Abstract
The mur4 mutant of Arabidopsis shows a 50% reduction in the monosaccharide l-Ara in leaf-derived cell wall material because of a partial defect in the 4-epimerization of UDP-d-Xyl to UDP-l-Ara. To determine the genetic lesion underlying the mur4 phenotype, the MUR4 gene was cloned by a map-based procedure and found to encode a type-II membrane protein with sequence similarity to UDP-d-Glc 4-epimerases. Enzyme assays of MUR4 protein expressed in the methylotropic yeast Pichia pastoris indicate that it catalyzes the 4-epimerization of UDP-d-Xyl to UDP-l-Ara, the nucleotide sugar used by glycosyltransferases in the arabinosylation of cell wall polysaccharides and wall-resident proteoglycans. Expression of MUR4–green fluorescent protein constructs in Arabidopsis revealed localization patterns consistent with targeting to the Golgi, suggesting that the MUR4 protein colocalizes with glycosyltransferases in the biosynthesis of arabinosylated cell wall components. The Arabidopsis genome encodes three putative proteins with >76% sequence identity to MUR4, which may explain why mur4 plants are not entirely deficient in the de novo synthesis of UDP-l-Ara.
INTRODUCTION
In plants, l-Ara is an important constituent of cell wall poly-saccharides, such as the pectic components rhamnoga-lacturonan I and II and the cellulose binding glucuronoara-binoxylans. Furthermore, l-Ara is a major component of Hyp-rich glycoproteins (HRGPs) and arabinogalactan proteins (AGPs). Rhamnogalacturonan I and the AGPs share common carbohydrate side chains consisting of a d-galactan framework substituted with terminal l-Ara (Carpita and Gibeaut, 1993). Whereas the Ara-containing pectic material plays a structural role in determining wall porosity and borate-mediated cross-linking (Baron-Epel et al., 1988; Fleischer et al., 1999; O'Neill et al., 2001), AGPs have been assigned various roles in plant development, including embryogenesis, cell–cell interactions, fertilization, cell proliferation, and cell expansion (for review, see Du et al., 1996; Gaspar et al., 2001). HRGPs contain numerous Ara side chains, which are linked to Hyp residues and stabilize the polyproline II conformation of the polypeptide backbone (Stafstrom and Staehelin, 1986). These glycoproteins are believed to play a structural role in strengthening the cell wall and often are expressed in response to pathogen attack (Showalter, 1993). An HRGP mutant of Arabidopsis was shown recently to display severe developmental abnormalities, such as incorrect positioning of the cell plate and distorted cell shapes, establishing a crucial role for HRGPs in plant growth and development (Hall and Cannon, 2002).
l-Ara is found much less frequently in other organisms, for example, in the lipopolysaccharide of the actinomycete Ampullariella digitata (Fan and Feingold, 1972) and in the cell wall of the purple sulfur bacterium Chromatium vinosum (Hurlbert et al., 1976). It also has been reported in hyaluronate peptides of brain tissue (Wardi et al., 1966; Varma et al., 1977), although this finding has been disputed (Katzman, 1974).
UDP-l-Ara, the activated sugar used by arabinosyltransferases, is known to be synthesized de novo from UDP-d-Glc via UDP-d-glucuronate and the 4-epimerization of UDP-d-Xyl (Figure 1). However, the enzyme catalyzing the final step in the de novo synthesis of UDP-l-Ara has not been cloned from any organism. A mutant of Arabidopsis (mur4) showing an ∼50% reduction in the l-Ara content of leaves was described recently and shown to be partially defective in the last step of the de novo synthesis of l-Ara (Burget and Reiter, 1999). We have now cloned the MUR4 gene and demonstrate that it encodes a Golgi-localized UDP-d-Xyl 4-epimerase (EC 5.1.3.5), which most likely is involved in the production of UDP-l-Ara at the site of polysaccharide synthesis.
Figure 1.
De Novo Pathway for the Synthesis of UDP-l-Ara and Other UDP Sugars from UDP-d-Glc via NAD(P)+-Dependent Dehydrogenation, Decarboxylation, and 4-Epimerization Reactions.
RESULTS
Identification of a Candidate Gene for MUR4
Because UDP-d-Xyl 4-epimerase and UDP-d-Glc 4-epimerase (EC 5.1.3.2) catalyze mechanistically identical reactions with structurally similar substrates, we hypothesized that UDP-d-Xyl 4-epimerases would show recognizable sequence similarities to UDP-d-Glc 4-epimerases, for which genes have been isolated from many organisms, including Arabidopsis (Dörmann and Benning, 1996). Although it was not clear whether the MUR4 gene encoded UDP-d-Xyl 4-epimerase or some factor regulating this enzymatic activity, the hypothesis described above opened the possibility to clone MUR4 by a combined positional and candidate gene approach.
