analyses, plants carrying a single kanamycin-resistant locus and homozygous for one of the T-DNA insertions described above were derived from the initial transformants EGS486 and CRZ9 after up to two and four backcrosses to wild-type plants, respectively. The homozygous mutant plants derived from EGS486 and CRZ9 were named pip2;2-1 and pip2;2-2, respectively.
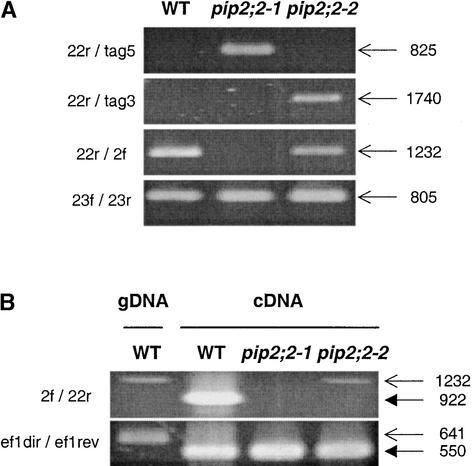
The genomic structure of PIP2;2 in the pip2;2-1 and pip2;2-2 mutant lines was analyzed by PCR, based on the genome sequence of this region (Figure 1). A reverse primer specific for the 3′ untranslated region of PIP2;2 (22r) was used in combination with primers specific for left border (tag5) or right border (tag3) sequences. Consistent with sequence data for the plant DNA/T-DNA junctions, these two combinations yielded specific amplification products with the genomic DNA of pip2;2-1 and pip2;2-2, respectively. The measured sizes of the products (∼800 and ∼1700 bp, respectively) were consistent with sizes expected from the genome sequence (825 and 1740 bp, respectively) (Figure 3A).
Figure 3.
PCR Analysis of the Genomic Structure and mRNA Expression of PIP2;2 in Wild-Type and Two Arabidopsis PIP2;2 Mutant Lines.
(A) Agarose gel separation of PCR products amplified from the genomic DNA of the wild type (WT), pip2;2-1, and pip2;2-2. The primer combinations used are indicated at left. The positions of the primers on the physical map of PIP2;2 and PIP2;3 are shown in Figure 1. The sizes (in bp) and calculated migrations (arrows) of the predicted PCR products are shown at right.
(B) Agarose gel separation of PCR products amplified from genomic DNA (gDNA) of the wild type and cDNA of the wild type, pip2;2-1, and pip2;2-2. The expression of Elongation Factor1α (primer combination ef1dir/ef1rev) is shown as a control. The sizes and calculated migrations of the PCR products predicted for amplification from gDNA and cDNA are shown by the top and bottom arrows in each gel, respectively. Amplification of gDNA products in the cDNA reactions is attributable to the presence of traces of gDNA (total RNA was not treated by RQ1 DNase). This establishes that the PCR conditions were suitable for the primer combinations used.
The integrity of the PIP2;2 coding sequence was probed using the primer combination 22r/2f (Figure 1). In our PCR conditions, the presence between two primer sequences of a >7-kb T-DNA insertion should prevent any DNA amplification. The results shown in Figure 3A confirm that the PIP2;2 coding sequence was disrupted in pip2;2-1 but was intact in wild-type and pip2;2-2 plants. Because T-DNA insertions in the plant genome can result in local but profound rearrangements, we also checked the integrity of the neighboring PIP2;3 coding region using the primer combination 23f/23r (Figure 1) and found this to be intact in the two transformed lines (Figure 3A).
The effects of T-DNA insertion on PIP2;2 mRNA expression in the two mutant lines were examined by reverse transcriptase–mediated PCR. The wild-type PIP2;2 cDNA was revealed using the primer combination 2f/22r (Figure 1) as a 922-bp PCR product that is distinctly smaller than the product amplified from genomic DNA using the same primer combination (Figure 3B, lanes 1 and 2). Figure 3B shows that a full-length PIP2;2 cDNA was detectable in the wild type but not in pip2;2-1 and pip2;2-2, whereas the Elongation Factor1α transcript used as a control was detected in all three lines.
Together, these results establish that T-DNA insertions in the coding sequence (pip2;2-1) and in the promoter (pip2;2-2) of PIP2;2 provide two independently isolated knockout alleles of this gene.
Tissue-Specific Expression of PIP2;2
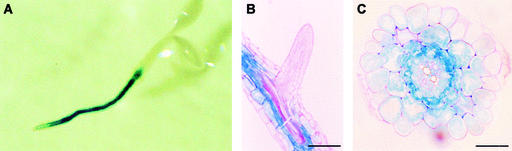
Phenotypic analysis of PIP2;2 mutant plants requires a precise knowledge of the tissue-specific expression profile of this gene. To establish this profile, a 3.7-kb fragment corresponding to the genomic region upstream of the PIP2;2 coding sequence was fused translationally to a β-glucuronidase (GUS) gene and transferred into Arabidopsis. In germinating seedlings containing the PIP2;2-GUS construct, GUS activity was localized primarily in the root, with predominant staining in the central part of the differentiated root tissues (Figure 4A). This pattern was maintained in hydroponically grown adult plants that showed no or faint GUS expression in the epidermis and in the root apices (dividing and elongating zones, emerging secondary roots). By contrast, GUS expression was observed in the root cortex, endodermis, and stele (Figures 4B and 4C), with the strongest staining in the endodermis and in the few outer cell layers of the stele (Figure 4C). In older root parts, GUS activity was lower but remained pronounced in the endodermis and in the stele. GUS expression also was revealed in the aerial parts of young or fully grown plants, with localized staining in the vascular tissues and hydathodes (data not shown).
Figure 4.
Expression Analysis of a PIP2;2-GUS Construct in Transgenic Arabidopsis.
(A) GUS staining of a 3-day-old seedling grown in vitro.
(B) Longitudinal section of a GUS-stained root from a 29-day-old plant grown in hydroponic conditions. Bar = 100 μm.
(C) Cross-section at 3 to 4 mm from the tip of a root of a 29-day-old plant grown in hydroponic conditions. Note that GUS staining was most intense in the endodermis and in the few outer cell layers of the stele. Bar = 25 μm.
Normal Growth and Development of pip2;2 Mutant Plants
Possible growth or developmental alterations of the pip2;2-1 and pip2;2-2 mutant lines with respect to the wild type were investigated in plants grown in vitro, in soil, or in hydroponic culture. Besides standard growth conditions, plants were subjected to a large variety of mild or severe water stresses imposed by progressive drought or sudden exposure of roots to a wide range of NaCl or polyethylene glycol concentrations. We also tested the effects of a prolonged deprivation or a changing supply of mineral nutrients (nitrogen). These treatments, which were performed under various light/dark cycles, represent typical conditions in which water channel regulation is supposed to occur (Javot and Maurel, 2002; Tyerman and Niemietz, 2002). Despite close anatomic inspection and quantitative measurements of root and shoot growth and of dry weight-to-fresh weight ratio, no growth or developmental phenotype was observed for the mutant plants in any of the conditions investigated. These negative results prompted us to probe aquaporin function by more precise techniques and to compare in closer detail the water relations of wild-type and mutant plants.
