Abstract
Auxin response factors (ARFs) are transcription factors that bind to TGTCTC auxin response elements in promoters of early auxin response genes. ARFs have a conserved N-terminal DNA binding domain (DBD) and in most cases a conserved C-terminal dimerization domain (CTD). The ARF CTD is related in amino acid sequence to motifs III and IV found in Aux/IAA proteins. Just C terminal to the DBD, ARFs contain a nonconserved region referred to as the middle region (MR), which has been proposed to function as a transcriptional repression or activation domain. Results with transfected protoplasts reported here show that ARFs with Q-rich MRs function as activators, whereas most, if not all other ARFs, function as repressors. ARF DBDs alone are sufficient to recruit ARFs to their DNA target sites, and auxin does not influence this recruitment. ARF MRs alone function as activation or repression domains when targeted to reporter genes via a yeast Gal4 DBD, and auxin does not influence the potency of activation or repression. ARF CTDs, along with a Q-rich MR, are required for an auxin response whether the MRs plus CTDs are recruited to a promoter by an ARF DBD or by a Gal4 DBD. The auxin response is mediated by the recruitment of Aux/IAA proteins to promoters that contain a DNA binding protein with a Q-rich MR and an attached CTD.
INTRODUCTION
The plant hormone auxin regulates a variety of genes, the most thoroughly studied of which include the Aux/IAA, GH3, and SAUR gene families (reviewed by Guilfoyle, 1999; Hagen and Guilfoyle, 2002). Most of the genes in these three families are primary/early response genes, which means that they are activated rapidly after auxin treatment and that protein synthesis is not required for their activation. At least one promoter DNA sequence that is involved with the auxin regulation of primary/early auxin response genes is the TGTCTC auxin response element (AuxRE). Auxin response factors (ARFs) are transcriptional activators and repressors that bind with specificity to TGTCTC AuxREs in promoters of primary/early auxin response genes (reviewed by Guilfoyle and Hagen, 2001). There are 22 ARF genes and 1 or more pseudogenes in Arabidopsis (Guilfoyle and Hagen, 2001; Liscum and Reed, 2002). ARFs contain an N-terminal DNA binding domain (DBD) and a middle region (MR) that is proposed to function as either an activation or a repression domain (Ulmasov et al., 1999a, 1999b). Five ARFs (ARF5, -6, -7, -8, and -19) have Gln-rich MRs and may function as transcriptional activators, whereas ARF1 appears to function as a transcriptional repressor (Ulmasov et al., 1999a).
All but two ARFs in Arabidopsis (ARF3/ETTIN and ARF17) contain a C-terminal dimerization domain (CTD), which is related in amino acid sequence to motifs III and IV found in Aux/IAA proteins (Guilfoyle and Hagen, 2001; Liscum and Reed, 2002). Aux/IAA proteins are short-lived nuclear proteins and are, in general, encoded by primary/early auxin response genes, some of which appear to contain TGTCTC AuxREs (reviewed by Reed, 2001; Rogg and Bartel, 2001). There are 24 genes in Arabidopsis that are predicted to encode Aux/IAA proteins and that contain four conserved motifs (which are referred to as motifs or domains I through IV). Five additional genes in Arabidopsis are predicted to encode proteins related to Aux/IAA proteins but lack one or more of the conserved motifs. Yeast two-hybrid experiments suggest that ARF CTDs and Aux/IAA proteins can homodimerize and heterodimerize with some selectivity (Kim et al., 1997; Ulmasov et al., 1997b; Ouellet et al., 2001). It has been hypothesized that Aux/IAA proteins are repressors that do not bind to TGTCTC AuxREs directly but are targeted to AuxREs by dimerizing with ARF transcriptional activators when auxin concentrations are low, resulting in the repression of primary/early auxin response genes (Guilfoyle et al., 1998; Guilfoyle and Hagen, 2001). When auxin concentrations are increased, the auxin response genes are postulated to be derepressed/activated after the dissociation (and proteolysis) of the Aux/IAA proteins from the ARF activators. An alternative to this hypothesis is that Aux/IAA proteins prevent ARF transcriptional activators from reaching their AuxRE target sites by dimerizing with ARFs at low auxin concentrations (i.e., Aux/IAA proteins effectively titrate out ARF transcriptional activators). In this latter hypothesis, Aux/IAA proteins would be degraded when auxin concentrations are high, allowing the dimerization of ARFs, their binding to AuxREs, and the subsequent activation of auxin response genes (Benfey, 2002; Hellmann and Estelle, 2002).
To further investigate how ARF proteins might function to regulate primary/early auxin response genes and to test the hypotheses described above, we have used protoplast transfection assays to functionally characterize the different domains in these transcription factors. Our results provide information regarding which ARFs are activators or repressors, which ARF domain(s) is required for their being targeted to AuxREs, which ARF domain(s) is required for transcriptional activation or repression, and which ARF domain(s) is required for an auxin response. Furthermore, our results provide information on whether ARFs must dimerize via their CTD to bind to AuxREs, whether auxin influences the binding of ARFs to AuxREs, and whether MR repression or activation is influenced by auxin. The role that Aux/IAA proteins play in bringing about the repression of auxin-responsive genes also is addressed.