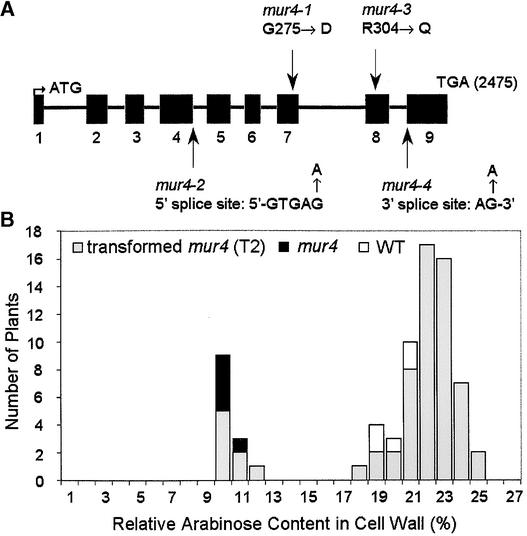
Using an F2 population established from a cross between mur4 plants in the Columbia background and Landsberg erecta plants, the mur4 mutation was mapped to chromosome 1 at ∼1–2 centimorgans above the UFO gene (one recombinant between mur4 and UFO within 60 plants of the mapping population). An evaluation of the Arabidopsis genome sequence within this region (Arabidopsis Genome Initiative, 2000) revealed that gene At1g30620 located ∼180 kb above the UFO gene showed substantial sequence similarity to known UDP-d-Glc 4-epimerases. An EcoRV restriction fragment length polymorphism within 10 kb of At1g30620 did not reveal any recombinants within a mapping population of 117 lines, suggesting that At1g30620 and MUR4 represent the same gene. To test this hypothesis, the entire At1g30620 coding region was amplified from the four known mur4 alleles, and the sequences of the PCR products were determined. All four mur4 alleles showed single point mutations within the gene, representing G-to-A transitions on the coding strand (Figure 2A). mur4-1 had a mutation in exon 7 leading to a Gly-to-Asp amino acid change, mur4-2 showed a mutation in intron 4 at the 5′ splice site (GTGAG to GTGAA), mur4-3 had a mutation in exon 8 leading to an Arg-to-Gln amino acid change, and mur4-4 showed a mutation in intron 8 at the 3′ consensus splice site (AG to AA) (Figure 2A).
Figure 2.
Molecular Cloning and Structure of the MUR4 Gene.
(A) Exon-intron structure of the MUR4 coding region based on a comparison of the genomic and cDNA sequences. The location of the translational initiation codon was inferred from the presence of a 419–amino acid open reading frame on the cDNA preceded by an in-frame stop codon. The positions of missense and splice site mutations within the four known mur4 alleles are indicated.
(B) Relative Ara content of wild-type (WT) plants (white bars), the mur4 mutant (black bars), and mur4 plants transformed with the wild-type allele (gray bars). The latter line represents a T2 population segregating for the MUR4 transgene. Leaf material from individual plants was analyzed for the Ara content of its cell walls by gas-liquid chromatography of alditol acetates. The height of the bars indicates the number of plants with a specific percentage of Ara content.
Genetic Complementation of the mur4 Mutation
Transgenic plants were generated by Agrobacterium-mediated transformation to confirm that the MUR4 gene had been identified. A 7.6-kb SalI fragment of the BAC clone F23C5 that contained the At1g30620 coding region was placed into the pCAMBIA1300 T-DNA binary vector and transformed into mur4-1 plants. The hygromycin-resistant T1 plants showed a restoration of the Ara content to wild-type or higher levels. The T2 generation of transformed mur4 plants segregated into phenotypically mur4 plants (50% reduction of the Ara content of leaves) and into plants with wild-type or higher levels of Ara (up to 2.4 times the Ara content of mur4 leaves) (Figure 2B). For most transgenic mur4 lines, less than one-fourth of the T2 plants were phenotypically mur4, suggesting the insertion of more than one transgene.
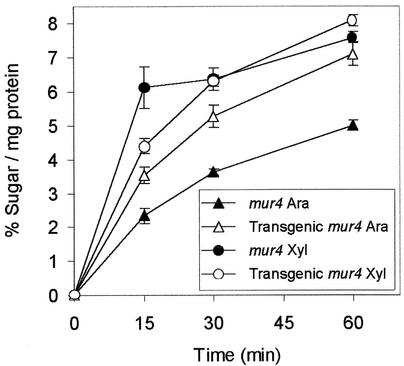
Two transgenic mur4 lines from the T3 generation were used for in vitro enzyme assays. Radiolabeled UDP-d-glucuronate was incubated with microsomal protein extracts of mur4 and transgenic mur4 lines. The conversion of UDP-d-Xyl to UDP-l-Ara was significantly higher in the two transgenic lines than in nontransformed mutant plants, confirming that the MUR4 gene had been cloned (Figure 3).
Figure 3.
Assay of the Microsomal Fractions from mur4 Plants and a mur4 Line Transformed with the Wild-Type MUR4 Allele for the Conversion of UDP-d-Glucuronate to UDP-d-Xyl and UDP-l-Ara.
Radiolabeled UDP sugars produced at different time points were hydrolyzed, and the monosaccharides were separated and quantified by thin layer chromatography. Data points represent means of three samples ± sd.
Isolation of MUR4 cDNAs
Partial cDNAs were obtained by screening a cDNA library with a 32P-labeled 500-bp PCR probe corresponding to the 3′ end of the gene. A 5′-anchored PCR method led to the isolation of a 5′ cDNA sequence. A full-length cDNA then was constructed by PCR with a mix of template DNAs consisting of the 5′ partial cDNA and the 3′ partial cDNA isolated from the λPRL-2 library.
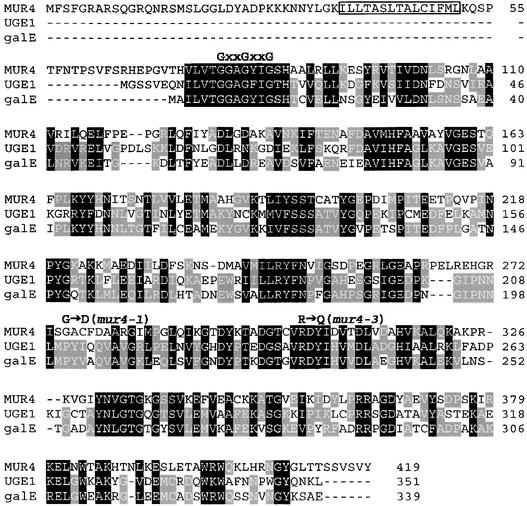
The cDNA sequence of MUR4 is predicted to encode a 419–amino acid protein with a molecular mass of 46 kD. The MUR4 gene product showed significant sequence similarities to UDP-d-Glc 4-epimerases such as the Arabidopsis UGE1 and the Bacillus subtilis galE gene products (Figure 4). Unlike the cytoplasmic UDP-d-Glc 4-epimerases, the MUR4 protein contained an N-terminal extension encompassing a potential transmembrane domain (Figure 4). An evaluation of the protein topology via the transmembrane hidden Markov model method (Krogh et al., 2001) suggests that the catalytic domain is located within the Golgi lumen (data not shown).