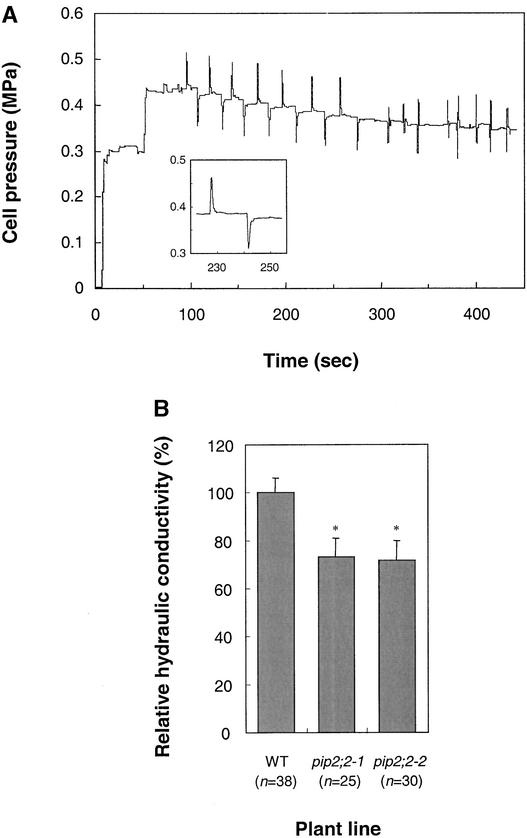
Reduced Cell Hydraulic Conductivity in the Root Cortex of pip2;2 Mutants
The PIP2;2 gene is expressed in root cortical cells (Figure 4). Consistent with the lack of a macroscopic growth phenotype in mutant plants, we first observed that cortical cells of wild-type and mutant plants had similar sizes (cell length of ∼170 μm; cell diameter of ∼23 μm) (Table 2). Because of their relatively large size, these cells are amenable to stable impalement with a cell pressure probe. Thus, in situ measurements in the root cortex of wild-type and mutant plants can be used to investigate the cellular function of PIP2;2.
Table 2.
Size and Water Relation Parameters of Root Cortex Cells of Wild-Type and PIP2;2 Mutant Lines
| Measurement | Wild Type | pip2;2-1 | pip2;2-2 |
|---|---|---|---|
| Cell sizea | n = 37 | n = 34 | n = 42 |
| Length (μm) | 169 ± 7 | 164 ± 9 | 172 ± 10 |
| Diameter (μm) | 23.0 ± 0.9 | 23.7 ± 1.0 | 22.6 ± 0.7 |
| Water relation parametersa,b | n = 38 | n = 25 | n = 30 |
| Stationary turgor pressure (Pe; MPa) | 0.32 ± 0.01 | 0.33 ± 0.01 | 0.31 ± 0.01 |
| Volumetric elastic modulus (ɛ; MPa) | 2.23 ± 0.14 | 3.15 ± 0.31c | 2.34 ± 0.21 |
| Half-time of water exchange (t1/2; s) | 0.62 ± 0.03 | 0.88 ± 0.10d | 1.03 ± 0.12d |
| Hydraulic conductivity (Lpcell; 10−6 m·s−1·MPa−1) | 2.69 ± 0.18 | 1.74 ± 0.19e | 1.92 ± 0.23e |
Data (means ± se) were compiled from individual measurements (n of cells), obtained in three sets of independently grown plants, in which pip2;2-1 and pip2;2-2 mutants were characterized in parallel with wild-type plants.
Number of cells and plants tested: wild type, n = 38 cells from a total of 35 root segments from 13 plants; pip2;2-1, n = 25 cells from a total of 21 root segments from 7 plants; pip2;2-2, n = 30 cells from a total of 27 root segments from 9 plants.
Values in the pip2;2-1 mutant lines are statistically different (P < 0.01) from values in both the wild-type and pip2;2-2 lines. However, values in the wild-type and pip2;2-2 lines are not statistically different (P > 0.7).
Values in the pip2;2-1 and pip2;2-2 mutant lines are not statistically different (P > 0.2) but are all distinct from control wild-type values (pip2;2-1, P = 0.035; pip2;2-2, P = 0.003).
Values in the pip2;2-1 and pip2;2-2 mutant lines are not statistically different (P > 0.5) but are all distinct from control wild-type values (pip2;2-1, P = 0.004; pip2;2-2, P = 0.009).
A typical recording of turgor pressure in the root cortex cells of wild-type plants, exemplifying hydrostatic pressure relaxations and cell wall elasticity measurements, is shown in Figure 5A. A comprehensive characterization of water relation parameters in wild-type and mutant cells was made by measurements on three sets of independently grown plants. In all plants tested, the half-time of water exchange (t1/2) observed upon hydrostatic pressure relaxations was extremely short, in the range of 1 s (for wild-type; Figure 5A). Each set of measurements indicated the same tendency, with the two PIP2;2 mutants showing a t1/2 increased by 30 to 60% and a cell hydraulic conductivity (Lpcell) decreased by 25 to 30% with respect to wild-type values. Mean values of water relation parameters compiled from measurements in the three sets of independently grown plants (n = 7 to 13 plants; n = 25 to 38 cells) are shown in Table 2. These data establish that the two mutants pip2;2-1 and pip2;2-2 have significant alterations in both t1/2 (P < 0.05) and Lpcell (P < 0.01). We also noticed that, although all plant lines had a similar stationary cell turgor (Pe), the pip2;2-1 mutant, but not pip2;2-2, exhibited a slightly increased volumetric elastic modulus (ɛ). This finding might be attributable to the fact that the pip2;2-1 homozygous plants used in these experiments had not been backcrossed, whereas the pip2;2-2 plants were obtained after four successive backcrosses. Nevertheless, the pip2;2-1 and pip2;2-2 mutants showed very similar reductions in relative Lpcell, by 27 and 28%, respectively (Figure 5B). The consistent Lpcell alterations observed in the two allelic mutants indicate that PIP2;2 is responsible for at least 28% of water transport in root cortical cells.
Figure 5.
Cell Pressure Probe Measurements in the Cortex Cells of Arabidopsis Roots.
(A) Typical recording trace of turgor pressure in a wild-type root. The first stationary pressure measured after penetration of the pipette into the root tissue corresponds to the impalement of an epidermal cell. Upon displacement of the pipette deeper into the root (at ∼50 s), a higher stationary turgor pressure (Pe) was measured in an adjacent cortical cell. After stabilization of the cell pressure, a rapid increase or decrease in cell volume can be imposed using the pressure probe, resulting in exosmotic (outward) or endosmotic (inward) water movements, respectively. The associated pressure relaxations were used to deduce the t1/2. On the right side of the recording (>300 s), quick changes in pressure were imposed and reverted before any significant pressure-induced water flow had occurred. These maneuvers allow the determination of the cell volumetric elastic modulus (ɛ). The inset shows a detail of the results from a hydrostatic experiment showing two opposite pressure relaxations. Mean water relations parameters deduced from the present recording are as follows: Pe = 0.40 ± 0.06 MPa; t1/2 = 0.40 ± 0.02 s; ɛ = 1.66 ± 0.06 MPa; calculated Lpcell = 4.34 × 10−6 m·s−1·MPa−1.