RESULTS
Full-Length ARF Proteins Function as Repressors or Activators in Protoplast Transfection Assays with Auxin-Responsive Reporter Genes
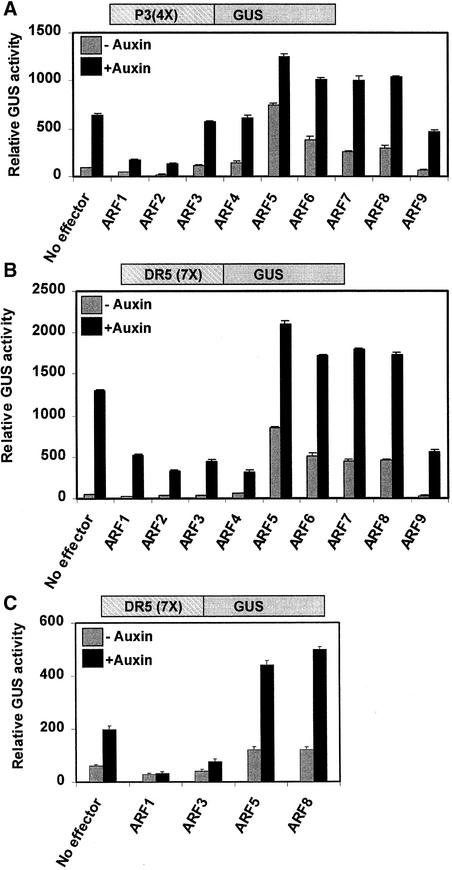
We have shown previously that effector genes that encode full-length Arabidopsis ARF6 and ARF1 bring about the activation and repression, respectively, of auxin-responsive β-glucuronidase (GUS) reporter genes in transfected carrot protoplasts (Ulmasov et al., 1999a). To determine if other full-length ARF proteins from Arabidopsis function as activators or repressors, ARF-2, -3, -4, -5, -6, -7, -8, and -9 (for ARF nomenclature, see Guilfoyle and Hagen 2001) were tested as effector genes with two different auxin-responsive GUS reporter genes in carrot protoplasts. With the auxin-responsive palindromic P3(4X)-GUS reporter gene, effector genes that encode full-length ARF1 and ARF2 repressed transcription with or without auxin treatment, whereas effector genes that encode ARF5, -6, -7, and -8 activated transcription (Figure 1A). With the P3(4X)-GUS reporter gene, effector genes that encode ARF9 repressed weakly, whereas effector genes that encode ARF3 and ARF4 had little if any effect on reporter gene expression. The small amount of activation observed with ARF3 and ARF4 in the absence of auxin was not reproducible with other carrot protoplast preparations. To determine if the same pattern of repressor and activator activity resulted with a different auxin-responsive reporter gene, an auxin-responsive direct repeat DR5(7X)-GUS reporter gene was cotransfected with ARF1 through ARF9 effector genes. With the DR5(7X)-GUS reporter gene, effector genes that encode ARF1, -2, -3, -4, and -9 repressed reporter gene expression, whereas effector genes that encode ARF5, -6, -7, and -8 activated transcription (Figure 1B). Thus, the repressor activity of selected ARFs shows some specificity for the auxin-responsive reporter gene used in the transfection assays.
Figure 1.
Activation and Repression by ARFs on Auxin-Responsive Reporter Genes.
Effector genes, which consisted of the CaMV 35S promoter encoding full-length ARF proteins, were cotransfected into protoplasts with a GUS reporter gene, which contained an auxin-responsive promoter. Reporter genes are diagrammed above the graphs. GUS activities were measured from protoplasts that were treated (+ auxin) or not treated (− auxin) with 10 μM 1-NAA.
(A) The P3(4X)-GUS reporter gene was cotransfected into carrot suspension cell protoplasts with the ARF effector genes indicated.
(B) Same as (A), but with a DR5(7X)-GUS reporter gene.
(C) Same as (B), but with Arabidopsis suspension cell protoplasts.
We have routinely used carrot suspension culture cells to study auxin-responsive transcription and auxin response transcription factors (Ulmasov et al., 1997a, 1997b, 1999a; Tiwari et al., 2001) because of the ease of carrot cell protoplast preparation, the robust yields of viable protoplasts, and the greater reporter gene activities. However, to confirm that ARFs function similarly in other plant cells, we tested a few effectors that encode full-length ARFs in protoplasts prepared from Arabidopsis (ecotype Columbia) suspension culture cells. Figure 1C shows that full-length ARF1 and ARF3 functioned as repressors and ARF5 and ARF8 functioned as activators in Arabidopsis suspension cell protoplasts assayed with a DR5(7X)-GUS reporter gene.
The protoplast transfection results indicate that full-length ARFs with Gln-rich MRs function as transcriptional activators on two different auxin-responsive promoter-GUS reporter genes and that full-length ARFs without Gln-rich MRs function as repressors. However, the capacity of ARFs to function as repressors is somewhat dependent on the auxin-responsive reporter gene tested.
ARF1 and ARF5 DBDs Are Targeted to AuxREs Independently of ARF MRs and CTDs
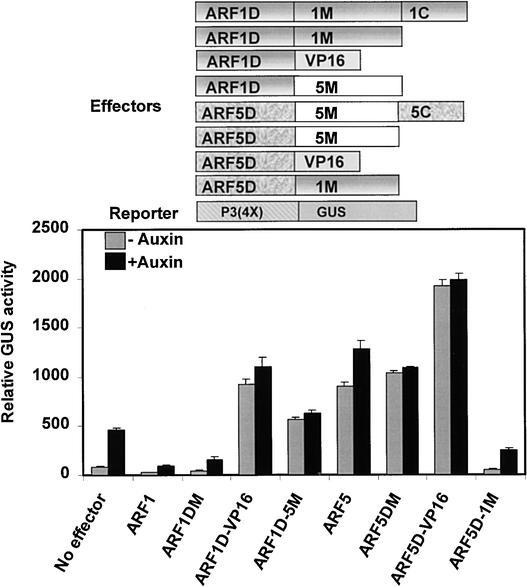
To determine if DBDs (defined as amino acids 1 to ∼350 by Ulmasov et al. [1999b]), derived from either an ARF repressor or an ARF activator, are sufficient to target AuxREs in vivo, effector genes that encode chimeric ARF1 and ARF5 proteins were tested in carrot suspension cell protoplasts with the P3(4X)-GUS reporter gene. Figure 2 shows that when the ARF1 DBD was fused to either the VP16 activation domain (ARF1D-VP16) or the ARF5 MR (ARF1D-5M), the chimeric ARF functioned as an activator, in contrast to full-length ARF1 and ARF1 lacking its CTD (ARF1DM), which functioned as repressors on the P3(4X)-GUS reporter gene. When the ARF5 DBD was fused to the VP16 activation domain (ARF5D-VP16), the chimeric ARF functioned as an activator that was stronger than the full-length ARF5 activator or the activator that consisted of ARF5 lacking its CTD (ARF5DM). By contrast, when the ARF5 DBD was fused to the ARF1 MR (ARF5D-1M), the chimeric ARF functioned as a repressor. To exclude the possibility that MRs or VP16 could function as activators or repressors of the auxin-responsive reporter gene in the absence of an ARF DBD, effector genes that encode chimeric proteins consisting of the yeast Gal4 DBD fused to the ARF1 MR, ARF5 MR, or VP16 were tested with the P3(4X)-GUS reporter gene. The activity of the reporter gene was similar to that seen with no effector gene when cotransfected with effector genes that encode Gal4 DBD-MR or Gal4 DBD-VP16 chimeric proteins (data not shown); however, these chimeric proteins were repressors or activators when assayed with reporter genes containing Gal4 DNA binding sites in the promoter (see below).