Figure 4.
Alignment of the Amino Acid Sequences of the MUR4 and UGE1 Proteins from Arabidopsis and the galE Gene Product from B. subtilis.
The putative transmembrane domain within MUR4 is boxed, and the highly conserved GxxGxxG motif involved in the binding of the NAD(P)+ cofactor is indicated above the sequence alignment. The amino acid substitutions in mur4-1 and mur4-3 also are shown.
Identification of Arabidopsis Genomic Sequences Similar to That of MUR4
Database searches with the MUR4 genomic and amino acid sequences identified three putative UDP-d-Xyl 4-epimerases from Arabidopsis with >76% amino acid sequence identity to MUR4: At2g34850 on chromosome 2, At5g44480 on chromosome 5, and At4g20460 on chromosome 4. Although at least some of these coding regions are presumed to represent membrane-bound UDP-d-Xyl 4-epimerases, their biochemical functions remain to be established.
Expression of the MUR4 Gene in Different Organs
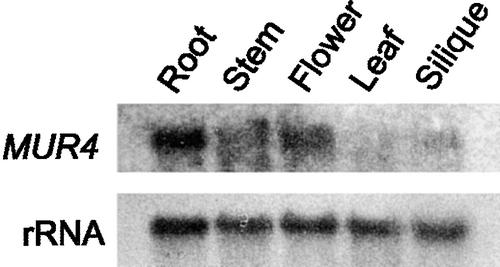
The expression pattern of the MUR4 gene was analyzed by RNA gel blot hybridization. RNA samples from roots, leaves, elongating stems, flowers, and siliques were probed with a 32P-labeled MUR4 probe. Hybridizing mRNAs were observed in all of these organs, with the highest level of expression in roots (Figure 5).
Figure 5.
RNA Gel Blot Analysis of Total RNA Extracted from Roots, Flowers, Leaves, Stems, and Siliques and Probed with a 32P-Labeled MUR4 Fragment.
Expression of MUR4 in Pichia pastoris
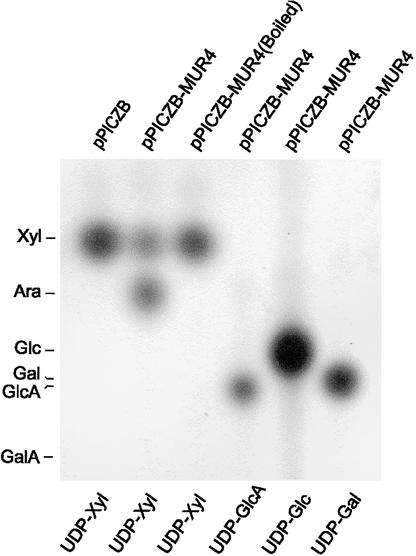
Expression of the full-length MUR4 cDNA in P. pastoris and assay of crude extracts for nucleotide sugar interconversion activities indicated that the MUR4 protein acted as a UDP-d-Xyl 4-epimerase but lacked both UDP-d-Glc and UDP-d-glucuronic acid 4-epimerase activities in vitro (Figure 6). Extracts from P. pastoris cells transformed with the vector alone were enzymatically inactive (Figure 6). At equilibrium, the ratio between UDP-l-Ara and UDP-d-Xyl was ∼1:1, which is in agreement with published values (Feingold and Avigad, 1980).
Figure 6.
Autoradiograph of a Thin Layer Chromatogram of the 14C-Labeled Reaction Products from Assays for 4-Epimerase Activities of the MUR4 Protein Expressed in P. pastoris.
The types of protein extracts used are indicated at top, and the nucleotide sugar substrates are indicated at bottom. Nucleotide sugars were hydrolyzed to monosaccharides before thin layer chromatography analysis. The only interconversion reaction observed was the reversible 4-epimerization of UDP-d-Xyl to UDP-l-Ara in the presence of native MUR4 protein.
To verify that the reaction products from the MUR4 assays with radiolabeled nucleotide sugar represented nucleotide sugars rather than monosaccharides or sugar phosphates, the reaction products were purified using anion-exchange chromatography (Bonin and Reiter, 2000). Ionic strengths required to elute the three sugar types were determined by loading and eluting d-Xyl, α-d-Xyl-1-phosphate, and a mixture of unlabeled and radiolabeled UDP-d-Xyl. d-Xyl eluted at 1 mM potassium phosphate, pH 6.0, whereas α-d-Xyl-1-phosphate eluted at 50 mM and UDP-d-Xyl eluted at 100 mM of this buffer. Once these conditions were established, the reaction products from the assays with MUR4 and UDP-d-U-14C-Xyl were loaded onto an anion-exchange column and eluted with two column volumes each of 10, 60, 120, and 200 mM potassium phosphate, pH 6.0. More than 90% of the radioactivity eluted with 120 mM of this buffer, indicating that the reaction products represented nucleotide sugars.