(B) Relative Lpcell from wild-type (WT) and PIP2;2 mutants. The raw data obtained from the indicated number of individual cells and compiled from measurements on three sets of independently grown plants are shown in Table 2. Because of slight variations in absolute
Altered Root Sap Exudation in pip2;2 Mutants
To investigate the function of PIP2;2 in whole root water transport, root systems excised from wild-type and mutant plants grown in hydroponic conditions were inserted in a pressure chamber. The steady state rate of sap flow was recorded for pressures between 0.16 and 0.32 MPa. A linear relationship was observed that was used to calculate hydrostatic root water conductivity (Lpr-h). Values in the range of 10−7 m·s−1·MPa−1 (Table 3) are consistent with values reported in other plant species (Steudle, 1994). Using this technique, however, we failed to detect a statistically significant difference in Lpr-h between wild-type and mutant plants (Table 3).
Table 3.
Hydrostatic Hydraulic Conductivity of Roots of Wild-Type and PIP2;2 Mutant Lines
| Measurement | Wild Type (n = 34) |
pip2;2-1 (n = 30) |
pip2;2-2 (n = 28) |
|---|---|---|---|
| Lpr-h (10−8 m·s−1·MPa−1) | 9.02 ± 0.32 | 8.47 ± 0.28a | 8.42 ± 0.36a |
Lpr-h was measured in a pressurized chamber on four sets of independently grown plants. Values are given as means ± se determined from the indicated number of plants.
Mean Lpr-h values in PIP mutant lines were not statistically different (P > 0.05) from Lpr-h values in wild-type plants.
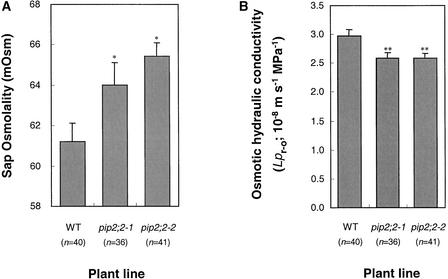
Hydrostatically induced water flow may favor apoplastic water transport (i.e., across cell walls), and the contribution of cell-to-cell water transport may be masked (Steudle, 1994; Steudle and Peterson, 1998; Javot and Maurel, 2002). Therefore, we reasoned that an entirely osmotic mode of tissue water transport may better reveal aquaporin dysfunction in roots of mutant plants. For this, plants grown in hydroponic conditions were decapitated, and both the rate of flow and the osmolality of spontaneously exuded sap were recorded over time. Sap exudation reached a close to steady state level within 60 min after stem excision, and the osmolality of sap collected in the subsequent 60 min was stable, in the range of 60 mosmol. In three independent experiments, we observed that sap osmolality in both pip2;2-1 and pip2;2-2 was increased significantly with respect to that in wild-type controls, by 5% (P = 0.027) and 7% (P = 0.003), respectively (Figure 6A). These data indicate that sap transport was driven by an increased osmotic driving force in mutant compared with wild-type roots. To determine whether this effect was caused by a primary defect of mutant roots in water transport capacity, osmotic water conductivity values (Lpr-o) were calculated from the ratio between the rate of sap flow and the osmotic driving force. Wild-type plants exhibited steady state Lpr-o values in the range of 2.5 to 3.0 × 10−8 m·s−1·MPa−1. A significant reduction (P < 0.005), by 14% with respect to the wild type, was observed in both the pip2;2-1 and pip2;2-2 mutants (Figure 6B). Together, these results establish the contribution of a single aquaporin, PIP2;2, to water transport and more specifically to water conductivity in the Arabidopsis root. They suggest a predominant function for PIP2;2 in root water uptake in conditions of reduced transpiration and, more generally, in osmotic fluid transport.
Figure 6.
Osmotic Water Transport in Roots of the Wild Type and PIP2;2 Mutants.
Data were pooled from measurements on three sets of independently grown plants, and the numbers of individual plants tested (n) are indicated in parentheses.
(A) Osmolality of sap exuded from roots of wild-type (WT), pip2;2-1, and pip2;2-2 plants. Asterisks indicate values that are statistically different from control wild-type values: pip2;2-1, P = 0.027; pip2;2-2, P = 0.003.
(B) Deduced osmotic hydraulic conductivity of roots (Lpr-o) of wild-type, pip2;2-1, and pip2;2-2 plants. Double asterisks indicate values that are statistically different from control wild-type values: pip2;2-1, P = 0.002; pip2;2-2, P = 0.006.
DISCUSSION
Genetics of Plant Aquaporins
Because of the numerous side effects and overall toxicity of mercury compounds (Zhang and Tyerman, 1999; Javot and Maurel, 2002), reverse genetics provides a more reliable approach than mercury treatment to explore the integrated function of aquaporins. Antisense inhibition of PIP1 aquaporin gene expression in Arabidopsis and tobacco has revealed the role of this group of aquaporins in water transport in isolated protoplasts and excised roots and in water stress attenuation in tobacco leaves (Kaldenhoff et al., 1998; Siefritz et al., 2002). However, as a result of the high isoform multiplicity of plant aquaporins, several factors, including the identity of all downregulated genes, their degrees of extinction, and the role of gene-specific expression patterns, remained largely unknown in these studies. Thus, the contribution of specific aquaporin genes to the phenotype observed in antisense plants could not be established.
In the present work, we investigated the function of an aquaporin of the PIP2 subgroup and report on aquaporin knockouts in plants. Two independently isolated Arabidopsis mutants were characterized, each with a T-DNA insertion that led to selective inactivation of PIP2;2. Because the two allelic mutants displayed similar alterations in their water transport properties, our observations indicate a causal relationship between these alterations and the inactivation of PIP2;2.
Function of PIP2;2 in the Arabidopsis Root
Expression analysis of a PIP2;2-GUS chimeric gene revealed the PIP2;2 promoter to be active predominantly in roots, with strong expression in the cortex, endodermis, and stele of elongated root segments. This information was used first to determine whether pip2;2 mutants displayed a primary defect in membrane water transport. Although technically challenging, cell pressure probe measurements in the root cortex were favored, assuming that an in situ approach might provide a more reliable estimate of aquaporin cell function than measurements on isolated protoplasts. Our results indicate for PIP2;2 a contribution of at least 25 to 30% to the hydraulic conductivity of root cortex cells. In tobacco, antisense suppression of aquaporins in the PIP1 subgroup resulted in a 55% decrease in the osmotic water permeability of root protoplasts (Siefritz et al., 2002). In comparison, our data reveal a significant role for a single PIP2 isoform in the Arabidopsis root.
