Abstract
The Arabidopsis ethylene-overproducing mutants eto1, eto2, and eto3 have been suggested to affect the post-transcriptional regulation of 1-aminocyclopropane-1-carboxylic acid synthase (ACS). Here, we present the positional cloning of the gene corresponding to the dominant eto3 mutation and show that the eto3 phenotype is the result of a missense mutation within the C-terminal domain of ACS9, which encodes one isoform of the Arabidopsis ACS gene family. This mutation is analogous to the dominant eto2 mutation that affects the C-terminal domain of the highly similar ACS5. Analysis of purified recombinant ACS5 and epitope-tagged ACS5 in transgenic Arabidopsis revealed that eto2 does not increase the specific activity of the enzyme either in vitro or in vivo; rather, it increases the half-life of the protein. In a similar manner, cytokinin treatment increased the stability of ACS5 by a mechanism that is at least partially independent of the eto2 mutation. The eto1 mutation was found to act by increasing the function of ACS5 by stabilizing this protein. These results suggest that an important mechanism by which ethylene biosynthesis is controlled is the regulation of the stability of ACS, mediated at least in part through the C-terminal domain.
INTRODUCTION
The simple gas ethylene has been recognized as a plant hormone since the turn of the last century (Neljubov, 1901; Crocker and Knight, 1908; Knight et al., 1910; Funke et al., 1938). It has been shown to influence a diverse array of plant growth and developmental processes, including germination, leaf and flower senescence and abscission, fruit ripening, nodulation, and the response to a wide variety of stresses (Abeles et al., 1992). There have been significant advances in our understanding of ethylene signaling, derived mainly from molecular genetic studies in Arabidopsis (Bleecker and Kende, 2000; Alonso and Ecker, 2001; Schaller and Kieber, 2002; Wang et al., 2002). Although elucidation of the signaling apparatus is important in understanding the function of a signal, it is equally important to understand how the production of that signal is regulated.
The biosynthetic pathway for ethylene has been determined in a series of elegant studies by a number of researchers (Yang and Hoffman, 1984; Kende, 1989, 1993; Zarembinski and Theologis, 1994). 1-Aminocyclopropane-1-carboxylic acid (ACC) synthase (ACS), which converts S-adenosyl-Met to ACC, is the first committed and generally rate-limiting step in ethylene biosynthesis. The level of ACS activity closely parallels the level of ethylene production in most plant tissues (Acaster and Kende, 1983; Yang and Hoffman, 1984; Mattoo and Suttle, 1991). ACC is converted to ethylene by the enzyme ACC oxidase, the production of which also is highly regulated.
In the plant species that have been examined, ACS is encoded by a multigene family, different members of which are expressed differentially in response to various developmental, environmental, and hormonal factors. In Arabidopsis, there are 12 ACS genes (ACS1 to ACS12). The ACS3 gene most likely is a pseudogene, and ACS1 encodes a nonfunctional ACS (Liang et al., 1995). ACS4 is an auxin primary response gene (Abel et al., 1995), ACS5 is responsive to cytokinin (Vogel et al., 1998), and ACS6 is induced by exposure to ozone and other stimuli (Vahala et al., 1998). A fusion of the ACS2 promoter to a β-glucuronidase reporter gene revealed that ACS2 expression is high in young tissues and is switched off as the tissue matures, and its expression also is correlated with lateral root formation (Rodrigues-Pousada et al., 1993). Thus, as in other plant species, the Arabidopsis ACS gene family displays differential transcriptional responses to various inducers.
Several reports have indicated that ACS also is regulated post-transcriptionally. In suspension-cultured cells of parsley and tomato, the increase of ACS activity observed in response to elicitor was insensitive to inhibitors of RNA transcription (Chappell et al., 1984; Felix et al., 1991). Protein phosphorylation has been implicated in the regulation of the turnover of ACS in elicitor-stimulated tomato suspension cells (Spanu et al., 1994); the addition of protein phosphatase inhibitors greatly enhanced the induction of ACS activity in response to elicitor, and the inhibition of Ser/The protein kinases blocked this induction. Furthermore, ACS from tomato fruit tissue has been shown to be phosphorylated by a calcium-dependent protein kinase (Tatsuki and Mori, 2001).
Three mutants affected in the regulation of ethylene biosynthesis were identified in Arabidopsis based on a constitutive triple-response phenotype that was the result of ethylene overproduction (Guzman and Ecker, 1990; Kieber et al., 1993). Etiolated seedlings of these eto (ethylene-overproducing) mutants produce from 10- to 40-fold more ethylene than wild-type seedlings. eto1 is inherited as a recessive mutation, and eto2 and eto3 are dominant. The eto2 mutation results from a 1-bp insertion in ACS5 that is predicted to disrupt the C-terminal 12 amino acids of ACS5 (Vogel et al., 1998). The eto2 mutation does not increase the steady state level of ACS5 mRNA, which suggests that the mutation acts post-transcriptionally (Vogel et al., 1998). Furthermore, in both eto1 and eto3, the steady state levels of all of the ACS transcripts examined were not significantly higher than those observed in wild-type seedlings, although the level of ACS activity from eto1 and eto3 etiolated seedlings was highly increased (Woeste et al., 1999). These results suggest that these mutants also are affected in the post-transcriptional regulation of ACS.
To further understand the mechanism by which the eto mutants and the phytohormone cytokinin regulate ethylene biosynthesis post-transcriptionally, we cloned ETO3 and found that a single amino acid change in the C terminus of ACS9 was responsible for the ethylene-overproducing phenotype. In addition, we analyzed the degradation rate of wild-type and eto2 ACS5 protein using epitope-tagged proteins expressed in wild-type and eto1 mutant Arabidopsis seedlings. Together, our results suggest that all three eto mutations, as well as cytokinin treatment, affect the half-life of ACS proteins.