Localization of MUR4–Green Fluorescent Protein
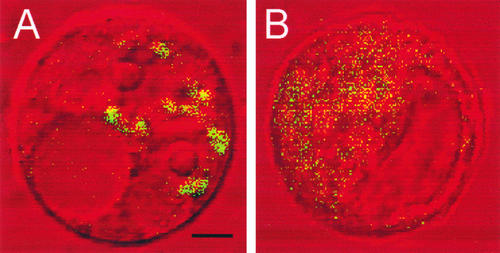
The subcellular localization of MUR4 was analyzed in protoplasts of Arabidopsis plants expressing a MUR4–green fluorescent protein (GFP) fusion protein. The protoplasts displayed a pattern of GFP fluorescence consistent with that of Golgi-localized protein (Figure 7A). To determine whether the fluorescent structures were in fact Golgi stacks, the effect of brefeldin A treatment was investigated (Figure 7B). Brefeldin A is a fungal metabolite known to cause vesiculation and disassembly or clustering of the Golgi apparatus, depending on the conditions and cell system (Staehelin and Driouich, 1997). When protoplasts were incubated with 100 μg/mL brefeldin A for 2 h, the pattern of GFP fluorescence changed to numerous small punctate bodies (Figure 7B), indicating that the MUR4-GFP fusion protein was localized in a brefeldin A–sensitive compartment, consistent with targeting to the Golgi.
Figure 7.
MUR4-GFP Localization in Arabidopsis Root Protoplasts.
(A) Confocal image of a single protoplast showing MUR4-GFP fluorescence consistent with a Golgi-localized protein.
(B) Confocal image of a single protoplast showing numerous punctate bodies indicating vesiculation and collapse of the Golgi stacks after brefeldin A treatment (2 h at 100 μg/mL).
Bar in (A) = 3 μm for (A) and (B).
DISCUSSION
To determine the genetic lesion underlying the mur4 phenotype, the MUR4 gene was cloned via a combined map-based and candidate gene approach. Taking advantage of the data released from the Arabidopsis genome sequencing project (Arabidopsis Genome Initiative, 2000), a gene that has some homology with known UDP-d-Glc 4-epimerases was identified and shown to be tightly linked to mur4. This candidate MUR4 gene was isolated from wild-type plants and all four known mur4 alleles. Nucleotide sequence determinations showed single point mutations within each mutant allele.
Two of the mur4 alleles showed mutations leading to nonconservative amino acid substitutions. These mutations were found in conserved regions of the protein. The Gly at position 275 of MUR4 corresponds to a Pro in Arabidopsis UGE1 and the bacterial galE gene product (Figure 4). This Gly was mutated to Asp in mur4-1, removing an α-helix–breaking amino acid. The Arg at position 304 of MUR4 is part of a large conserved domain within UDP-d-Glc 4-epimerases; it was mutated to Gln in mur4-3. The other two mur4 alleles showed mutations in introns. mur4-4 was altered in a 3′ acceptor splice site, changing the consensus nucleotides AG to AA, which is known to affect the correct splicing of pre-mRNAs. The mur4-2 mutation affected a 5′ splice site, changing the nucleotide sequence GTGAG to GTGAA. A similar G-to-A change at position 5 within an intron was reported to lead to a splicing defect in the Arabidopsis det1-1 and det1-3 mutant alleles (Pepper et al., 1994). Because the mur4-2 allele is leaky (Reiter et al., 1997), some correct splicing appears to take place or there is some missplicing, leading to a partially functional protein.
Correct identification of the MUR4 gene was verified by complementation of mutant plants by the wild-type allele. Wild-type or higher than wild-type levels of Ara were observed in the transgenic mur4 plants. Increased levels of cell wall Ara also were observed in wild-type plants containing extra copies of the MUR4 gene (E.G. Burget and W.-D. Reiter, unpublished results), suggesting that MUR4 activity is rate limiting for the synthesis of arabinosylated glycans in Arabidopsis. The growth of wild-type plants in the presence of l-Ara caused increased incorporation of this monosaccharide into cell wall material (Burget and Reiter, 1999), which supports this conclusion.
The activity of the MUR4 gene product was demonstrated by expression in P. pastoris. The recombinant protein catalyzed the interconversion between UDP-d-Xyl and UDP-l-Ara but did not act on UDP-d-Glc or UDP-d-glucuronate. The enzyme also was not inhibited by UDP-d-Glc or UDP-d-Gal, suggesting that it cannot bind these substrates in its active site. Although d-Glc is structurally very similar to d-Xyl (Figure 1), its hydroxymethyl group may cause steric hindrance, preventing binding to the catalytic center.
The Ara content in the pectic material and in the arabinogalactan component of AGPs is reduced in the mur4 mutant (Burget and Reiter, 1999). Although the mur4 mutation affects these carbohydrates, it does not appear to have a physiological effect on plant growth and fertilization processes. It was shown that the AGP-like TTS proteins must be reduced to very low levels in transgenic plants before a reduction in the pollen tube growth rate or reduced fertility is manifested (Cheung et al., 1995). Because the mur4 mutation does not lead to any obvious visible phenotype, the residual Ara in mur4 plants appears to be sufficient for plant growth and development, at least under laboratory conditions. Although mur4 plants show a wild-type growth habit, the mur4 mutation greatly exacerbates the dwarfed appearance of mur1 plants, which are defective in the de novo synthesis of GDP-l-Fuc (Reiter et al., 1997). The slightly dwarfed growth habit of Arabidopsis mur1 has been traced to Fuc deficiency in the pectic component rhamnogalacturonan II, which is cross-linked by borate within the cell wall (O'Neill et al., 2001). Because this polysaccharide contains both l-Fuc and l-Ara, we speculate that the extreme dwarfism of mur1 mur4 double mutants is caused by a severe defect in rhamnogalacturonan II, which might be expected as a result of the reduced availability of both GDP-l-Fuc and UDP-l-Ara. This hypothesis is supported by our observation that the visible phenotype of mur1 mur4 double mutants can be rescued by growth in the presence of millimolar concentrations of borate (M. Mølhøj and W.-D. Reiter, unpublished results), which compensates for the reduced borate binding ability of structurally altered rhamnogalacturonan II (O'Neill et al., 2001).