Plant aquaporin mutants are particularly valuable materials if a link between altered cell functions and a phenotype at the tissue or organism level can be established. Altered osmotic transport properties in the roots of pip2;2 mutants were revealed from free exudation experiments. A decrease in Lpr-o of 14% with respect to the wild type was calculated. By contrast, the overall solute pumping activity of the root, as indicated by the product of sap flow rate and osmolality, was not altered significantly in the two mutants (data not shown). Together, these results indicate that in our experimental conditions, mutation of PIP2;2 did not alter the ability of roots to generate an osmotic driving force; rather, a genuine reduction in their water transport capacity was induced. Thus, the 5 to 7% increase in sap osmolality in pip2;2 mutants mechanically follows from a reduction in Lpr-o. These interpretations are justified by the fact that the root anatomy of wild-type and mutant plants was indistinguishable.
The phenotype observed in the roots of pip2;2 mutants is similar to the phenotypes observed in the salivary glands and airway submucosal glands of mice disrupted for AQP5 (Ma et al., 1999; Krane et al., 2001; Song and Verkman, 2001). In these two organs, knockout mice showed a reduced rate of fluid transport accompanied by increased fluid osmolality, suggesting that AQP5 was critical in favoring near isosmolar transport. A similar function for AQP1 in the renal proximal tubule also has been inferred from analyses in knockout mice (Schnermann et al., 1998).
Our results suggest that in plants, water channel function might be crucial for root water uptake in conditions of reduced transpiration, in which cell membranes are an obligatory path for osmotically driven water transport. More generally, high membrane water permeability might be necessary to maintain a net sap flow in quasi-isosmolar conditions. In certain cell types (e.g., root endodermis and xylem parenchyma) and under certain physiological conditions (onset of light period), high rates of osmotic transport are likely to be needed. A high membrane water permeability would avoid the requirement of a high driving force—that is, of a significant water (or osmotic) potential gradient. Such a gradient may result in turn in a diffusional back flow of solutes and therefore in a reduction in net solute pumping. We note that, although PIP2;2 was one of the abundantly expressed PIPs in roots, the decrease in Lpr-o shown by the pip2;2 mutants was of modest amplitude. Yet, in view of the extremely high isoform multiplicity of aquaporins in plants (Arabidopsis has 35 aquaporin homologs) (Chaumont et al., 2001; Johanson et al., 2001; Quigley et al., 2001), we believe these alterations to be highly significant. They support the idea that together the complement of aquaporins play a major role in water transport across tissues.
The finding that pip2;2 mutants showed an altered Lpr-o is in contrast to our failure to detect any significant reduction in Lpr-h. It has been suggested that hydrostatic measuring conditions may favor apoplastic water transport and thus a bypass of transcellular water movement along the cell wall continuum (Steudle and Peterson, 1998; Javot and Maurel, 2002). In support of this notion, we found that in our experimental conditions, the Lpr-h of wild-type plants was systematically higher than their Lpr-o. Thus, a modest contribution of aquaporins, and of PIP2;2 in particular, to overall Lpr-h may have been masked. On the other hand, recent data from our group indicate that both mercury treatment and stress imposition induce a significant (>50%) inhibition in Lpr-h, suggesting that even under hydrostatic measuring conditions, water channel–mediated transport still may be important in the Arabidopsis root (data not shown). A significant (−55%) reduction in Lpr-h has been found in PIP1 antisense tobacco plants, whereas the effects on root cell water transport were only twofold greater than those found in pip2;2 mutants. Thus, PIP isoforms may participate differentially in the distinct modes of root water uptake. A functional distinction between aquaporins of the PIP1 and PIP2 subgroups has not been established in plants, but the latter show more efficient functional expression in Xenopus oocytes (Kammerloher et al., 1994; Chaumont et al., 2000). PIP2;2 showed the strongest expression in the endodermis and in the few outer cell layers of the stele (Figure 4C). These cells may contribute critically to osmotically driven water uptake, which in turn supplies xylem sap flow. Our presumption is that knockouts of PIP aquaporins that belong to a subgroup distinct from PIP2;2 and/or that show a significantly different expression pattern in the root might show another profile of alterations in osmotic and hydrostatic water transport in this organ.
Reverse Genetics Analyses of Tandem Duplicated Genes
There have been concerns that the presence in plants of large gene families, and of duplicated gene clusters in particular, might prevent analyses by gene inactivation (Krysan et al., 1999; Bouché and Bouchez, 2001). The PIP subgroup comprises 13 members in Arabidopsis, and PIP2;2 has a close homolog, PIP2;3, that shares the same promoter region along with >96% amino acid identity. Despite this similarity, the phenotypic alterations observed at the cell and tissue levels in pip2;2 mutants demonstrate directly that PIP2;2 and PIP2;3 have evolved with functions that do not overlap fully. This idea differs from that raised regarding some flower development genes of the MADS box family, for which genetic combinations of individual knockouts for two to three homologs were needed to reveal a phenotype (Liljegren et al., 2000; Pelaz et al., 2000).
Thus, our studies establish the adaptive value of a single aquaporin gene that allows plants to optimize both water and solute transport in roots. Although the PIP2;2 mutation did not cause any macroscopic alterations in growth and development in our experimental conditions, slight differences might exist and result in an altered fitness of mutant plants. We realize, however, that the PIP2;2 and PIP2;3 aquaporins share such a high structural similarity that close functional properties can be expected and may prevent the detection of a more pronounced phenotype in pip2;2 mutants. Nevertheless, it appears that the functional divergence between PIP2;2 and PIP2;3 relies in part on distinct expression properties in roots. This was first suggested by matrix-assisted laser desorption/ionization time-of-flight analyses (this work) and is supported further by recent macroarray analyses of aquaporin gene expression, which showed that PIP2;2 had a higher mRNA expression level than PIP2;3 in roots (S. Chen and C. Maurel, unpublished data). These analyses also indicated that the disruption of PIP2;2 was not compensated for by enhanced expression of either of the other PIP homologs.
In conclusion, the present work provides experimental evidence for the contribution of a single aquaporin gene to water transport in planta. Our study also demonstrates that, despite their small size, Arabidopsis plants are amenable to in-depth biophysical analyses. Thus, Arabidopsis is a fully relevant model for the reverse genetics of plant aquaporins. In the future, novel integrated functions should be revealed by the analyses of knockouts for aquaporin isoforms that are expressed in organs other than roots.