Figure 2.
The DBD Is Sufficient for Targeting ARF1 and ARF5 to an Auxin-Responsive P3(4X)-GUS Reporter Gene in Protoplast Transfection Assays, and This Targeting Is Independent of Auxin.
Effector and reporter genes (diagrammed at top) were cotransfected into carrot suspension cell protoplasts and assayed in the presence (+ auxin) or absence (− auxin) of 10 μM 1-NAA. ARF1 and ARF5 were tested as full-length constructs, as constructs that lacked a CTD dimerization domain, and as constructs that lacked both their normal MR and CTD but contained a heterologous MR or VP16 activation domain.
These results showed that ARF1 and ARF5 DBDs were interchangeable when assayed with a P3(4X)-GUS reporter gene in transfected protoplasts and that the ARF MR or a heterologous activation domain (i.e., VP16) dictated whether the chimeric ARF functioned as a repressor or an activator. Furthermore, our results indicate that the ARF1 or ARF5 CTD was required neither for ARF binding to DNA target sites nor to activate or repress transcription on an auxin-responsive reporter gene. Thus, dimerization of ARF1 and ARF5 via the CTD was not required for these ARFs to bind an AuxRE or to function in activation or repression. Because the chimeric ARF proteins were targeted to AuxREs regardless of whether auxin was withheld or included in the assays (i.e., equivalent levels of activation or repression were observed with or without auxin treatment), it is unlikely that auxin influences the binding of ARFs to their DNA target sites.
ARF MRs Function as Repression or Activation Domains in an Auxin-Independent Manner
To determine if ARF MRs in isolation from ARF DBDs and CTDs can function as activation or repression domains, the MRs of the ARF repressors (Figure 1) (ARF1, -2, -3, -4, and -9) and the ARF activators (ARF5, -6, -7, and -8) were fused to the yeast Gal4 DBD and tested as effector genes in transfected carrot protoplasts along with a constitutively expressed reporter gene containing four Gal4 DNA binding sites, Gal4(4X)-D1-3(4X)-GUS (Tiwari et al., 2001), or a GUS reporter gene containing only a minimal −46 promoter of Cauliflower mosaic virus (CaMV) with four Gal4 DNA binding sites, Gal4(4X)-GUS (Tiwari et al., 2001). Each of the ARF repressors contains a MR that is enriched for P and/or S. ARF1 and ARF4 contain MRs rich in both S and P (i.e., SP rich), ARF2 and ARF9 contain MRs rich in S (i.e., S rich), and ARF3 contains a MR rich in both S and G (i.e., SG rich) (Guilfoyle and Hagen, 2001). ARF5, -6, -7, and 8 contain MRs that are enriched for Q, and some of these contain polymeric stretches of Q residues.
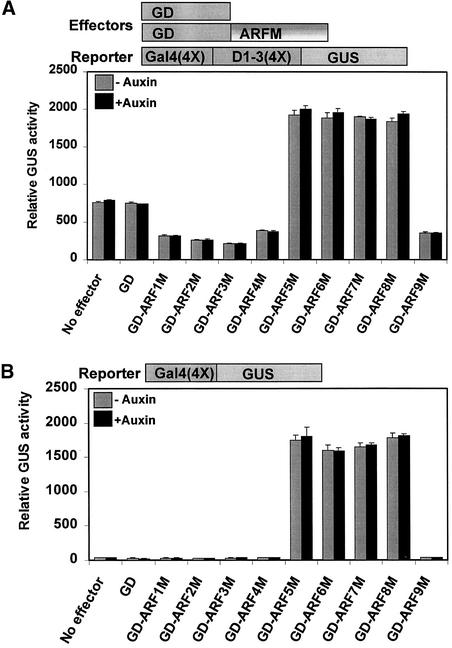
Figure 3A shows that with the Gal4(4X)-D1-3(4X)-GUS reporter gene, cotransfection of effector genes that encoded the Gal4 DBD fused to the MR of ARF1, -2, -3, -4, and -9 resulted in repression of the reporter gene compared with transfections that contained no effector or an effector that encoded only the Gal4 DBD (GD). Cotransfection of effector genes that encoded the Gal4 DBD fused to the MR of ARF5, -6, -7, and -8 resulted in enhanced expression of the reporter gene (i.e., approximately twofold greater activity than transfections with the reporter gene alone or with a GD effector gene). Figure 3B shows that the minimal promoter GUS reporter gene, Gal4(4X)-GUS, was inactive when transfected with no effector gene or cotransfected with an effector gene that encoded only the Gal4 DBD (GD). Effector genes that encoded the Gal4 DBD fused to the MR of ARF1, -2, -3, -4, and -9 had no effect on GUS expression compared with transfections that contained no effector or the GD effector gene. By contrast, cotransfection of effector genes that encoded the Gal4 DBD fused to the MR of ARF5, -6, -7, and -8 resulted in at least 50-fold activation of the minimal promoter gene. The addition of auxin to the protoplasts had little if any effect on reporter gene expression with all of the effector genes tested.
Figure 3.
ARF MRs Function as Repression or Activation Domains in an Auxin-Independent Manner.
Effector genes containing a yeast Gal4 DBD (GD) fused in frame to ARF MRs are diagrammed at top. ARF1, -2, -3, -4, and -9 MRs are enriched for P and S, and ARF5, -6, -7, and -8 MRs are enriched for Q. The GD effector gene contained only the yeast Gal4 DBD. Carrot suspension cell protoplasts cotransfected with a reporter gene and an effector gene were assayed in the presence (+ auxin) or absence (− auxin) of 10 μM 1-NAA.
(A) To measure repression by ARF effector genes, transfection assays were performed with a constitutive Gal4(4X)-D1-3(4X)-GUS reporter gene, which contains four Gal4 DNA binding sites upstream of four tandem copies of the constitutive D1-3(4X) element (Tiwari et al., 2001).