RESULTS
The eto3 Mutation Results from a Single Amino Acid Change in the C Terminus of ACS9
Previous studies of the ethylene-overproducing mutants eto1 and eto3 suggested that these mutations affect the post-transcriptional regulation of ACS (Woeste et al., 1999). To learn more about the mechanism by which ACS is regulated at the post-transcriptional level, the ETO3 gene was cloned using a map-based approach. The eto3 mutation was mapped to chromosome 3 between the cleaved amplified polymorphic sequence markers ALS and PUR5 using a mapping population generated from a cross of eto3 (Columbia [Col] ecotype) to the Wassilewskija (Ws) ecotype (Figure 1A). A database search of this region revealed that a predicted gene with strong similarity to ACS (At3g49700; ACS9) is located on a BAC clone (T16K5) that lies at a position consistent with the map position of eto3 relative to these two markers. The eto3 mutation is inherited as a single-gene dominant mutation, similar to the eto2 mutation, which was found to be the result of an alteration of the C terminus of ACS5 (Vogel et al., 1998). Thus, ACS9 was a strong candidate for ETO3.
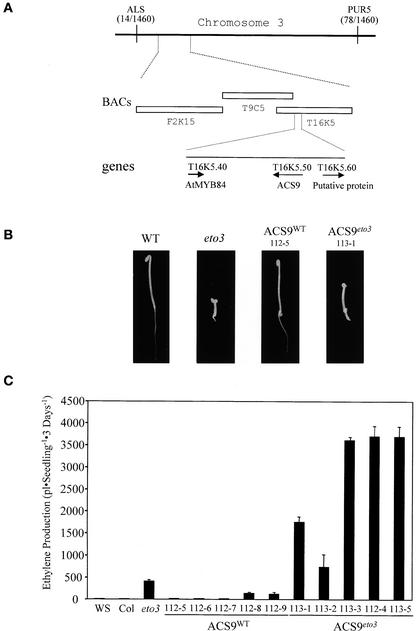
Figure 1.
Cloning of the ETO3 Gene.
(A) Physical map of the eto3 locus. A mapping population of a backcross to the Ws ecotype was used to map eto3 between the ALS and PUR5 markers on chromosome 3. A total of 730 F2 seedlings were analyzed for these markers, and the number of crossovers to each marker is indicated in parentheses. The three BACs in the region corresponding to this position are indicated, as is the position of the ACS9 gene.
(B) Seedling phenotypes of various lines. Seedlings were grown for 3 days in the dark at 23°C on Murashige and Skoog (1962) (MS) medium in the absence of exogenous ethylene, and representative seedlings were picked and photographed. Genotypes are noted at top. ACS9WT and ACS9eto3 refer to transgenic lines transformed with the wild-type (WT) and eto3 versions of the ACS9 genomic region, respectively (see Methods).
(C) Ethylene production from various etiolated seedlings. Wild-type (Ws), eto3 mutant, and independent transgenic (112-x = ACS9WT and 113-x = ACS9eto3) seedlings were grown for 3 days on MS agar in capped gas chromatography (GC) vials, and the accumulated ethylene was measured as described in Methods. Values shown are means ± sd (n = 3) of ethylene produced.
Sequencing of genomic DNA revealed that the eto3 mutant had a T-to-A transversion within the ACS9 coding region that results in a predicted change of Val-457 to an Asp residue. To determine whether this single amino acid change in the C terminus of ACS9 is responsible for the ethylene-overproducing phenotype of eto3, 3.2-kb genomic DNA fragments containing either the wild-type ACS9 (ACS9WT) or the eto3 ACS9 (ACS9eto3) coding region and associated flanking sequences were cloned into a binary plant transformation vector and introduced into wild-type Arabidopsis. Five of five independent transformants with ACS9eto3 displayed a constitutive triple-response phenotype as etiolated seedlings (Figure 1B). Ethylene production by these ACS9eto3 etiolated seedlings was highly increased compared with that of nontransgenic, wild-type seedlings and was twofold to ninefold higher than that of homozygous eto3 mutants (Figure 1C). Several of these lines displayed an adult morphology suggestive of a constitutive ethylene response (i.e., similar to the phenotype of the ctr1 mutant) (Kieber et al., 1993), indicating that they also are likely to overproduce ethylene as adults. By contrast, seven of nine independent transgenic ACS9WT seedlings displayed a wild-type phenotype as etiolated seedlings and ethylene production levels that were similar to those of wild-type, nontransgenic plants (Figure 1C). Two of the ACS9WT transgenic lines (112-8 and 112-9) displayed a constitutive triple-response phenotype as etiolated seedlings and produced eightfold and ninefold more ethylene than wild-type, nontransgenic plants, which still was significantly less than the level of ethylene produced by homozygous eto3 mutant seedlings. These overproducing ACS9WT transgenic lines likely are the result of increased ACS9 copy number and/or an increased basal level of ACS9 expression as a result of a position effect. Together, these results indicate that the V457D change in ACS9 is the cause of the ethylene-overproducing phenotype of eto3 etiolated seedlings.