METHODS
Plant Material and Culture Conditions
All genetic and physiological experiments were performed using Arabidopsis thaliana ecotype Wassilewskija. Expression analyses of a PIP2;2–β-glucuronidase (GUS) construct were performed with transgenic Arabidopsis ecotype C24. Seeds were surface-sterilized, kept for 2 to 3 d at 4°C, and grown in clear polystyrene culture boxes (12 × 12 cm) on standard culture medium [5 mM KNO3, 2 mM MgSO4, 1 mM Ca(NO3)2, 50 μM FeEDTA, microelements according to Murashige and Skoog (1962), 2.5 mM K2HPO4 + KH2PO4, 1 mM Mes, 10 g/L Suc, and 7 g/L agar, pH 6.1, adjusted with KOH]. The boxes were incubated vertically at 25°C in the light for 10 days before seedlings were transplanted into hydroponic culture. Plants were mounted on a 35- × 35- × 0.6-cm polystyrene raft floating on a basin filled with 8 L of culture medium [1.25 mM KNO3, 0.75 mM MgSO4, 1.5 mM Ca(NO3)2, 0.5 mM KH2PO4, 50 μM FeEDTA, 50 μM H3BO3, 12 μM MnSO4, 0.70 μM CuSO4, 1 μM ZnSO4, 0.24 μM MoO4Na2, and 100 μM Na2SiO3]. The culture medium was replaced each week. For GUS assay, pressure chamber, and pressure probe experiments, the culture room was maintained at 28°C at a RH of 70% with a 16-h-light (100 μE·m−2·s−1)/8-h-dark cycle. For exudation experiments, the culture room was maintained at 21°C at a RH of 70% with a 16-h-light (180 μE·m−2·s−1)/8-h-dark cycle.
Mass Spectrometry Analysis of Plasma Membrane Proteins
Plasma membrane vesicles were purified from roots of 6-week-old plants by aqueous two-phase partitioning, essentially as described by Gerbeau et al. (2002), in a mixture of PEG 3350 and Dextran T-500, 6.4% (w/w) each, in the presence of 5 mM KCl. Extrinsic membrane proteins were stripped using a procedure adapted from that of Fotiadis et al. (2001). Briefly, membranes were incubated in 5 mM EDTA, 5 mM EGTA, 4 M urea, and 5 mM Tris-HCl, pH 9.5, for 5 min on ice. After centrifugation for 20 min at 100,000g, the membranes were washed successively in 20 mM NaOH and in 2 mM EDTA, 2 mM EGTA, 100 mM NaCl, and 5 mM Tris-HCl, pH 8. Finally, membranes were resuspended in 9 mM KCl, 300 mM Suc, 5 mM Na2EDTA, 5 mM Na2EGTA, 50 mM NaF, 5 mM DTT, 2 μg/mL leupeptin, and 10 mM Tris-borate, pH 8.3, and stored at −80°C before analysis.
Electrophoretically separated proteins were excised from SDS-PAGE gels, washed, and analyzed as described previously (Santoni et al., 2000b). Routinely, the gel pieces were reswollen with 0.25 to 0.5 μg of sequence-grade modified porcine trypsin (Promega, Madison, WI) at 37°C for 16 h. The supernatant of the tryptic digest was collected, and the remaining peptides were extracted twice in 0.1% trifluoroacetic acid dissolved in 60% acetonitrile. Supernatants were pooled, and the final volume was reduced to 5 μL under vacuum. Protein digests were desalted with ZipTip μC18 (Millipore, Bedford, MA) before mass spectrometry analysis. The matrix-assisted laser desorption/ionization mass spectrometry spectra were acquired with a Biflex (Bruker, Billerica, MA) matrix-assisted laser desorption/ionization time-of-flight apparatus according to Santoni et al. (2000b). Analysis of the digest by tandem mass spectrometry was performed on a nano-electrospray ionization quadrupole time-of-flight hybrid mass spectrometer (Micromass, Manchester, UK). The sample was loaded into nanospray metallized glass capillaries (MDS Protana, Odense, Denmark). The capillary voltage was set at 1000 V, the cone voltage was set at 45 V, and collision energies were on the order of 10 to 40 eV. The instrument was calibrated with Glu-Fibrinopeptide fragmentation products (Sigma, Saint Louis, MO). Argon was used as the collision gas. Instrument operation, data acquisition, and analysis were performed using MassLynx/Biolynx software (Micromass).
Reverse Genetics Screening
A collection of 35,328 Arabidopsis lines independently transformed by Agrobacterium tumefaciens strain C58(pMP90) containing the binary vector pGKB5 (Bechtold et al., 1993; Bouchez et al., 1993) has been prepared at the Institut National de la Recherche Agronomique (Versailles, France). Part of this collection is available at the Nottingham Arabidopsis Stock Centre or through the flanking sequence tag database FLAGdb (http://flagdb-genoplante-info.infobiogen.fr/projects/fst/). The collection was screened using a PCR-based strategy similar to that described by Geelen et al. (2000). Primers specific for right border (tag3) and left border (tag5) T-DNA sequences were as follows: tag3, 5′-CTGATACCAGACGTTGCCCGCATAA-3′; tag5, 5′-CTGCAAATTGCCTTTTCTTATCGAC-3′.
The CRZ9 transformed line was identified in a first screening using the degenerate primer mip1f (corresponding to the first of the two Asn-Pro-Ala motifs conserved in aquaporins) in combination with tag3. The EGS486 line was identified in a second screening using either a forward (2f) or a reverse (2r) primer specific for PIP2 genes combined with tag3 or tag5, respectively. For subsequent genetic characterization and cloning of the plant DNA/T-DNA fusion sequences, we used primers specific for PIP2;2 (22r) or PIP2;3 (23f and 23r).
The sequences of the aquaporin-specific primers were as follows: mip1f, 5′-GGIGGICA(C/T)(A/G)TIAA(C/T)CCIGCIGTIACITT(C/T)GG-3′; 2f, 5′-GCCGAGTTCGTAGCCACTCTCCTCTTC-3′; 2r, 5′-GCAGCT-ATCGCAGCTCCAATGAATGGTC-3′; 22r, 5′-CCGAGTACACAAACA-TTGGCATTGG-3′; 23f, 5′-CTATTTGTGGAGTTGGGTTTGTGAAG-3′; and 23r, 5′-CACCAAACTTACATACGTTG-3′.
For all analyses, PCR was performed in 25-μL reactions containing 75 to 125 ng of genomic DNA, 50 pmol of each primer, 1.5 mM MgCl2, 0.2 mM deoxynucleotide triphosphates, and 0.75 units of Taq polymerase (EuroBio, Les Ulis, France) in a full-strength reaction buffer (EuroBio). The following PCR program was used: 94°C for 3 min; 25 to 35 cycles of 94°C for 45 s, 55 to 57°C for 45 s, and 72°C for 60 s; and 72°C for 7 min. PCR products were separated on 1 to 2% agarose gels, transferred onto nylon membranes, and hybridized with aquaporin or T-DNA probes as described by Geelen et al. (2000).