(B) To measure activation by ARF effector genes, transfection assays were performed with the minimal promoter Gal4(4X)-GUS reporter gene, which contains four Gal4 DNA binding sites upstream of the CaMV −46 minimal promoter element (Tiwari et al., 2001).
These results showed that ARF MRs that are enriched for P and S amino acids functioned as repression domains, and ARF MRs that are enriched in Q residues functioned as activation domains, in transfected carrot suspension cell protoplasts. Furthermore, ARF MR repression and activation domains did not appear to be a target for auxin action, at least when isolated from the ARF DBD and/or CTD.
The ARF CTD Is Required for an Auxin Response
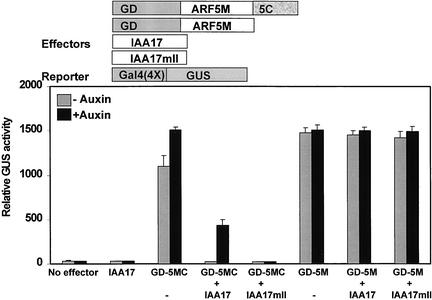
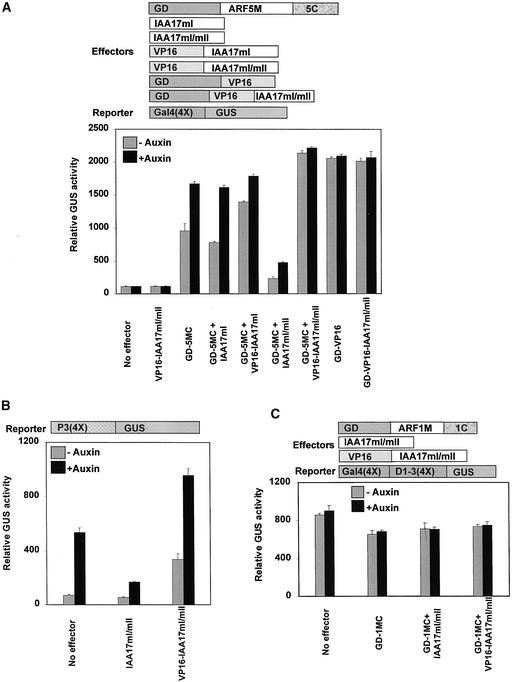
Ulmasov et al. (1999a) reported previously that the Gal4(4X)-GUS reporter gene showed a small but significant induction by auxin when cotransfected with effector genes that encode the yeast Gal4 DBD fused in frame to ARF5, -6, -7, and -8 MRs plus CTDs (i.e., lacking only the ARF DBD). To confirm that there was an auxin response that was dependent on the ARF CTD, Gal4 DBD fusion proteins containing the ARF5 MR (GD-ARF5M) and the ARF5 MR plus CTD (GD-ARF5MC) were compared directly as effector genes in carrot protoplasts that were cotransfected with the Gal4(4X)-GUS reporter gene. Figure 4 shows that although transfections with the GD-ARF5M effector gene showed no auxin response, there was a modest but significant (25 to 30%) increase in GUS reporter activity with the GD-ARF5MC effector gene when auxin was present. These results indicated that when the CTD was attached to the ARF MR activation domain, the MR was not fully active unless auxin was present.
Figure 4.
The ARF CTD Dimerization Domain Confers Auxin Responsiveness to an ARF5 MR.
Effector and reporter genes (diagrammed at top) were cotransfected into carrot suspension cell protoplasts and assayed in the presence (+ auxin) or absence (− auxin) of 10 μM 1-NAA. The IAA17 and IAA17mII effector genes encode a full-length wild-type IAA17 and a full-length box II mutant IAA17mII protein, respectively. The IAA17mII mutant protein is more stable and is a stronger repressor than the wild-type IAA17 protein (Tiwari et al., 2001).
The most likely explanation for the lower activity observed with GD-ARFMC effector genes in the absence of auxin treatment is that endogenous Aux/IAA proteins, which function as active repressors at low auxin concentrations (Tiwari et al., 2001), were able to dimerize with GD-ARFMC proteins but not with GD-ARFM proteins. In this scenario, the addition of auxin would result in more rapid degradation of the Aux/IAA proteins (Rogg and Bartel, 2001; reviewed by Kepinski and Leyser, 2002), resulting in derepression (i.e., increased activation) with the GD-ARFMC effector genes. This scenario is supported by results from experiments in which a second effector that encodes the Aux/IAA protein, Arabidopsis IAA17, was cotransfected with either a GD-ARF5M or a GD-ARF5MC effector gene. Figure 4 shows that cotransfection with GD-ARF5MC plus IAA17 effector genes into carrot protoplasts along with the Gal4(4X)-GUS reporter gene resulted in a strong reduction of GUS expression compared with protoplasts transfected with only the GD-ARF5MC effector gene. In the absence of the GD-ARF5MC effector gene, the IAA17 effector gene had no effect on reporter gene activity. A further reduction in GUS expression was observed if the GD-ARFMC effector gene was cotransfected with the more stable IAA17mII effector gene. The IAA17mII protein contains a site-specific mutation in conserved motif II of Aux/IAA proteins, which increases its life span in cells and blocks its auxin-induced destabilization (Gray et al., 2001; Ouellet et al., 2001), thus increasing its capacity to repress auxin-responsive gene expression (Tiwari et al., 2001). By contrast, cotransfection of the effector gene that encodes GD-ARF5M, which lacks the CTD dimerization domain, with either an IAA17 or IAA17mII effector gene resulted in reporter gene expression that was equivalent to that observed with the GD-ARF5M effector gene alone.