To determine if increased ACS9 mRNA levels contribute to the increased ethylene biosynthesis observed in eto3 etiolated seedlings, ACS9 transcript levels in wild-type and eto3 seedlings were analyzed by real-time reverse transcriptase–mediated PCR. The cycle threshold (CT) values were determined with ACS9 gene-specific primers and were normalized to a product derived from actin-specific primers. The CT value is defined as the PCR cycle number at which the fluorescence signal of amplified PCR products crosses a specific threshold; the lower the CT value, the more templates are present for that specific primer pair (Dhar et al., 2002). Each dissociation curve displayed a single peak at the expected melting temperature for each product, and successful amplification of ACS9 and actin genes was monitored using agarose gel electrophoresis (data not shown). From three independent assays, the average normalized CT of ACS9 from eto3 was comparable to that of the wild type, indicating that the steady state level of ACS9 mRNA is not increased in the eto3 mutant etiolated seedlings (Table 1). Thus, like the eto2 mutation, eto3 affects the post-transcriptional control of the mutated ACS.
Table 1.
Levels of ACS9 mRNA in Wild-Type and eto3 Seedlings
| Sample | CTnor (Experiment 1) a | CTnor (Experiment 2) | CTnor (Experiment 3) | CTnor (mean ± sd) | ACS9 mRNAb |
|---|---|---|---|---|---|
| Wild type | 6.95 | 6.36 | 6.53 | 6.61 ± 0.30 | 1.0 |
| eto2 | 7.23 | 6.99 | 6.07 | 6.76 ± 0.61 | 0.9 |
CTnor is the cycle threshold value of the ACS9 product normalized to the CT value of an actin control.
Relative mRNA levels were determined by normalizing average CTnor of eto3 to that of the wild type.
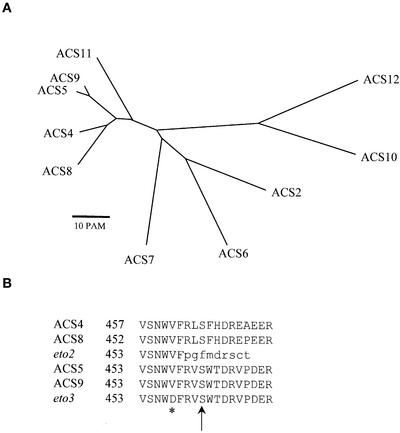
A computer-assisted analysis of the complete Arabidopsis genomic sequence revealed that the ACS gene family consists of 10 genes (Liang et al., 1995), each of which is associated with a corresponding cDNA sequence (Figure 2A). Comparison of predicted amino acid sequences of the ACS genes revealed that ACS9 is most similar to ACS5 (91% identity at the amino acid level). Phylogenetic analysis indicates that ACS9, ACS5, ACS4, and ACS8 form a distinct clade within the ACS family and are 45 to 65% identical to the other ACS proteins (Figure 2A). Although the C-terminal regions of other ACS isogenes are not highly conserved, the sequences of the C-terminal 18 amino acids of ACS4, ACS5, ACS8, and ACS9 are very similar (Figure 2B). The mutant phenotypes of eto2 and eto3 both result from alterations within the C-terminal regions of ACS proteins from this clade (Figure 2B). The analysis of eto3 further supports the notion that the conserved C termini of this group of ACS proteins play an important role in the regulation of ACS function and suggests that similar regulatory mechanisms could be involved in the regulation of these proteins.
Figure 2.
Comparison of Arabidopsis ACS Genes.
(A) Phylogenetic analysis of Arabidopsis ACS genes. An unrooted phylogenetic tree was derived from the predicted amino acid sequences of the Arabidopsis ACS proteins using the AllAll program of the Molecular Biology Computational Resource at the Baylor College of Medicine (http://cbrg.inf.ethz.ch/subsection3_1_1.html). This program uses a least-squared heuristic method to calculate trees. The lengths of the branches correspond to the point accepted mutation (1 point accepted mutation = 1 change per 100 residues) distances between the sequences, and the length of each branch is proportional to the evolutionary distance between the nodes.
(B) Alignment of the C-terminal domains of ACS4, ACS8, ACS5, ACS9, and mutants eto2 and eto3. The altered residue in eto3 is marked with an asterisk, and the affected amino acids predicted by the eto2 mutation are indicated in lowercase letters. The predicted site of phosphorylation is shown with an arrow (Tatsuki and Mori, 2001).
The eto2 Mutation Does Not Affect the Specific Activity of ACS5
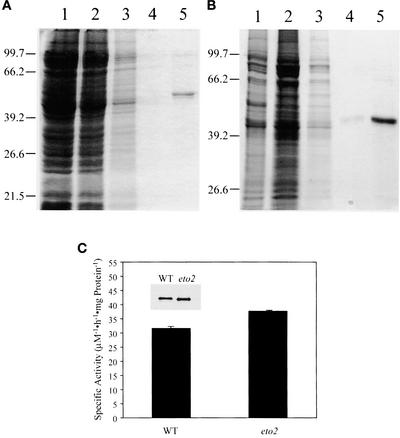
One explanation for the increased ethylene production in eto2 and eto3 mutant seedlings is that the C terminus of these ACS enzymes acts as an autoinhibitory domain; thus, a mutation in this domain would lead to increased ACS function by increasing the intrinsic activity of the enzyme. To test this possibility, wild-type and eto2 ACS5 were expressed in Escherichia coli, purified, and analyzed. Both proteins were made using the IMPACT system (New England Biolabs, Beverly, MA). The apparent molecular mass of purified, recombinant, wild-type ACS5 as determined by SDS-PAGE was ∼55 kD, in good agreement with its predicted mass (Figure 3A). The eto2 ACS5 is slightly smaller as a result of the truncation of the C terminus. The specific activity of purified eto2 ACS5 was very similar (119%) to that of wild-type ACS5 (Figure 3C). Because ethylene production of eto2 etiolated seedlings is ∼20-fold higher than that of wild-type seedlings, the small increase in specific activity of eto2 ACS5 observed in vitro cannot account for the increase in ACS5 function observed in eto2 mutant seedlings. Thus, either the C-terminal domain does not autoinhibit the catalytic activity of ACS5 or the eto2 mutant has only a minor effect on this function.