Genetic Analysis
PCR-based segregation analysis of T-DNA insertions in the progeny of the EGS486 and CRZ9 transformants yielded isolates that showed segregation of kanamycin resistance according to a 3:1 ratio (n = 100 to 150 plants) and with a T-DNA insertion in all kanamycin-resistant plants tested (n = 10). These isolates were backcrossed to wild-type plants, up to two and four times, for EGS486 and CRZ9, respectively, and used to generate plants homozygous for a single T-DNA insertion and named pip2;2-1 and pip2;2-2, respectively. At each step of the backcross procedure, we determined by PCR that plants whose progeny was 100% kanamycin resistant carried the expected T-DNA insertion and lacked the wild-type allele. Cell pressure probe measurements were made on pip2;2-1 and pip2;2-2 mutants obtained without or after four backcrosses, respectively. For root exudation experiments, the pip2;2-1 and pip2;2-2 mutants had undergone two and four backcrosses, respectively.
DNA and RNA Extraction and RNA Expression Analysis
For genomic DNA extraction, leaf tissues (0.1 to 0.2 mg fresh weight) were ground with a mortar and incubated for 30 min at 60°C in extraction buffer (1.4 M NaCl, 20 mM EDTA, 2% cetyltrimethylammonium bromide, and 100 mM Tris-HCl, pH 8.0). After chloroform extraction, DNA was precipitated with isopropanol, washed with 70% ethanol, and treated with 0.1 mg/mL RNase A for 10 min at 37°C. DNA then was ethanol precipitated, rinsed with 70% ethanol, and finally resuspended in distilled water. Total mRNA was extracted from roots according to the procedure of Höfte et al. (1993). For RNA expression studies, 3 μg of total RNA was used for first-strand synthesis by reverse transcriptase from Moloney murine leukemia virus (Promega) according to the instructions of the supplier. A 1/25th fraction of this reaction was used as a matrix for PCR amplification in the conditions mentioned above. Expression of the Elongation Factor1α gene (Axelos et al., 1989) was studied as a control.
GUS Reporter Assay Construction and Histochemical Analyses
A translational fusion between the 3700-bp DNA sequence upstream of the PIP2;2 coding sequence and the GUS coding sequence was made by introducing an NcoI restriction site at the PIP2;2 start codon and using a version of GUS that also bears an NcoI site at its start codon followed by nopaline synthase poly(A) signal sequences. This construct was transferred into the vector backbone pGPTV-BAR (Becker et al., 1992) and used for Arabidopsis transformation by the floral-dip method (Clough and Bent, 1998).
Histochemical staining for GUS enzyme activity was performed on 3-day-old in vitro plantlets or on roots of 29-day-old plants grown in hydroponic conditions. Plant materials were transferred into prefixation buffer (1.5% formaldehyde and 100 mM phosphate buffer, pH 7.0) for 15 min at room temperature under vacuum and rinsed three times in 100 mM phosphate buffer, pH 7.0. Staining was allowed to proceed by incubating the tissues in revelation buffer (5 mM EDTA, 0.5 mM K-ferricyanure, 0.5 mM K-ferrocyanure, 1 mM 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid [Euromedex, Mundolsheim, France], and 100 mM phosphate buffer, pH 7.0) in the dark for 5 min at room temperature under vacuum and at 37°C for 90 min. Finally, samples were fixed for 15 min at room temperature under vacuum and then at 4°C for 24 h in fixation buffer A (3.7% formaldehyde, 60% ethanol, and 5% acetic acid) or B (2% paraformaldehyde, 1% glutaraldehyde, 1% caffeine, 0.05% SDS, and 100 mM phosphate buffer, pH 7.0) for plantlets and root systems, respectively. Tissues were dehydrated and cleared of chlorophyll by incubation in increasing (70 to 100%) ethanol concentrations. For additional cross-sections, roots were embedded in Technovit 7100 resin (Kulzer, Wehrheim, Germany) according to the supplier's instructions. A microtome UltracutE (Reichert, Heidelberg, Germany) was used to generate 4-μm transverse sections at a distance of 3 to 4 mm from the root apex. Sections were submitted to a standard Schiff reaction before observation. Plantlets and root cross-sections were observed by microscopy and photographed using Kodak Elite Chrome 100 film.
Cell Pressure Probe Measurements
Cell pressure probe measurements were performed essentially as described previously (Gerbeau et al., 2002). Pulled micropipettes were beveled to a tip diameter of 4 to 5.5 μm, filled with mineral oil, and mounted vertically on a pressure probe. Root tip segments with a length of 30 to 70 mm were excised from plants grown in hydroponic conditions, laid horizontally on filter paper, and partially submerged with a gentle perfusion of bathing solution containing 10 mM KCl and 5 mM Mes-Tris, pH 6.0. Pressure probe measurements were performed at a distance of 2.0 to 4.0 mm from the root apex, with no significant effects of cell position along the root axis on water relation parameters. Upon progressive impalement of the root tissues with the oil-filled micropipette, a typical sequence of two pressure increases was observed. A first unstable measurement at ∼0.2 MPa probably corresponds to the pipette being inserted in the epidermis. Further progression of the pipette into the root tissue (to a depth of 15 to 25 μm) resulted in a sudden increase to a more stable value (pressure of ∼0.3 MPa) in the second cell layer (i.e., cortex cells). The measurement of cell hydraulic parameters (half-time of water exchange [t1/2], cell volumetric elastic modulus [ɛ], and stationary turgor pressure [Pe]) was performed as described. Data were recorded and analyzed with a personal computer using an AD converter device (Sensor-interface 92101; Burster, Gernsbacht, Germany) and specially designed software (Pfloek; Department of Plant Ecology, University of Bayreuth, Germany). This setup allowed us to acquire data at a rate of 50 values per second at a normal resolution of 2 × 10−4 MPa. For each cell, pressure probe measurements were completed in <7 min within the 40 min after root excision. Cell hydraulic conductivity (Lpcell) was calculated according to the following equation:
where t1/2 and ɛ are as defined above and V and A are the cell volume and surface, respectively. Πi is the intracellular osmotic pressure estimated from Pe and the external osmotic pressure (Πe):
Assuming that cortex cells are cylinders with a mean radius (R) and length (L), Equation 2 yields
L and R were measured independently on the same set of plants, using an inverted microscope, at a distance of 2.0 to 4.0 mm from the apex. No significant effects of cell position along the root axis were observed. For ɛ and Lpcell determination in wild-type and mutant plants, similar values of R = 11.5 μm and L = 168 μm were used.