Considering that three of the four known mur4 alleles show the same reduction in Ara content, it appears unlikely that the residual Ara is caused by leakiness of the mutant alleles. We speculate that one or several of the MUR4 paralogs in the Arabidopsis genome contributes to the de novo synthesis of UDP-l-Ara in vivo.
MUR4 represents the first nucleotide sugar interconversion enzyme known to be targeted to the plant Golgi. This raises the question of whether other enzymes in nucleoside diphospho sugar interconversion pathways are directed similarly to the Golgi apparatus. It has been noted that putative UDP-d-Xyl synthases and UDP-d-glucuronate 4-epimerases in Arabidopsis contain a predicted transmembrane domain at the N terminus similar to that of MUR4 (Reiter and Vanzin, 2001). This finding suggests that the colocalization of nucleoside diphospho sugar interconversion enzymes and glycosyltransferases is a widespread phenomenon in higher plants, particularly for those activated monosaccharides that are needed primarily or exclusively in the Golgi for the synthesis of cell wall material and other complex glycans. It will be interesting to determine whether MUR4 is present in the same Golgi cisternae as arabinosyltransferases in cell wall synthesis; however, this will require the identification of coding regions for arabinosyltransferases, which has not yet been accomplished.
The mur4 mutant provides a means to change the cell wall without affecting the growth habit of a plant, which could have practical applications in the area of biotechnology. Changes in arabinosylated glycans could lead to alterations in covalent cross-linking between different cell wall polymers. For instance, covalent ester-ether bridges between polysaccharides and lignins are formed by ferulic acids on arabinoxylans in wheat straw (Iiyama et al., 1990), and these ferulic acids are esterified to pectins via Ara residues in spinach and sugar beet (Lam et al., 1990). Thus, the practical implications could be the easier processing of plant cell wall material for the paper industry, which requires the breaking of polysaccharide/lignin cross-links, and an increased digestibility of plant cell wall material for the cattle industry.
METHODS
Plant Material
Arabidopsis thaliana plants were grown at 23°C and 70% RH under continuous fluorescent light (60 to 70 μmol·m−2·s−1). Wild-type plants of the Columbia and Landsberg erecta ecotypes, and mutant plants of the Columbia ecotype carrying the mur4-1, mur4-2, mur4-3, and mur4-4 alleles (Reiter et al., 1997), were used.
Genetic Mapping
To map the mur4 locus, the mur4-1 mutant in the Columbia background of Arabidopsis was crossed to Landsberg erecta plants, and the resulting F1 progeny were selfed to obtain an F2 population. Approximately 1000 plants from this population were screened for reduced Ara content by gas-liquid chromatography of alditol acetates (Reiter et al., 1993), leading to the identification of 120 mutants. These mur4 plants were selfed, and the F3 plants were analyzed by gas-liquid chromatography to confirm the mur4 phenotype. Genomic DNA was extracted from the pooled leaf material of the F3 generation as described (Dellaporta et al., 1983). The mur4-1 mutation was mapped relative to simple sequence length polymorphism markers (Bell and Ecker, 1994) and cleaved amplified polymorphic sequence markers (Konieczny and Ausubel, 1993) using oligonucleotide primers from Research Genetics (Huntsville, AL).
Cloning of a MUR4 cDNA
Two million plaques from the λPRL-2 library (Newman et al., 1994) were screened for hybridization of cDNAs using the 3′ part of the MUR4 gene as a probe. Because no full-length clones were obtained using this approach, a 5′-anchored PCR procedure was used to obtain the 5′ end of the MUR4 transcript. For this purpose, total RNA was extracted from wild-type floral organs, and poly(A+) RNA was isolated from 150 μg of total RNA using the Oligotex mRNA kit (Qiagen, Valencia, CA). To synthesize first-strand cDNA, the mRNA in 11 μL of water was incubated at 65°C for 3 min, cooled on ice, and then incubated for 1.5 h at 37°C in the presence of 1 × first-strand buffer (Gibco BRL, Rockville, MD), 0.1 mg/mL BSA, 1 mM deoxynucleoside triphosphates, 50 μg/mL actinomycin D, 1.25 μg of the gene-specific primer 5′-CCATACGTTGCACAAGTGCTTG-3′, 10 mM DTT, and 200 units of reverse transcriptase from Moloney murine leukemia virus (Gibco BRL) in a total volume of 25 μL. After phenol extraction and ethanol precipitation, the pellet was resuspended in 5 μL of water, boiled for 2 min, and cooled on ice before the addition of a poly(dA) tail. The cDNA was incubated at 37°C for 45 min in the presence of 1 × terminal transferase buffer (0.25 mM CoCl2, 0.1 mM dATP, and 25 units of terminal transferase; New England Biolabs, Beverly, MA) in a final volume of 10 μL. The enzyme was inactivated by heating at 70°C for 10 min. After ethanol precipitation, a first PCR procedure was performed with 50 pmol of the gene-specific primer described above and a 20-mer oligo(dT) primer in the presence of 1.5 mM MgCl2 at an annealing temperature of 42°C. A second PCR procedure was performed with the nested cDNA-specific primer 5′-CCCGTGATAGATTGTCCAC-3′ and a 5′ primer representing positions −38 to −16 upstream of the predicted start codon of MUR4 (5′-CATCTAGTGTTTGTTTCCGCCAG-3′) in the presence of 2 mM MgCl2 at an annealing temperature of 59°C. The PCR products were used to determine the nucleotide sequence of the 5′ end of the cDNA.