Figure 3.
Purification and Analysis of Wild-Type and eto2 ACS5 Proteins.
(A) SDS-PAGE analysis of the purification of ACS5WT. E. coli cells harboring the IMPACT-ACS5WT expression construct were induced, and the protein was extracted and applied to a chitin agarose column as described in Methods. Various extracts were analyzed by SDS-PAGE, and the proteins were visualized by Coomassie blue staining. Lane 1, crude extract; lane 2, column flow through; lane 3, column wash; lane 4, first fraction after the activation of intein protease; lane 5, second fraction after the activation of intein protease. The positions of migration of the molecular mass markers are indicated at left.
(B) SDS-PAGE analysis of the purification of ACS5eto2. Lanes are as in (A).
(C) Specific activity of purified recombinant ACS5 proteins. The fractions analyzed in lanes 5 from (A) and (B) were assayed for ACS activity as described in Methods. The activity was normalized to the amount of protein present. Values shown are means of three assays ± sd. The inset shows an immunoblot of proteins used in the assay probed with an anti-ACS5 antibody. WT, wild type.
Expression of Epitope-Tagged, Dexamethasone-Inducible ACS5
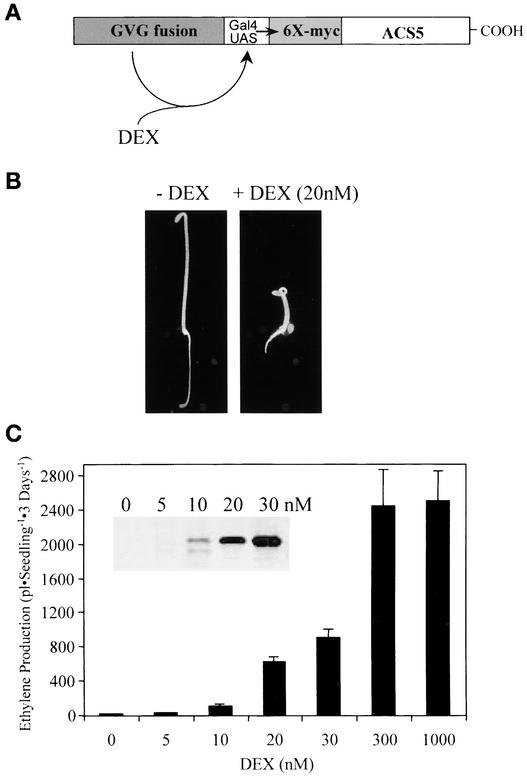
There are two remaining models to explain the increased ACS5 function in eto2 mutants: either the C terminus of ACS5 is the target of a negative modification in vivo, or the half-life of the eto2 ACS5 protein is increased compared with that of the wild type. To distinguish between these models, we expressed an epitope-tagged (6 × myc) wild-type version of ACS5 (myc-ACS5WT) and an eto2 version (myc-ACS5eto2) from a dexamethasone-inducible promoter in transgenic plants (Figure 4A). This system allows the quantification of the level of the ACS5 fusion protein in these transgenic plants using an anti-myc monoclonal antibody, which would have been difficult using anti-ACS5 antibodies because the Arabidopsis ACS proteins are highly conserved.
Figure 4.
System for Inducible Expression of Epitope-Tagged ACS5.
(A) Diagram of the inducible system used to express ACS5. The DEX-inducible system (Aoyama and Chua, 1997) uses a tripartite fusion transcription factor (GVG fusion) composed of the GAL4 DNA binding domain, the VP16 activation domain, and the glucocorticoid-responsive domain to drive DEX-inducible expression of the target gene. We placed the ACS5 coding region fused to a 6 × myc epitope tag downstream of the GAL4 operator sequences to which the GVG fusion protein binds, which results in DEX-responsive expression of the fusion protein.
(B) Phenotypes of 3-day-old etiolated seedlings from a transgenic line harboring the myc-ACS5WT transgene grown in the presence (+) and absence (−) of DEX. Seedlings were grown on MS medium, and representative seedlings were photographed.
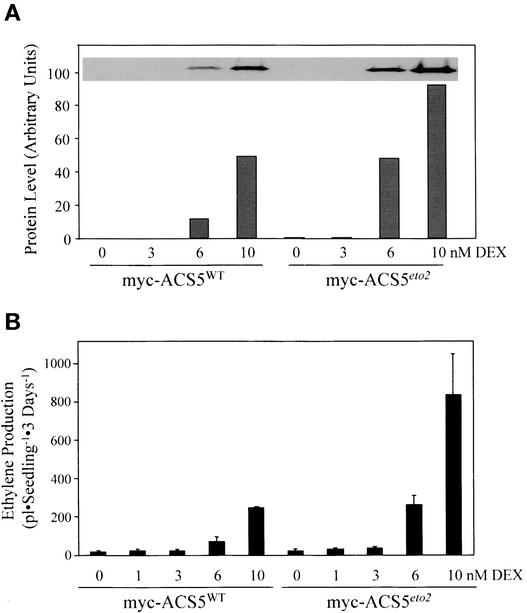
(C) Measurement of ethylene produced by 3-day-old etiolated seedlings of a myc-ACS5WT transgenic line grown on MS medium in the presence of increasing concentrations of DEX. The inset shows an immunoblot using the anti-myc monoclonal antibody as a probe of protein extracts from 3-day-old etiolated seedlings of the same line grown at the indicated concentrations of DEX.