Measurement of Root Hydrostatic Water Conductivity in a Pressurized Chamber
The root system of a freshly detopped Arabidopsis plant was inserted into the container of a pressure chamber filled with hydroponic culture medium. The hypocotyl was threaded carefully through the soft plastic washer of the metal lid. Tight sealing was performed using a low-viscosity dental paste (President Light; Coltene, Altstätten, Switzerland). Pressure (P) was applied to the chamber using a cylinder of nitrogen gas. The rate of sap flow (Jv) was determined by collecting the expressed sap into a graduated glass micropipette. A linear relationship between Jv and P was observed for P values up to 400 kPa. In routine measurements, excised roots were subjected to a pretreatment at 320 kPa for 15 min to attain equilibration, and Jv was measured successively at 160 and 310 kPa for ∼5 min. After flow measurements, root dry weight was measured. An independent set of calibration experiments was necessary to determine a relationship between root dry weight and overall root surface area (Ar) estimated from measurements of the length and number of fine, medium, and coarse roots in the root system. For these experiments, the root system was spread on a grid and the numbers of roots in each of three classes of diameter (mean values of 160, 109, and 77 μm) were estimated at intervals of 1 cm. From this calculation, we derived an estimated root length for each class, which was used to integrate Ar. A linear regression was used to establish a relationship between root dry weight (mg) and Ar (m2) for each growth condition (at 28°C, Ar = 5.15 × 10−4 root dry weight + 9.76 × 10−6; at 21°C, Ar = 4 × 10−4 root dry weight + 8.10−5). A similar relationship was observed for the different genotypes examined. The apparent hydrostatic water conductivity of an individual root system (Lpr-h; m·s−1·MPa−1) was calculated from the following equation:
Sap Exudation and Measurements of Osmotic Root Water Conductivity
Plants grown in hydroponic conditions were decapitated, and the entire root systems were maintained in the original culture. Root systems were incubated for 60 min in culture medium supplemented with 10 mM Glc. This procedure allowed spontaneous sap exudation to reach a steady state level and to equilibrate solute concentrations in xylem vessels. Exuded sap then was collected into a graduated glass micropipette, and flow rate (Jv) was measured over the next 60 min. The osmolalities of bath medium (Πb) and sap exuded from individual plants (Πs) were measured by freezing-point depression osmometry. The apparent osmotic root water conductivity (Lpr-o) was deduced from the following equation:
where Ar is the overall root surface area as described above.
Statistics
The effects of the PIP2;2 mutation on each of the water relation parameters investigated were tested in at least three sets of measurements made on independently grown plants, with each mutant line being compared at least twice with the wild type (n = 3 to 20 plants of each genotype in each individual experiment). A putative difference between wild-type and mutant plants was considered if the same tendency was observed in each independent set of measurements. Data pooled from all sets of measurements then were analyzed using analysis of variance to test for the effects of plant genotype on the water relation parameter investigated. When a significant effect was detected (P < 0.01), post hoc comparisons using Statistica software (StatSoft, Tulsa, OK) with a Newman-Keuls test were run on the same set of data to identify which genotypes were statistically different (P < 0.05). All data are presented as means ± se, and the statistical significance (P value) is reported in the legends of the figures and tables.
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes.
Accession Number
The accession number for PIP2;2 is AC006260.
Acknowledgments
We thank Susanna Malmström for the gift of primers, Mathias Knauer for kindly providing the Pfloek software, Sheng Chen for helping with RNA extraction, Tobias Henzler for discussions and critical reading of the manuscript, and Claude Grignon for help with the statistical analyses. This work was supported in part by the Centre National de la Recherche Scientifique (Action Thématique Incitative sur Programme et Equipe “Function and Regulation of Plant Aquaporins”), the Commission of the European Union (Contract BIO4-CT98-0024), and the Deutsche Forschungsgemeinschaft (Scha 454/5-2).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.008888.
References
- Agre, P. (1998). Aquaporin null phenotypes: The importance of classical physiology. Proc. Natl. Acad. Sci. USA 95, 9061–9063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agre, P., Bonhivers, M., and Borgnia, M.J. (1998). The aquaporins, blueprints for cellular plumbing systems. J. Biol. Chem. 273, 14659–14662. [DOI] [PubMed] [Google Scholar]
- Axelos, M., Bardet, C., Liboz, T., Le Van Thai, A., Curie, C., and Lescure, B. (1989). The gene family encoding the Arabidopsis thaliana translation elongation factor EF-1α: Molecular cloning, characterization and expression. Mol. Gen. Genet. 219, 106–112. [DOI] [PubMed] [Google Scholar]
- Barrieu, F., Morillon, R., and Chrispeels, M. (2000). Modulation of aquaporin gene expression in Arabidopsis leads to altered membrane water permeability. In Molecular Biology and Physiology of Water and Solute Transport, S. Hohmann and S. Nielsen, eds (New York: Kluwer Academic/Plenum Publishers), pp. 255–259.
- Bechtold, N., Ellis, J., and Pelletier, G. (1993). In planta Agrobacterium-mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C. R. Acad. Sci. Paris 316, 1194–1199. [Google Scholar]
- Becker, D., Kemper, E., Schell, J., and Masterson, R. (1992). New plant binary vectors with selectable markers located proximal to the left T-DNA border. Plant Mol. Biol. 20, 1195–1197. [DOI] [PubMed] [Google Scholar]
- Bouché, N., and Bouchez, D. (2001). Arabidopsis gene knockout: Phenotypes wanted. Curr. Opin. Plant. Biol. 4, 111–117. [DOI] [PubMed] [Google Scholar]
- Bouchez, D., Camilleri, C., and Caboche, M. (1993). A binary vector based on Basta resistance for in planta transformation of Arabidopsis thaliana. C. R. Acad. Sci. Paris 316, 1188–1193. [Google Scholar]
- Chaumont, F., Barrieu, F., Jung, R., and Chrispeels, M.J. (2000). Plasma membrane intrinsic proteins from maize cluster in two sequence subgroups with differential aquaporin activity. Plant Physiol. 122, 1025–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaumont, F., Barrieu, F., Wojcik, E., Chrispeels, M.J., and Jung, R. (2001). Aquaporins constitute a large and highly divergent protein family in maize. Plant Physiol. 125, 1206–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Deen, P.M.T., Verdijk, M.A.J., Knoers, N.V.A.M., Wieringa, B., Monnens, L.A.H., van Os, C.H., and van Oost, B.A. (1994). Requirement of human renal water channel aquaporin-2 for vasopressin-dependent concentration of urine. Science 264, 92–95. [DOI] [PubMed] [Google Scholar]