To obtain a full-length cDNA, PCR was performed with a mix of DNA from the 5′-anchored PCR (first PCR product) and DNA from the cloned partial cDNA (3′ region) from the λPRL-2 library screen in the presence of 4 mM MgCl2 at an annealing temperature of 59°C using the following primers: 5′-TAGGATCCGCCAGCTGAAGTGAATC-3′ (forward primer representing positions −23 to −4 relative to the ATG start codon) and 5′-GCGGTACCATCTGGAATCAAAATC-3′ (reverse primer representing positions 30 to 49 downstream of the TGA stop codon) (BamHI and KpnI sites are underlined). After digestion with BamHI and KpnI, the PCR product was cloned into pESC-TRP (Stratagene, La Jolla, CA) to generate pESC-TRP-MUR4.
RNA Gel Blot Analysis
Total RNA was isolated from shoot tissues of 3-week-old plants and from roots of axenically grown plants by grinding in liquid nitrogen and homogenization in Trizol reagent (Gibco BRL) according to the manufacturer's protocol. Ten micrograms of RNA was treated with formamide and separated by formaldehyde gel electrophoresis (Sambrook et al., 1989). Blotting and hybridization were as described previously (Bonin and Reiter, 2000) using a MUR4 probe generated by PCR from pESC-TRP-MUR4 and random primer 32P labeling.
Plant Transformation
For complementation of the mur4 mutation, a 7.6-kb SalI fragment from BAC F23C5 was cloned into the plant transformation vector pCAMBIA1300 (CAMBIA, Canberra, Australia) and introduced by electroporation into Agrobacterium tumefaciens strain GV3101 (pMP90). mur4 plants then were transformed with this construct by vacuum infiltration (Bechtold et al., 1993). Transgenic plants were selected on agar medium containing 1% (w/v) Suc, 25 μg/mL hygromycin B (Calbiochem, La Jolla, CA), and 500 mg/L vancomycin (Wako Chemicals, Richmond, VA). Resistant T1 seedlings were transferred to soil, and their leaves were analyzed for their monosaccharide composition by gas-liquid chromatography of alditol acetates (Reiter et al., 1993). The segregation pattern of the T2 generation of transformed mur4 plants was analyzed by gas-liquid chromatography, and the transgenic lines showing the highest Ara levels were used in the T3 generation for biochemical assays. Extraction of microsomal proteins and assay of the conversion of UDP-d-glucuronate to UDP-d-Xyl and UDP-l-Ara were as described previously (Burget and Reiter, 1999).
Heterologous Expression of MUR4 in Pichia pastoris
Recombinant MUR4 protein was produced using the Pichia pastoris expression system from Invitrogen (Carlsbad, CA). The MUR4 cDNA sequence was amplified by PCR from pESC-TRP-MUR4 using the oligonucleotides 5′-CAGGTACCAATAATGTTTAGTTTTGGCCGAG-3′ and 5′-CATCTAGATAAACCGAGACCGATGAGGTTG-3′ (KpnI and XbaI sites are underlined) and cloned into the pPICZB expression vector to generate a fusion of MUR4 with the c-myc epitope and polyhistidine tag. After verification of the insert sequence, the resulting plasmid, pPICZB-MUR4, was linearized with SacI before transformation of P. pastoris KM71 by electroporation. Transformants were selected on plates with yeast extract peptone dextrose sorbitol medium and zeocin (100 μg/mL). Several individual transformants were screened for high expression of the intracellular MUR4 protein as follows: 20 mL of buffered complex glycerol medium supplemented with 100 μg/mL zeocin in 50-mL flasks was inoculated with a single colony and grown in a shaking incubator (28°C at 280 rpm) for 24 h until the OD600 reached a value between 2 and 6. Cells were harvested by centrifugation and resuspended in 30 mL of buffered complex methanol medium to OD600 = 1. The cultures were incubated for another 96 h at 28°C.
Every 24 h, 4-mL aliquots were removed and methanol (0.5% final concentration) was added to the remaining culture. Cells were harvested by centrifugation and stored at −80°C. Protein extracts were prepared in 200 μL of breaking buffer (50 mM Tris-HCl, pH 7.6, 5% [v/v] glycerol, 1 mM EDTA, 2 mM DTT, 4% 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid, and 1 × complete protease inhibitor cocktail [Roche Diagnostics, Mannheim, Germany]) by adding an equal volume of acid-washed glass beads (425 to 600 μm; Sigma, St. Louis, MO), rupturing the cells by sequential vortexing for 30 s, and placing them on ice for a total of eight cycles. After centrifugation for 10 min at 15,000g, the cleared supernatants were analyzed by an immunoblot procedure using the Bio-Dot apparatus from Bio-Rad (Hercules, CA), an Immobilon-P transfer membrane (Millipore, Bedford, MA), a primary rabbit anti-c-myc antibody (Santa Cruz Biotechnology, Santa Cruz, CA), and a secondary anti-rabbit-IgG antibody conjugated to alkaline phosphatase (Sigma). Two transformants with the highest levels of recombinant protein after 2 days of induction were chosen for large-scale expression.