Aux/IAA Proteins Bring about Repression by Dimerizing with ARF Activators on AuxRE Target Sites
To further address whether Aux/IAA proteins function as repressors by being recruited to AuxREs through interactions with bound ARF activators rather than by simply titrating out ARF activators and preventing them from binding to AuxREs, the IAA17 repressor was converted to an activator. We took advantage of the fact that the IAA17mI/mII double mutant protein was stabilized by the mII mutation but that some of the repressor activity was lost as a consequence of the mI mutation (Tiwari et al., 2001). The VP16 activation domain was fused to the N terminus of the IAA17mI/mII repressor, creating the effector gene VP16-IAA17mI/mII. When VP16-IAA17mI/mII was transfected as the lone effector gene with the Gal4(4X)-GUS reporter gene, GUS activity was no greater than that observed with no effector gene (Figure 5A). By contrast, when the VP16-IAA17mI/mII effector gene was cotransfected with the GD-ARF5MC effector gene (GD-ARF5MC + VP16-IAA17mI/mII), GUS expression was increased in both the absence and the presence of auxin compared with that in protoplasts transfected with only the GD-ARF5MC effector gene. An IAA17mI/mII effector plasmid that lacked the VP16 activation domain repressed GUS expression when cotransfected with GD-5MC (GD-ARF5MC + IAA17mI/mII), showing that fusion of the VP16 activation domain to the IAA17mI/mII protein converted the mutant IAA17 protein from a repressor to an activator. To demonstrate that VP16-IAA17mI/mII was a transcriptional activator in its own right, this construct was fused to the Gal4 DBD (GD-VP16-IAA17mI/mII) and cotransfected with the Gal4(4X)-GUS reporter gene. When targeted to the promoter of the reporter gene by the Gal4 DBD, the GD-VP16-IAA17mI/mII effector gene activated transcription of the GUS reporter gene to approximately the same level as was observed with an effector gene that encoded the Gal4 DBD fused in frame with the VP16 activation domain (GD-VP16) or the cotransfected GD-ARF5MC + VP16-IAA17mI/mII effector genes.
Figure 5.
Dimerization of ARF and Aux/IAA Proteins Occurs on the Promoter to Bring about Repression.
Effector genes (diagrammed at top) were cotransfected along with a reporter gene into carrot suspension cell protoplasts and assayed in the presence (+ auxin) or absence (− auxin) of 10 μM 1-NAA. The GD-5MC and GD-1MC effector genes contain the yeast Gal4 DBD fused in frame to the ARF5 MR + CTD and the Gal4 DBD fused in frame to the ARF1 MR + CTD, respectively. The IAA17mI effector gene encodes an IAA protein with a mutation in conserved motif I, resulting in an unstable protein that is a weaker repressor than wild-type IAA17 (Tiwari et al., 2001). The VP16-IAA17mI effector gene contains a VP16 activation domain fused in frame to IAA17mI. The IAA17mI/mII effector gene encodes an IAA protein with a mutation in both conserved motifs I and II, resulting in a protein that is more stable than the wild-type IAA17 protein but a weaker repressor than the motif II mutant protein, IAA17mII (Tiwari et al., 2001). The GD-VP16-IAA17mI/mII effector gene encodes a protein containing a yeast Gal4 DBD fused in frame to the VP16 activation domain, which in turn is fused in frame to IAA17mI/mII. The VP16-IAA17mI/mII effector gene is identical to the GD-VP16-IAA17mI/mII effector gene but lacks the Gal4 DBD.
(A) Transfection assays were performed with a minimal promoter Gal4(4X)-GUS reporter gene (diagrammed at top)
(B) Transfection assays were performed with an auxin-responsive P3(4X)-GUS reporter gene (diagrammed at top).
(C) Transfection assays were performed with a constitutive promoter Gal4(4X)-D1-3(4X)-GUS reporter gene (diagrammed at top).
Two additional IAA17 effector constructs with mutations only in motif I (IAA17mI and VP16-IAA17mI) were tested along with the effector gene GD-ARF5MC for effects on reporter gene expression. These effector genes were predicted to encode proteins that are less stable than the motif I and II double mutant proteins and subject to auxin-regulated degradation. Figure 5A shows that, as predicted, in the absence of auxin, IAA17mI was a less effective repressor than IAA17mI/mII and VP16-IAA17mI was a less effective activator than VP16-IAA17mI/mII. Unlike IAA17mI/mII, repression by IAA17mI was relieved in the presence of auxin. Furthermore, activation by VP16-IAA17mI was reduced (i.e., nearly equivalent to that observed with GD-ARFMC alone) compared with activation by VP16-IAAmI/mII in the presence of auxin. Weaker activation in the presence of auxin by VP16-IAAmI probably resulted from the auxin-promoted degradation of the protein that lacks a motif II mutation.
To determine if the VP16-IAA17mI/mII effector gene also was able to activate an auxin-responsive reporter gene, the VP16-IAA17mI/mII effector gene was cotransfected into carrot protoplasts with the auxin-responsive P3(4X)-GUS reporter gene. Figure 5B shows that cotransfection of this effector gene resulted in enhanced expression of the auxin-responsive reporter gene in both the presence and absence of auxin. The IAA17mI/mII effector gene, which lacks the VP16 activation domain, repressed reporter gene expression. With the auxin-responsive reporter gene, it is likely that the overexpressed modified or mutant IAA17 proteins are targeted to endogenous ARFs that occupy AuxREs in the promoter of the GUS reporter gene (Tiwari et al., 2001).
To determine whether ARF repressors, such as ARF1, also might interact with IAA17 in carrot protoplasts, an effector gene that encodes the Gal4 DBD fused in frame to the ARF1 MR plus CTD (GD-ARF1MC) was cotransfected with either an IAA17mI/mII or a VP16-IAA17mI/mII effector gene along with a constitutive Gal4(4X)-D1-3(4X)-GUS reporter gene. Figure 5C shows that neither the IAA17mI/mII repressor nor the VP16-IAA17mI/mII activator affected the expression of the reporter gene compared with the expression observed with GD-ARF1MC alone, whether in the presence or the absence of auxin. In each case, an equivalent amount of repression occurred relative to the no-effector control. This repression likely resulted from the ARF1 MR, and the results suggest the IAA17 does not dimerize with ARF1 to bring about additional repression in the case of IAA17mI/mII or activation in the case of VP16-IAA17mI/mII.
The results with the VP16-IAA17mI/mII effector construct are consistent with a model in which ARF activators bound to AuxREs recruit Aux/IAA proteins to the promoter, bringing about the repression of auxin response genes when auxin concentrations are low. Furthermore, the results suggest that Aux/IAA proteins do not simply titrate out ARF activators, preventing them from reaching their DNA target sites (see Discussion).