Application of dexamethasone (DEX; a synthetic glucocorticoid) to multiple independent transgenic lines resulted in increased ethylene production from both myc-ACS5WT and myc-ACS5eto2 transgenic plants (Figures 4 and 5 and data not shown). We chose a single transgenic line for each of these two constructs that had a relatively low level of expression of the transgene to mimic the low level of endogenous ACS5.
Figure 5.
The eto2 Mutation Does Not Affect the in Vivo Specific Activity of ACS5.
(A) Quantification of myc fusion protein levels from myc-ACS5WT and myc-ACS5eto2 transgenic lines. Three-day-old etiolated seedlings were grown in the presence of various concentrations of DEX, and the proteins were extracted and analyzed by immunoblotting using an anti-myc monoclonal antibody. The relative level of myc antigen in each sample was quantified using a densitometer as described in Methods and is plotted as arbitrary units. The inset shows an image of the original immunoblot.
(B) Ethylene production from seedlings grown at various concentrations of DEX. The myc-ACS5WT and myc-ACS5eto2 transgenic lines were grown as in (A), and the amount of ethylene produced during the course of 3 days was measured as described in Methods.
The growth of the myc-ACS5WT transgenic line in the presence of DEX led to a constitutive triple-response phenotype in etiolated seedlings (Figure 4B). The level of ethylene produced in transgenic plants was increased by exogenous DEX in a dose-dependent manner, with a maximum at ∼300 nM DEX (Figure 4C). This finding is consistent with the observation that ACS is the rate-limiting step in ethylene biosynthesis in etiolated Arabidopsis seedlings (Woeste et al., 1999). There was a corresponding increase in the level of the myc-ACS5WT protein in response to DEX (Figure 4). For further analysis, low DEX concentrations were used, which resulted in an ∼3- to 10-fold increase in ethylene biosynthesis compared with that in wild-type etiolated seedlings, to obtain expression levels of ACS5 as low as possible but still allowing detection of the protein by immunoblot analysis.
The eto2 Mutation Affects the Stability of the ACS5 Protein
To determine if the increased ethylene production by eto2 seedlings was the result of an increase in the specific activity of ACS5 in vivo, we compared the level of ethylene produced by myc-ACS5WT and myc-ACS5eto2 transgenic plants and quantified the levels of the fusion proteins using immunoblot analysis (Figure 5). At comparable levels of expression of the fusion proteins, the myc-ACS5WT and myc-ACS5eto2 transgenic plants produced approximately equal levels of ethylene (Figure 5, cf. 10 and 6 nM DEX for wild type and eto2, respectively). This finding indicates that the specific activity of myc-ACS5eto2 is not significantly different from that of myc-ACS5WT in vivo and suggests that the stability of ACS5 may be affected by the eto2 mutation.
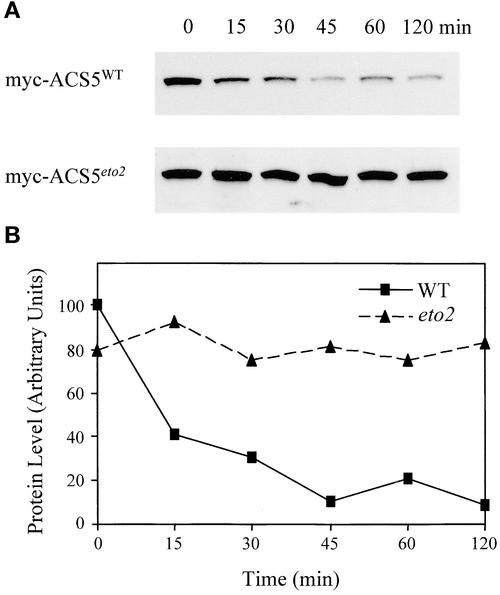
We determined the half-life of myc-ACS5WT and myc-ACS5eto2 in vivo by measuring the level of the fusion proteins at various times after the inhibition of protein synthesis. In brief, transgenic seedlings expressing approximately equal levels of myc-ACS5WT or myc-ACS5eto2 were washed with MS medium without DEX and then incubated in cycloheximide-containing MS medium to stop protein synthesis. Total proteins were extracted at various times, and the level of myc-ACS protein was determined by immunoblot analysis (Figure 6). Using this assay, the level of myc-ACS5WT was found to decline rapidly in the presence of cycloheximide, with a half-life of ∼15 min (Figure 6). The myc-ACS5WT protein reached a minimal level at 45 min after the application of cycloheximide and then remained stable. The failure to degrade the protein completely may reflect residual translation under the conditions used or may be the consequence of potentially ectopic expression of the fusion protein. By contrast, there was little if any decrease in the level of myc-ACS5eto2 protein even after 2 h of cycloheximide treatment, indicating that myc-ACS5eto2 had a much longer half-life compared with the wild-type protein. Thus, we conclude that the change in eto2 increases ACS5 function by means of an increased stability of the protein.
Figure 6.
Effect of the eto2 Mutation on ACS5 Protein Stability.
(A) Immunoblots of myc-ACS5WT and myc-ACS5eto2 transgenic lines after inhibition of protein synthesis. The indicated transgenic lines were grown for 3 days in the dark on MS plates containing 15 nM DEX for myc-ACS5WT and 10 nM DEX for myc-ACS5eto2. The seedlings were washed in liquid MS medium lacking DEX and then suspended in liquid MS lacking DEX but containing the protein synthesis inhibitor cycloheximide at time 0. At various times (indicated in minutes above each lane), the seedlings were harvested, and protein extracts were analyzed by immunoblotting using an anti-myc monoclonal antibody probe.