Enzyme Assays
Measurements of protein concentration were performed using a bicinchoninic acid kit (Sigma). Protein extracts (25 to 100 μg) in a final volume of 50 μL were incubated separately with 50 nCi each of UDP-d-U-14C-Xyl (238 mCi/mmol), UDP-d-U-14C-Gal (300 mCi/mmol), UDP-d-U-14C-glucuronic acid (300 mCi/mmol), or UDP-d-U-14C-Glc (300 mCi/mmol) at room temperature for 1 h. Boiled protein extracts were incubated with the radiolabeled substrates as controls. All of the 14C-labeled nucleotide sugars were purchased from American Radiolabeled Chemicals (St. Louis, MO). The resulting UDP sugars were hydrolyzed by the addition of 300 μL of 2 M trifluoroacetic acid and incubation at 98°C for 30 min. The trifluoroacetic acid was removed in vacuo, and sugars were analyzed by silica thin layer chromatography as described (Burget and Reiter, 1999). UDP-d-glucuronate 4-epimerase and UDP-d-Glc 4-epimerase from Arabidopsis (Reiter and Vanzin, 2001) were used as positive controls for assays of these enzymatic activities.
Anion-Exchange Chromatography
Three separate Superclean LC-SAX solid-phase columns (Supelco, Bellefonte, PA) were used to determine the ionic strengths required to elute 200 μg of d-Xyl, 200 μg of α-d-Xyl-1-phosphate, and a mixture of 200 μg of unlabeled UDP-d-Xyl and 100 nCi of UDP-d-U-14C-Xyl. Nucleotide sugars were identified by liquid scintillation counting, and the monosaccharides and sugar phosphates were identified by total carbohydrate assays of the eluted fractions (Fry, 1988).
Expression and Localization of a MUR4–Green Fluorescent Protein Fusion Protein
The MUR4 cDNA was amplified by PCR using the oligonucleotides 5′-CATTCCATGGCTATGTTTAGTTTTGGCCGAGCAAG-3′ and 5′-CCATTACTAGTGTAAACCGAGACCGATGAGGTTG-3′ (NcoI and SpeI sites are underlined) and cloned in frame with the mgfp5 reporter gene in pCAMBIA1302, giving pCAMBIA1302-MUR4. The correct sequence of the construct was verified before transformation into Arabidopsis ecotype Columbia. Protoplasts obtained from roots were used to analyze the localization of MUR4–green fluorescent protein. These protoplasts were generated from 3-week-old plants grown on vertical Murashige and Skoog (1962) medium plates using the protocol described by Altmann et al. (1992). Confocal images were obtained with a MRC600 confocal laser scanning microscope (Bio-Rad) with argon/krypton laser excitation at 488 nm, emission at 505 to 545 nm, and optical sections of 0.45 μm. Combined images were obtained using merging software.
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes.
Acknowledgments
We thank Raymond Washington for technical assistance with confocal microscopy and CAMBIA (Center for the Application of Molecular Biology to International Agriculture) for plant transformation vectors. Support for this work from the Department of Energy's Energy Biosciences Program (Grant DE-FG02-95ER20203) is gratefully acknowledged. M.M. was financially supported by Grant SJVF 23000237 from the Danish Agricultural and Veterinary Research Council.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.008425.
References
- Altmann, T., Damm, B., Halfter, U., Willmitzer, L., and Morris, P.-C. (1992). Protoplast transformation and methods to create specific mutants in Arabidopsis thaliana. In Methods in Arabidopsis Research, C. Koncz, N.-H. Chua, and J. Schell, eds (Singapore: World Scientific Publishing), pp. 310–330.
- Arabidopsis Genome Initiative (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. [DOI] [PubMed] [Google Scholar]
- Baron-Epel, O., Gharyal, P.K., and Schindler, M. (1988). Pectins as mediators of wall porosity in soybean cells. Planta 175, 389–395. [DOI] [PubMed] [Google Scholar]
- Bechtold, N., Ellis, J., and Pelletier, G. (1993). In planta Agrobacterium-mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C. R. Acad. Sci. Paris. 316, 1194–1199. [Google Scholar]
- Bell, C.J., and Ecker, J.R. (1994). Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics 19, 137–144. [DOI] [PubMed] [Google Scholar]
- Bonin, C.P., and Reiter, W.-D. (2000). A bifunctional epimerase-reductase acts downstream of the MUR1 gene product and completes the de novo synthesis of GDP-l-fucose in Arabidopsis. Plant J. 21, 445–454. [DOI] [PubMed] [Google Scholar]
- Burget, E.G., and Reiter, W.-D. (1999). The mur4 mutant of Arabidopsis is partially defective in the de novo synthesis of uridine diphospho l-arabinose. Plant Physiol. 121, 383–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpita, N.C., and Gibeaut, D.M. (1993). Structural models of primary cell walls in flowering plants: Consistency of molecular structure with the physical properties of the walls during growth. Plant J. 3, 1–30. [DOI] [PubMed] [Google Scholar]
- Cheung, A.Y., Wang, H., and Wu, H.-M. (1995). A floral transmitting tissue-specific glycoprotein attracts pollen tubes and stimulates their growth. Cell 82, 383–393. [DOI] [PubMed] [Google Scholar]
- Dellaporta, S.L., Wood, J., and Hicks, J.B. (1983). A plant DNA minipreparation: Version 2. Plant Mol. Biol. Rep. 1, 19–22. [Google Scholar]
- Dörmann, P., and Benning, C. (1996). Functional expression of uridine 5′-diphospho-glucose 4-epimerase (EC 5.1.3.2) from Arabidopsis thaliana in Saccharomyces cerevisiae and Escherichia coli. Arch. Biochem. Biophys. 327, 27–34. [DOI] [PubMed] [Google Scholar]
- Du, H., Clarke, A.E., and Bacic, A. (1996). Arabinogalactan-proteins: A class of extracellular matrix proteoglycans involved in plant growth and development. Trends Cell Biol. 6, 411–414. [DOI] [PubMed] [Google Scholar]
- Fan, D.-F., and Feingold, D.S. (1972). Biosynthesis of uridine diphosphate d-xylose. V. UDP-d-glucuronate and UDP-d-galacturonate carboxy-lyase of Ampullariella digitata. Arch. Biochem. Biophys. 148, 576–580. [DOI] [PubMed] [Google Scholar]
- Feingold, D.S., and Avigad, G. (1980). Sugar nucleotide transformations in plants. In The Biochemistry of Plants: A Comprehensive Treatise, Vol. 3, P.K. Stumpf and E.E. Conn, eds (New York: Academic Press), pp. 101–170.