- Fotiadis, D., Jenö, P., Mini, T., Wirtz, S., Müller, S.A., Fraysse, L., Kjellbom, P., and Engel, A. (2001). Structural characterization of two aquaporins isolated from native spinach leaf plasma membranes. J. Biol. Chem. 276, 1707–1714. [DOI] [PubMed] [Google Scholar]
- Geelen, D., Lurin, C., Bouchez, D., Frachisse, J.-M., Lelièvre, F., Courtial, B., Barbier-Brygoo, H., and Maurel, C. (2000). Disruption of putative anion channel gene AtCLC-a in Arabidopsis suggests a role in the regulation of nitrate content. Plant J. 21, 259–267. [DOI] [PubMed] [Google Scholar]
- Gerbeau, P., Amodeo, G., Henzler, T., Santoni, V., Ripoche, P., and Maurel, C. (2002). The water permeability of Arabidopsis plasma membrane is regulated by divalent cations and pH. Plant J. 30, 71–81. [DOI] [PubMed] [Google Scholar]
- Höfte, H., et al. (1993). An inventory of 1152 expressed sequence tags obtained by partial sequencing of cDNAs from Arabidopsis thaliana. Plant J. 4, 1051–1061. [DOI] [PubMed] [Google Scholar]
- Ikeda, S., Nasrallah, J.B., Dixit, R., Preiss, S., and Nasrallah, M.E. (1997). An aquaporin-like gene required for the Brassica self-incompatibility response. Science 276, 1564–1566. [DOI] [PubMed] [Google Scholar]
- Javot, H., and Maurel, C. (2002). The role of aquaporins in root water uptake. Ann. Bot. 90, 301–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johanson, U., Karlsson, M., Gustavsson, S., Sjovall, S., Fraysse, L., Weig, A.R., and Kjellbom, P. (2001). The complete set of genes encoding major intrinsic proteins in Arabidopsis provides a framework for a new nomenclature for major intrinsic proteins in plants. Plant Physiol. 126, 1358–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson, I., Karlsson, M., Johanson, U., Larsson, C., and Kjellbom, P. (2000). The role of aquaporins in cellular and whole plant water balance. Biochim. Biophys. Acta 1465, 324–342. [DOI] [PubMed] [Google Scholar]
- Kaldenhoff, R., Grote, K., Zhu, J.-J., and Zimmermann, U. (1998). Significance of plasmalemma aquaporins for water-transport in Arabidopsis thaliana. Plant J. 14, 121–128. [DOI] [PubMed] [Google Scholar]
- Kammerloher, W., Fischer, U., Piechottka, G.P., and Schäffner, A.R. (1994). Water channels in the plant plasma membrane cloned by immunoselection from a mammalian expression system. Plant J. 6, 187–199. [DOI] [PubMed] [Google Scholar]
- Krane, C.M., Melvin, J.E., Nguyen, H.-V., Richardson, L., Towne, J.E., Doetschman, T., and Menon, A.G. (2001). Salivary acinar cells from Aquaporin 5-deficient mice have decreased membrane water permeability and altered cell volume regulation. J. Biol. Chem. 276, 23413–23420. [DOI] [PubMed] [Google Scholar]
- Krysan, P.J., Young, J.C., and Sussman, M.R. (1999). T-DNA as an insertional mutagen in Arabidopsis. Plant Cell 11, 2283–2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liljegren, S.J., Ditta, G.S., Eshed, Y., Savidge, B., Bowman, J.L., and Yanofsky, M.F. (2000). SHATTERPROOF MADS-box genes control seed dispersal in Arabidopsis. Nature 404, 766–770. [DOI] [PubMed] [Google Scholar]
- Ma, T., Song, Y., Gillespie, A., Carlson, E.J., Epstein, C.J., and Verkman, A.S. (1999). Defective secretion of saliva in transgenic mice lacking aquaporin-5 water channels. J. Biol. Chem. 274, 20071–20074. [DOI] [PubMed] [Google Scholar]
- Ma, T., Song, Y., Yang, B., Gillespie, A., Carlson, E.J., Epstein, C.J., and Verkman, A.S. (2000). Nephrogenic diabetes insipidus in mice lacking aquaporin-3 water channels. Proc. Natl. Acad. Sci. USA 97, 4386–4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manley, G.T., Fujimura, M., Ma, T., Noshita, N., Filiz, F., Bollen, A.W., and Verkman, A.S. (2000). Aquaporin-4 deletion in mice reduces brain edema after acute water intoxication and ischemic stroke. Nat. Med. 6, 159–163. [DOI] [PubMed] [Google Scholar]
- Maurel, C., Javot, H., Lauvergeat, V., Gerbeau, P., Tournaire, C., Santoni, V., and Heyes, J. (2002). Molecular physiology of aquaporins in plants. Int. Rev. Cytol. 215, 105–148. [DOI] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 15, 473–497. [Google Scholar]
- Nejsum, L.N., Kwon, T.-H., Jensen, U.B., Fumagalli, O., Frokiaer, J., Krane, C.M., Menon, A.G., King, L.S., Agre, P.C., and Nielsen, S. (2002). Functional requirement of aquaporin-5 in plasma membranes of sweat glands. Proc. Natl. Acad. Sci. USA 99, 511–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelaz, S., Ditta, G.S., Baumann, E., Wisman, E., and Yanofsky, M.F. (2000). B and C floral organ identity functions require SEP-ALLATA MADS-box genes. Nature 405, 200–203. [DOI] [PubMed] [Google Scholar]
- Preston, G.M., Smith, B.L., Zeidel, M.L., Moulds, J.J., and Agre, P. (1994). Mutations in aquaporin-1 in phenotypically normal humans without functional CHIP water channels. Science 265, 1585–1587. [DOI] [PubMed] [Google Scholar]
- Quigley, F., Rosenberg, J.M., Shachar-Hill, Y., and Bohnert, H.J. (2001). From genome to function: The Arabidopsis aquaporins. Genome Biol. 3, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoni, V., Gerbeau, P., Javot, H., and Maurel, C. (2000. a). The high diversity of aquaporins reveals novel facets of plant membrane functions. Curr. Opin. Plant. Biol. 3, 476–481. [DOI] [PubMed] [Google Scholar]
- Santoni, V., Kieffer, S., Masson, F., Desclaux, D., and Rabilloud, T. (2000. b). Membrane proteomics: Use of additive main effects with multiplicative interaction model to classify plasma membrane proteins according to their solubility and electrophoretic properties. Electrophoresis 21, 3329–3344. [DOI] [PubMed] [Google Scholar]
- Schnermann, J., Chou, C.-L., Ma, T., Traynor, T., Knepper, M.A., and Verkman, A.S. (1998). Defective proximal tubular fluid reabsorption in transgenic aquaporin-1 null mice. Proc. Natl. Acad. Sci. USA 95, 9660–9664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siefritz, F., Tyree, M.T., Lovisolo, C., Schubert, A., and Kaldenhoff, R. (2002). PIP1 plasma membrane aquaporins in tobacco: From cellular effects to function in plants. Plant Cell 14, 869–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, Y., and Verkman, A.S. (2001). Aquaporin-5 dependent fluid secretion in airway submucosal glands. J. Biol. Chem. 276, 41288–41292. [DOI] [PubMed] [Google Scholar]
- Steudle, E. (1994). The regulation of plant water at the cell, tissue, and organ level: Role of active processes and of compartmentation. In Flux Control in Biological Systems: From Enzymes to Populations and Ecosystems, E.D. Schultze, ed (San Diego, CA; Academic Press), pp. 237–299.
- Steudle, E., and Peterson, C.A. (1998). How does water get through roots? J. Exp. Bot. 49, 775–788. [Google Scholar]
- Tyerman, S.D., and Niemietz, C.M. (2002). Plant aquaporins: Multifunctional water and solute channels with expanding roles. Plant Cell Environ. 25, 173–194. [DOI] [PubMed] [Google Scholar]
- Zhang, W.-H., and Tyerman, S.D. (1999). Inhibition of water channels by HgCl2 in intact wheat root cells. Plant Physiol. 120, 849–857. [DOI] [PMC free article] [PubMed] [Google Scholar]