(B) Quantification of the myc-ACS5 fusion protein levels from (A). The level of myc fusion protein in each sample was quantified as described in Methods and plotted as a function of time after inhibition of protein synthesis. The half-life of the wild-type (WT) protein is estimated to be 15 min, and that of the eto2 protein is estimated to be >120 min.
Cytokinin Increases the Steady State Level of ACS5
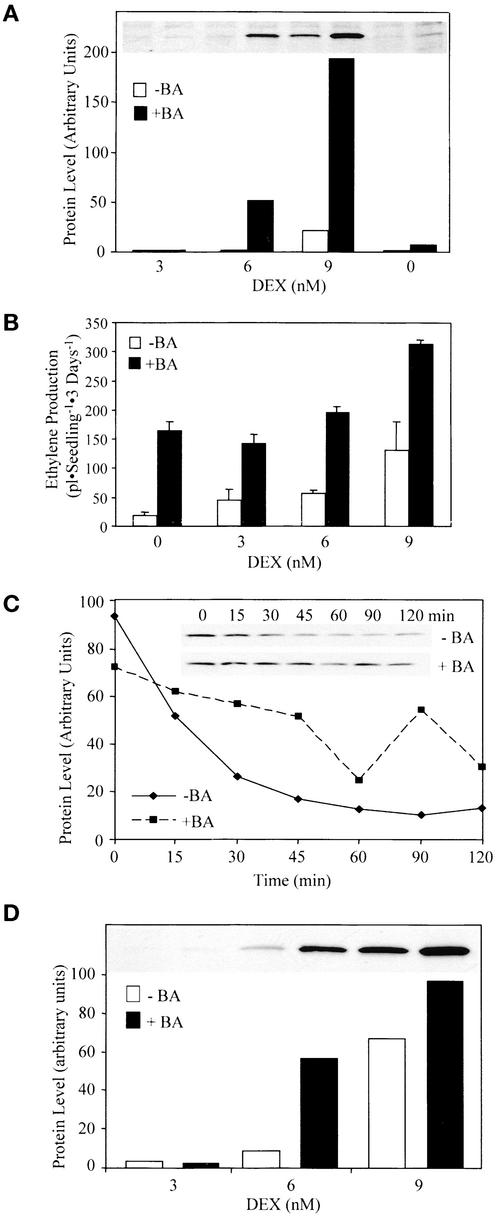
Cytokinin increases ethylene biosynthesis in etiolated Arabidopsis seedlings by increasing ACS5 function, which likely occurs by means of increased ACS5 protein activity or levels (Vogel et al., 1998). To characterize this effect, the level of the myc-ACS5WT fusion protein was measured in transgenic seedlings grown in the presence or absence of cytokinin (Figure 7A). Cytokinin caused an increase in the steady state level of myc-ACS5WT protein in etiolated seedlings (approximately ninefold at 9 nM DEX). The increased amount of myc-ACS5WT protein was partially reflected in the increase of ethylene biosynthesis (approximately twofold at 9 nM) in response to cytokinin (Figure 7B).
Figure 7.
Cytokinin Increases the Stability of the ACS5 Protein.
(A) Cytokinin causes an increase in the steady state level of myc-ACS5WT. Seedlings harboring the myc-ACS5WT transgene were grown in the dark for 3 days on MS medium containing the indicated amount of DEX in the presence of 5 μM benzyladenine (+BA) or a DMSO vehicle control (−BA) as indicated. Proteins were extracted from the seedlings and analyzed by immunoblotting using an anti-myc monoclonal antibody. The inset shows an image of the original film, and the graph is a depiction of the quantification of each signal. The lanes in the blot correspond to those indicated in the graph.
(B) Measurement of ethylene production from a myc-ACS5WT transgenic line. Seedlings were grown on MS medium containing the indicated amount of DEX for 3 days in the dark in the presence (closed bars) or absence (open bars) of 5 μM benzyladenine in capped GC vials, and the level of ethylene accumulated was measured using GC analysis.
(C) Cytokinin causes an increase in the half-life of ACS5. Myc-ACS5WT transgenic seedlings were grown for 3 days in the dark on MS plates containing 5 μM benzyladenine plus 15 nM DEX or a DMSO vehicle control plus 20 nM DEX. The seedlings were washed in liquid MS medium lacking DEX and then suspended in liquid MS medium lacking DEX plus either benzyladenine or DMSO and containing the protein synthesis inhibitor cycloheximide at time 0. At various times (indicated in minutes above each lane), the seedlings were harvested, and protein extracts were analyzed by immunoblotting using an anti-myc monoclonal antibody probe. The inset shows an image of the immunoblot that was quantified and the level of protein plotted versus time after the inhibition of protein synthesis.
(D) Cytokinin causes an increase in the steady state level of myc-ACS5eto2. Transgenic myc-ACS5eto2 seedlings were grown for 3 days in the dark on MS medium supplemented with 5 μM benzyladenine or a DMSO vehicle control and various concentrations of DEX as indicated. The proteins were extracted and analyzed by immunoblotting using an anti-myc monoclonal antibody probe. The inset shows an image of the protein blot that was quantified.
The rate of degradation of the myc-ACS5WT fusion protein in the presence or absence of cytokinin was measured as described above. As with the eto2 mutation, cytokinin treatment resulted in an increase in the half-life of myc-ACS5WT, suggesting that this hormone acts by increasing the stability of ACS5 (Figure 7C).