- Fleischer, A., O'Neill, M., and Ehwald, R. (1999). The pore size of non-graminaceous plant cell walls is rapidly decreased by borate ester cross-linking of the pectic polysaccharide rhamnogalacturonan II. Plant Physiol. 121, 829–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry, S.C. (1988). The Growing Plant Cell Wall: Chemical and Metabolic Analysis. (New York: Wiley).
- Gaspar, Y., Johnson, K.L., McKenna, J.A., Bacic, A., and Schultz, C.J. (2001). The complex structures of arabinogalactan-proteins and the journey towards understanding function. Plant Mol. Biol. 47, 161–176. [PubMed] [Google Scholar]
- Hall, Q., and Cannon, M.C. (2002). The cell wall hydroxyproline-rich glycoprotein RSH is essential for normal embryo development in Arabidopsis. Plant Cell 14, 1161–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurlbert, R.E., Weckesser, J., Mayer, H., and Fromme, I. (1976). Isolation and characterization of the lipopolysaccharide of Chromatium vinosum. Eur. J. Biochem. 68, 365–371. [DOI] [PubMed] [Google Scholar]
- Iiyama, K., Lam, T.B.T., and Stone, B.A. (1990). Phenolic acid bridges between polysaccharides and lignin in wheat internodes. Phytochemistry 29, 733–737. [Google Scholar]
- Katzman, R.L. (1974). Absence of arabinose in bovine brain hyaluronic acid as analysed by gas-liquid chromatography and mass spectrometry. Biochim. Biophys. Acta 372, 53–54. [PubMed] [Google Scholar]
- Konieczny, A., and Ausubel, F.M. (1993). A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J. 4, 403–410. [DOI] [PubMed] [Google Scholar]
- Krogh, A., Larsson, B., von Heijne, G., and Sonnhammer, E.L. (2001). Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 305, 567–580. [DOI] [PubMed] [Google Scholar]
- Lam, T.B.T., Iiyama, K., and Stone, B.A. (1990). Primary and secondary walls of grasses and other forage plants: Taxonomic and structural considerations. In Microbial and Plant Opportunities to Improve Lignocellulose Utilization by Ruminants, D.E. Akin, M.G. Ljundahl, R.J. Wilson, and P.J. Harris, eds (New York: Elsevier), pp. 43–69.
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473.–497. [Google Scholar]
- Newman, T., DeBruijn, F.J., Green, P., Keegstra, K., Kende, H., McIntosh, L., Ohlrogge, J., Raikhel, N., Somerville, S., Thomashow, M., Retzel, E., and Somerville, C.R. (1994). Genes galore: A summary of methods for accessing results from large-scale partial sequencing of anonymous Arabidopsis cDNA clones. Plant Physiol. 106, 1241–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill, M.A., Eberhard, S., Albersheim, P., and Darvill, A.G. (2001). Requirement of borate cross-linking of cell wall rhamnogalacturonan II for Arabidopsis growth. Science 294, 846–849. [DOI] [PubMed] [Google Scholar]
- Pepper, A., Delaney, T., Washburn, T., Poole, D., and Chory, J. (1994). DET1, a negative regulator of light-mediated development and gene expression in Arabidopsis, encodes a novel nuclear-localized protein. Cell 78, 109–116. [DOI] [PubMed] [Google Scholar]
- Reiter, W.-D., Chapple, C.C.S., and Somerville, C.R. (1993). Altered growth and cell walls in a fucose-deficient mutant of Arabidopsis. Science 261, 1032–1035. [DOI] [PubMed] [Google Scholar]
- Reiter, W.-D., Chapple, C.C.S., and Somerville, C.R. (1997). Mutants of Arabidopsis thaliana with altered cell wall polysaccharide composition. Plant J. 12, 335–345. [DOI] [PubMed] [Google Scholar]
- Reiter, W.-D., and Vanzin, G.F. (2001). Molecular genetics of nucleotide sugar interconversion pathways in plants. Plant Mol. Biol. 47, 95–113. [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Showalter, A.M. (1993). Structure and function of plant cell wall proteins. Plant Cell 5, 9–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staehelin, L.A., and Driouich, A. (1997). Brefeldin A effects in plants: Are different Golgi responses caused by different sites of action? Plant Physiol. 114, 401–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stafstrom, J.P., and Staehelin, L.A. (1986). The role of carbohydrate in maintaining extensin in an extended conformation. Plant Physiol. 81, 242–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma, R., Vercellotti, J.R., and Varma, R.S. (1977). On arabinose as a component of brain hyaluronate: Confirmation by chromatographic, enzymatic and chemical ionization-mass spectrometric analyses. Biochim. Biophys. Acta 497, 608–614. [DOI] [PubMed] [Google Scholar]
- Wardi, A.H., Allen, W.S., Turner, D.L., and Stary, Z. (1966). Isolation of arabinose-containing hyaluronate peptides and xylose-containing chondroitin sulfate peptides from protease-digested brain tissue. Arch. Biochem. Biophys. 117, 44–53. [DOI] [PubMed] [Google Scholar]