DISCUSSION
Protoplast transfection experiments presented in this study have provided new insight into which domains within ARF proteins are required for repression or activation, which ARF domains are required for ARFs to be targeted to AuxREs, and which ARF domains are required for an auxin response. The major findings of this study include the following results. ARF MRs function as activation or repression domains independently of ARF DBDs and CTDs. Q-rich MRs function as activation domains, whereas S-rich (i.e., ARF2 and ARF9), SP-rich (i.e., ARF1 and ARF4), and SG-rich (i.e., ARF3) MRs function as repression domains when targeted to an auxin-responsive promoter by an ARF DBD or to a promoter that is unresponsive to auxin by a heterologous (yeast Gal4) DBD. As isolated domains, ARF MRs are not responsive to auxin (i.e., the amount of activation or repression is equivalent regardless of whether protoplasts are treated or not treated with auxin). ARF DBDs are sufficient to target at least some ARFs to AuxREs, and neither an ARF MR nor a CTD dimerization domain is required for this targeting. As isolated domains, ARF DBDs are not responsive to auxin (i.e., homologous and heterologous activation domain activities are unaffected by auxin when these domains are targeted to AuxREs by ARF DBDs). With auxin-responsive reporter genes, ARF CTDs confer auxin responsiveness to ARF Q-rich MRs that are fused to ARF DBDs. With reporter genes that contain promoters with the heterologous DBD DNA binding sites and no AuxREs, ARF CTDs confer auxin responsiveness to ARF Q-rich MRs that are fused to a heterologous DBD (i.e., Gal4 DBD). In total, our results support a model for auxin-responsive gene expression that involves active repression by Aux/IAA proteins that are targeted to AuxREs by dimerizing with ARF activators located on those AuxREs (Figure 6).
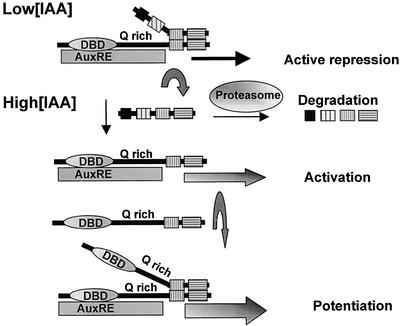
Figure 6.
Model for the Repression and Activation of Auxin Response Genes.
When auxin concentrations are low or below a threshold, early auxin response genes containing TGTCTC AuxREs are actively repressed, because Aux/IAA repressor proteins are dimerized to ARF transcriptional activators, preventing gene transcription. When auxin concentrations are increased, Aux/IAA proteins turn over more rapidly as a result of their being degraded more rapidly through the proteasome pathway (Rogg and Bartel, 2001; reviewed by Kepinski and Leyser, 2002). This more rapid degradation of Aux/IAA proteins effectively relieves the repression of early auxin response genes, resulting in gene activation. Gene activation might be enhanced further by the dimerization of ARF transcriptional activators to ARFs that are bound to AuxRE target sites. In this model, the auxin-sensitive target is the CTD dimerization domain, and the ARF DNA binding domain and activation/repression domain function independently of auxin.
Our previous analysis suggested that some ARFs appeared to be neither repressors nor activators when tested in carrot protoplast transfection assays (Ulmasov et al., 1999a); however, these assays were conducted with only the P3(4X)-GUS reporter gene, which contains repeats of a simple palindromic AuxRE. The results reported here indicate that ARF2, -3, -4, and -9, along with ARF1, function as repressors when assayed with the synthetic DR5(7X)-GUS reporter gene, suggesting that a given auxin-responsive reporter gene may influence whether an ARF with a S-rich, SG-rich, or SP-rich MR does or does not display repressor activity. It is possible that ARF3 and ARF4 efficiently bind to direct-repeat AuxREs, such as those found in DR5(7X)-GUS, but bind inefficiently to palindromic AuxREs, such as those found in P3(4X)-GUS. There are additional ARFs with S-rich, SL-rich, SP-rich, or SPL-rich MRs, which include ARF10 through ARF18 and ARF20 through ARF22 (for nomenclature, see Guilfoyle and Hagen, 2001), that have not been tested for repressor or activator function. Because these ARFs are related more closely in amino acid sequence to ARF1, -2, -3, -4, and -9, it is likely that they function as repressors on one or more AuxRE promoter-reporter genes. Seven of these ARFs (ARF12, -13, -14, -15, -20, -21, and -22) have highly similar amino acid sequences (i.e., most show >85% identity) and are related most closely to ARF9 (Liscum and Reed, 2002). One other ARF, ARF19, contains a Q-rich MR with poly-Q stretches and is most similar in overall amino acid sequence to ARF7 (Liscum and Reed, 2002). It is likely that ARF19 is a transcriptional activator like ARF5, -6, -7, and -8.
Using gel mobility shift assays, Ulmasov et al. (1997b) showed that the CTD of some ARFs (not including ARF1) or an artificial dimerization domain in the absence of the CTD facilitates their capacity to form stable complexes (i.e., distinct bands as opposed to smeared bands) on palindromic AuxREs in vitro. Furthermore, DNase I footprinting revealed that ARF1 bound as a dimer to a palindromic AuxRE, with each monomer occupying a half-site of the palindrome (i.e., each half-site being TGTCTC or its inverse, GAGACA). These results suggested that ARF dimers may bind palindromic AuxREs cooperatively in vitro with greater affinity than ARF monomers. The in vivo results presented here, however, indicate that the CTD is not required for ARF1 or ARF5 to function as a repressor or activator on a palindromic AuxRE. Although the CTD may promote the dimerization of ARFs and facilitate their binding to palindromic AuxREs that contain two TGTCTC binding sites (Ulmasov et al., 1999b), it is unlikely that ARFs must dimerize to bind AuxREs that consist of only a single TGTCTC element (i.e., composite AuxREs such as those found in the soybean GH3 promoter [Ulmasov et al., 1995]). Furthermore, selected ARF CTDs have been shown to facilitate the binding of ARFs to palindromic AuxREs only in vitro (i.e., in gel mobility shift assays [Ulmasov et al., 1999b]). In vivo conditions (e.g., in which accessory nuclear proteins may increase the affinity and/or stability of ARFs bound to AuxREs or in which some other aspect of the cellular environment contributes to ARF binding efficiency) may be sufficiently different from in vitro conditions that the CTD dimerization domain is not required for ARFs to bind stably to a palindromic AuxRE or other types of AuxREs.