Because the C-terminal domain has been implicated in ACS5 turnover, we determined whether cytokinin increased ACS5 protein stability by acting through the C-terminal regulatory domain, as is the case with the eto2 mutation. If this model is correct, cytokinin treatment should not affect the level of myc-ACS5eto2 protein. To test this possibility, we examined the level of myc-ACS5eto2 protein in seedlings grown in the presence or absence of cytokinin. As with the wild-type protein, cytokinin caused an increase in the steady state level of myc-ACS5eto2 (Figure 7D). Furthermore, treatment of eto2 etiolated seedlings with cytokinin resulted in an increase in the level of ethylene produced, although to a lesser extent than the increase observed in wild-type seedlings (Table 2). Together, these data suggest that the eto2 mutation and cytokinin both act to stabilize the ACS5 protein. The observations that cytokinin still stabilizes the ACS5eto2 protein and increases ethylene production in eto2 mutant seedlings suggest that cytokinin acts by a mechanism that is partly independent of the C-terminal domain or that the eto2 mutation only partially affects the C-terminal signal targeting ACS5 for proteolysis.
Table 2.
Levels of Ethylene Produced by Wild-Type and eto2 Seedlings in Response to Cytokinin
| Ethylene Produced (pL·seedling−1·3 days−1)
|
|||
|---|---|---|---|
| Sample | Control | + 5 μM Benzyladenine | Fold Induction |
| Wild type | 15.7 ± 3.9 | 90.0 ± 7.8 | 5.7 |
| eto2 | 651 ± 104 | 1715 ± 124 | 2.6 |
The eto1 Mutation Increases Ethylene Biosynthesis Partially by Increasing the Half-Life of the ACS5 Protein
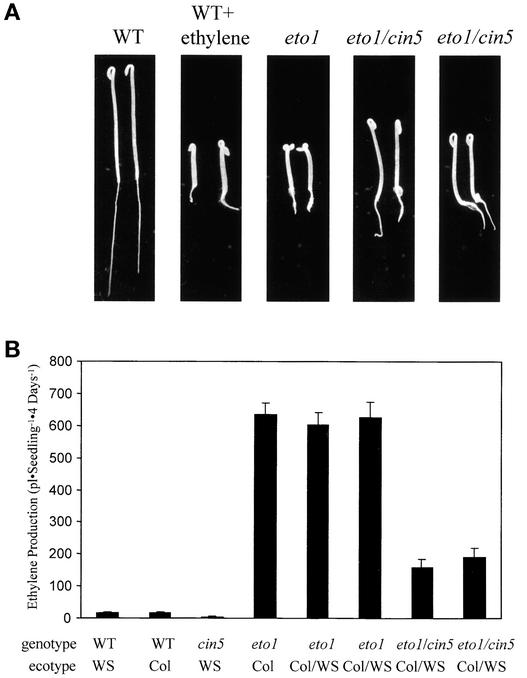
Previous studies suggested that the recessive eto1 mutation affected ethylene biosynthesis by means of a post-transcriptional regulation of ACS (Woeste et al., 1999). To determine if eto1 acts through the ACS5 isoform, we constructed a homozygous eto1 cin5 double mutant and analyzed its phenotype. The cin5 mutation is a loss-of-function allele of the ACS5 gene that reduces the level of ethylene produced in response to exogenous cytokinin (Vogel et al., 1998). The cin5 mutation significantly reduces the amount of ethylene produced by etiolated eto1 seedlings, resulting in a partial suppression of the constitutive triple-response phenotype (Figure 8). Thus, the eto1 mutant increases ethylene biosynthesis partly by increasing ACS5 function.
Figure 8.
The eto1 Mutation Acts Partially through the ACS5 Isoform.
(A) Seedling phenotypes of the wild type (WT), eto1, and eto1 cin5 double mutants (two different F2 lines). Seedlings were grown for 3 days in the dark on MS medium in air or ethylene as indicated, and representative seedlings were picked and photographed. Note that the double mutant lines have a phenotype intermediate between the wild-type and the eto1 parental lines.
(B) Ethylene production from various lines. Seedlings of the indicated genotypes were grown for 4 days in the dark on MS medium in capped GC vials, and the amount of accumulated ethylene was measured as described in Methods. The lines all are homozygous for the indicated mutations. The ecotype of each line is noted below the graph (Col/Ws indicates that the line is the F3 generation of a cross between Col and Ws parents). The original eto1 allele is in the Col ecotype, and cin5 is in Ws. Two independent F3 lines from a backcross of eto1 to a Ws line also are shown (eto1 Col/Ws).
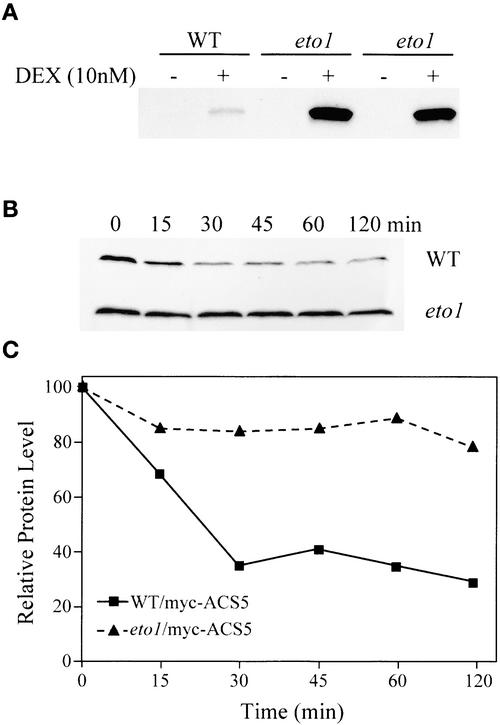
We examined the effect of the eto1 mutation on the level and stability of the myc-ACS5WT fusion protein by crossing the transgene into an eto1 mutant background. At equal levels of DEX, the eto1 line harboring the DEX-inducible myc-ACS5WT transgene produced up to 10-fold more ethylene than the wild-type parental transgenic line (data not shown). Because the eto1 mutation is unlikely to affect the transcription of this transgene, these data are consistent with a model in which the eto1 mutation affects the post-transcriptional regulation of ACS5. We sought to determine if the eto1 mutation increased ACS5 activity or protein levels. Immunoblot analysis indicated that in the presence of DEX, the eto1 line harboring the myc-ACS5WT transgene had a significantly higher level of the fusion protein than the wild-type parental transgenic line (Figure 9A). This was the result of the increased half-life of the myc-ACS5WT fusion protein in the eto1 mutant compared with the parental transgenic lines (Figures 9B and 9C). Thus, as is the case with cytokinin and the eto2 mutation, eto1 increases ACS5 function by increasing the stability of the protein.