Rather than the ARF CTD playing a role in ARFs reaching or binding to their DNA target sites, the CTDs likely function by providing a dimerization domain that regulates the auxin response (i.e., motifs III and IV in Aux/IAA proteins facilitate dimerization with related motifs in ARF proteins when auxin concentrations are low). Because Aux/IAA proteins can function as active repressors (Tiwari et al., 2001), dimerization between an ARF transcriptional activator bound to an AuxRE and an Aux/IAA protein would result in the repression of the auxin response gene. When auxin concentrations are increased, Aux/IAA repressors are degraded more rapidly via the proteasome pathway, resulting in the dissociation of Aux/IAA proteins from the ARF activators and the derepression of the auxin response gene (Gray et al., 2001; Tiwari et al., 2001; Zenser et al., 2001). We suggested previously that at high auxin concentrations, derepression of an auxin response gene may be activated further or potentiated by ARF activators being recruited to an ARF activator that is bound to the AuxRE. This latter interaction was proposed to occur by dimerization of the CTDs in two ARF proteins. Although published results are consistent with such ARF-ARF dimers potentiating gene activation (Ulmasov et al., 1999a), this type of dimerization has not been proven to occur on auxin response genes.
It is unclear if or how ARF repressors function in auxin-responsive gene expression. Perhaps another level of auxin-responsive gene regulation has not yet been revealed, or perhaps ARF repressors function on genes that are downregulated in response to auxin. Our results (Figure 5C), which indicate that ARF1 (and also ARF2 [data not shown]) does not interact effectively with IAA17 in carrot cells, suggests that ARF repressors, unlike ARF activators, may not be targets for Aux/IAA dimerization in plant cells. In our hands, ARF1 interacted poorly with IAA17 and several other Aux/IAA proteins in yeast two-hybrid assays as well (S. Tiwari, S. Doke, G. Hagen, and T. Guilfoyle, unpublished results). However, other results with yeast two-hybrid assays indicate that ARF1 does interact at least to some degree with IAA17 and other Aux/IAA proteins (Ulmasov et al., 1997b; Ouellet et al., 2001).
Our results that support the model described above (Figure 6) for auxin-responsive gene expression are based on transient transfection assays with plant protoplasts prepared from suspension cultured cells. These results may not reveal the entirety of the regulation of early auxin response genes because of the cell types used for transfections. It remains possible that in untransfected cells (i.e., normal plant cells that contain only endogenous ARFs), ARF-ARF dimerization might affect binding to natural promoters; however, because most natural TGTCTC promoters do not appear to be palindromic, it is unclear what role dimerization would play in the targeting of ARFs to a TGTCTC DNA binding site. In addition, protoplast transfection assays rely on reporter genes that are not integrated into chromosomal DNA and may not possess a chromatin structure identical to that of natural genes. Thus, it is possible that additional layers of regulation, which might involve the ARF DBD or MR directly, could be discovered. Nevertheless, our results indicate that the ARF CTD plays a pivotal role in conferring an auxin response on promoters of auxin response genes.
METHODS
Reporter and Effector Plasmids
The β-glucuronidase reporter genes used in this study have been described previously (Liu et al., 1994; Ulmasov et al., 1995, 1997a; Tiwari et al., 2001). Effector genes were placed under the control of the 35S double enhancer promoter of Cauliflower mosaic virus (CaMV) followed by the translational enhancer from the 5′ leader of Tobacco mosaic virus (Skuzeski et al., 1990). The 3′ untranslated region was derived from the nopaline synthetase gene (Skuzeski et al., 1990). Full-length Arabidopsis thaliana auxin response factor (ARF) effector genes contained a hemagglutinin-epitope tag at their N termini (Ulmasov et al., 1999a). For effector genes that encode truncated versions of Arabidopsis ARF proteins, domains in ARF proteins are referred to with a D for N-terminal DNA binding domain (DBD), M for the middle region (MR), and C for the C-terminal dimerization domain (CTD). Effector genes that contain truncated ARF proteins were generated using domain-specific oligonucleotides and PCR, and all of the plasmids were sequenced to confirm that no errors were introduced during PCR.
The effector genes ARF1DM and ARF5DM had their CTDs (including motifs III and IV) deleted after amino acids 512 and 766, respectively, from their N termini. The effector genes ARF1D-VP16 and ARF1D-5M contained the ARF1 DBD (amino acids 1 to 398) fused in frame to the VP16 activation domain of Herpes virus (amino acids 413 to 490) and the ARF5 MR (amino acids 349 to 766), respectively. The effector genes ARF5D-VP16 and ARF5D-1M contained the ARF5 DBD (amino acids 1 to 432) fused in frame to the VP16 activation domain (amino acids 413 to 490) and the ARF1 MR (amino acids 328 to 512), respectively. The effector genes GD-ARF1MC and GD-ARF5MC consisted of the Saccharomyces cerevisiae Gal4 DNA binding domain (amino acids 1 to 147 of the Gal4 protein; referred to as GD) cloned in frame to ARF1 and ARF5 proteins that had their N-terminal DBD, as defined by Ulmasov et al. (1997a)(1999b), deleted completely. The first amino acid from the N terminus for ARF1MC was residue 325 and that for ARF5MC was residue 349. The effector genes GD-ARF1M through GD-ARF9M had both their DBDs and CTDs deleted completely. The first and last amino acids from the N terminus for each ARFM truncation were as follows: ARF1M, 328 and 512; ARF2M, 398 and 764; ARF3M, 387 and 608; ARF4M, 409 and 676; ARF5M, 349 and 766; ARF6M, 351 and 797; ARF7M, 359 and 1041; ARF8M, 350 and 702; and ARF9M, 331 and 528.
IAA17 effector genes consisted of full length wild-type IAA17, IAA17mI, and IAA17mI/mII as described by Tiwari et al. (2001). The effector genes that encoded the VP16 activation domain fused in frame to the N terminus of mutated versions of IAA17 are referred to as VP16-IAA17, VP16-IAA17mI, and VP16-IAA17mI/mII. The GD-VP16-IAA17mI/mII effector gene contained an in-frame fusion of the Gal4 DNA binding domain at the N terminus. The GD-VP16 effector gene contained only the Gal4 DNA binding domain fused in frame to the VP16 activation domain.
Protoplast Transfection Assays
Protoplast isolation and transfection assays with carrot (Daucus carota) cells have been described previously (Liu et al., 1994; Ulmasov et al., 1995). Similar protocols were used for Arabidopsis protoplast isolation and transfection assays with the following modifications. After treatment of Arabidopsis (ecotype Columbia) suspension cells (Doelling and Pikaard, 1993) with cell wall–digesting enzymes, protoplasts were filtered through a Spectra/Mesh 200-μm nylon membrane (Spectrum Laboratories, Rancho Dominguez, CA). After the transfection assays, Arabidopsis protoplasts were suspended in 1 × Gamborg's B5 basal medium, 2 × Gamborg's vitamin solution, and 4% Suc (all from Sigma, St. Louis, MO).
Reporter and effector plasmid DNA was prepared using the EndoFree Plasmid Maxi Kit (Qiagen, Valencia, CA). Ten micrograms each of effector and reporter plasmids was cotransfected into protoplasts unless indicated otherwise, in which case either construct CaMV 35S promoter–chloramphenicol acetyltransferase (Ulmasov et al., 1999a) or construct CaMV 35S promoter–Gal4 DNA binding domain was used to equalize the amount of DNA used for the transfections. The efficiency of transfection was standardized with a CaMV 35S promoter–luciferase construct, which showed no response to auxin (Liu et al., 1994). All transfections were performed in triplicate, and at least two independent transfection assays were performed with protoplasts.
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes.
Acknowledgments
This work was supported by National Science Foundation Grant MCB 0080096 to T.G. and G.H.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.008417.
References
- Benfey, P.N. (2002). Auxin action: Slogging out of the swamp. Curr. Biol. 12, R389–R390. [DOI] [PubMed] [Google Scholar]
- Doelling, J.H., and Pikaard, C.S. (1993). Transient expression in Arabidopsis thaliana protoplasts derived from rapidly established cell suspension cultures. Plant Cell Rep. 12, 241–244. [DOI] [PubMed] [Google Scholar]
- Gray, W.M., Kepinski, S., Rouse, D., Leyser, O., and Estelle, M. (2001). Auxin regulates SCF TIR1-dependent degradation of Aux/IAA proteins. Nature 414, 271–276. [DOI] [PubMed] [Google Scholar]
- Guilfoyle, T.J. (1999). Auxin-regulated genes and promoters. In Biochemistry and Molecular Biology of Plant Hormones, P.J.J. Hooykaas, M.A. Hall, and K.R. Libbenga, eds (Leiden, The Netherlands: Elsevier), pp. 423–459.
- Guilfoyle, T.J., and Hagen, G. (2001). Auxin response factors. J. Plant Growth Regul. 10, 281–291. [Google Scholar]
- Guilfoyle, T.J., Hagen, G., Ulmasov, T., and Murfett, J. (1998). How does auxin turn on genes? Plant Physiol. 118, 341–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen, G., and Guilfoyle, T. (2002). Auxin-responsive gene expression: Genes, promoters and regulatory factors. Plant Mol. Biol. 49, 373–385. [PubMed] [Google Scholar]
- Hellmann, H., and Estelle, M. (2002). Plant development: Regulation by protein degradation. Science 297, 793–797. [DOI] [PubMed] [Google Scholar]
- Kepinski, S., and Leyser, O. (2002). Ubiquitination and auxin signaling: A degrading story. Plant Cell 14 (suppl.), S81.–S95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J., Harter, K., and Theologis, A. (1997). Protein-protein interactions among the Aux/IAA proteins. Proc. Natl. Acad. Sci. USA 94, 11786–11791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liscum, E., and Reed, J.W. (2002). Genetics of Aux/IAA and ARF action in plant growth and development. Plant Mol. Biol. 49, 387–400. [PubMed] [Google Scholar]
- Liu, Z.-B., Ulmasov, T., Shi, X., Hagen, G., and Guilfoyle, T.J. (1994). The soybean GH3 promoter contains multiple auxin-inducible elements. Plant Cell 6, 645–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouellet, F., Overvoorde, P.J., and Theologis, A. (2001). IAA17/AXR3: Biochemical insight into an auxin mutant phenotype. Plant Cell 13, 829–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed, J.W. (2001). Roles and activities of Aux/IAA proteins in Arabidopsis. Trends Plant Sci. 6, 420–425. [DOI] [PubMed] [Google Scholar]
- Rogg, L.E., and Bartel, B. (2001). Auxin signaling: Derepression through regulated proteolysis. Dev. Cell 1, 595–604. [DOI] [PubMed] [Google Scholar]
- Skuzeski, J.M., Nichols, L.M., and Gesteland, R.F. (1990). Analysis of leaky viral translation termination codons in vivo by transient expression of improved β-glucuronidase vectors. Plant Mol. Biol. 15, 65–79. [DOI] [PubMed] [Google Scholar]
- Tiwari, S.B., Wang, X.-J., Hagen, G., and Guilfoyle, T.J. (2001). Aux/IAA proteins are active repressors and their stability and activity are modulated by auxin. Plant Cell 13, 2809–2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov, T., Hagen, G., and Guilfoyle, T.J. (1997. a). ARF1, a transcription factor that binds auxin response elements. Science 276, 1865–1868. [DOI] [PubMed] [Google Scholar]
- Ulmasov, T., Hagen, G., and Guilfoyle, T.J. (1999. a). Activation and repression of transcription by auxin response factors. Proc. Natl. Acad. Sci. USA 96, 5844–5849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov, T., Hagen, G., and Guilfoyle, T.J. (1999. b). Dimerization and DNA binding of auxin response factors. Plant J. 19, 309–319. [DOI] [PubMed] [Google Scholar]
- Ulmasov, T., Liu, Z.-B., Hagen, G., and Guilfoyle, T.J. (1995). Composite structure of auxin response elements. Plant Cell 7, 1611–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov, T., Murfett, J., Hagen, G., and Guilfoyle, T.J. (1997. b). Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9, 1963–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenser, N., Ellsmore, A., Leasure, C., and Callis, J. (2001). Auxin modulates the degradation rate of Aux/IAA proteins. Proc. Natl. Acad. Sci. USA 98, 11795–11800. [DOI] [PMC free article] [PubMed] [Google Scholar]