Figure 9.
The eto1 Mutation Increases the Stability of the ACS5 Protein.
(A) The myc-ACS5WT transgene was crossed into the eto1 mutant, and lines doubly homozygous for the transgene and the eto1 mutation were isolated. Seedlings from the parental myc-ACS5WT transgenic line (WT) and two different F3 eto1/myc-ACS5WT mutant lines (eto1) were grown for 3 days in the dark on MS medium in the presence (+) or absence (−) of 10 nM DEX. The extracted proteins were analyzed by immunoblotting using an anti-myc monoclonal antibody probe.
(B) The eto1 mutation increases the half-life of the myc-ACS5WT fusion protein. Wild-type or eto1 mutant seedlings (indicated at right) harboring the myc-ACS5WT transgene (as in [A]) were grown for 4 days on MS medium containing either 10 nM DEX (WT) or 7.5 nM DEX (eto1) in the dark. The seedlings were washed in liquid MS medium lacking DEX and then suspended in liquid MS medium lacking DEX but containing the protein synthesis inhibitor cycloheximide at time 0. At various times (indicated in minutes above each lane), the seedlings were harvested, and protein extracts were analyzed by immunoblotting using an anti-myc monoclonal antibody probe.
(C) Quantification of the immunoblot shown in (B).
DISCUSSION
Previous studies suggested that the eto1, eto2, and eto3 mutants, as well as treatment with the phytohormone cytokinin, increased ethylene biosynthesis in etiolated Arabidopsis seedlings via a post-transcriptional mechanism (Vogel et al., 1998; Woeste et al., 1999). Here, we confirm this post-transcriptional control and show that in all cases this is the result of an increase in the stability of the ACS protein. The DEX myc-ACS5 system that was used here bypasses the normal transcriptional control of ACS5 and allows the specific quantification of this isoform using a fused epitope tag. Our results suggest that the stability of ACS enzymes plays an important role in regulating ethylene biosynthesis and that this stability is regulated in part by the C-terminal domain.
The C-terminal domain of recombinant tomato ACS has been shown to affect its activity and dimerization: deletion of the 46 to 52 C-terminal amino acids resulted in a monomeric enzyme that was reported to have fourfold higher specific activity than the full-length enzyme (Li and Mattoo, 1994). However, more recent analysis suggests that this deletion increases the specific activity of LE-ACS2 by only 20%, the previously reported larger difference being attributable to a decreased affinity of the antibody used to quantify the enzymes for the truncated form relative to the wild-type enzyme (Li and Mattoo, 1994; Tarun and Theologis, 1998). Interestingly, this increase is very close to the slight increase in specific activity that we observed in eto2 ACS5 (19%) and is consistent with our conclusion that the C-terminal domain does not have a major effect on the intrinsic catalytic activity of the enzyme.
Our results suggest that the rate of degradation of the ACS protein is an important aspect of its control. Several reports have demonstrated that ACS turns over rapidly in vivo. The half-life of ACS activity (as determined by the treatment of tissues with cycloheximide) has been reported to be 20 min in tomato leaves (Spanu et al., 1990), 25 min in mung bean hypocotyls (Yoshii and Imaseki, 1982), 40 min in tomato cell suspension cultures (Spanu et al., 1990), and 40 min and 2 h in green and pink tomato fruits, respectively (Kende and Boller, 1981). Kim and Yang (1992) examined the turnover of ACS protein (rather than activity) in tomato tissue using pulse-chase analysis and determined half-lives of 48 and 58 min in two separate experiments. This finding likely reflects an underestimation of the turnover rate (i.e., the actual half-life is likely to be shorter), because it would take some time to chase the relatively large pool of labeled Met that had accumulated in the plant cells. The half-life that we observed for ACS5 in Arabidopsis is shorter than that observed in most other systems. There are several possible explanations for this finding. First, in all other systems, the authors almost certainly measured a pool of ACS isoforms, whereas we examined only a single isoform in our system. Second, it is possible that the half-life of ACS in etiolated Arabidopsis seedlings is very short compared with that in other systems, or perhaps the myc tag increases the rate of degradation of the fusion protein. Another possibility is that the removal of the inducer from the DEX-inducible system, in conjunction with cycloheximide treatment, may allow a more rapid and efficient shutoff of de novo synthesis of ACS. In any case, it is clear that ACS5 is degraded rapidly in wild-type etiolated Arabidopsis seedlings.
The mechanism by which ACS is degraded in vivo is unknown. Several studies have indicated that ACS enzymes undergo mechanism-based inactivation, in which the substrate S-adenosyl-Met forms a covalent linkage to the Lys residue present in the active site in a fraction of the catalytic reactions, irreversibly inactivating the enzyme (Satoh and Esashi, 1986; Sato and Yang, 1988; Satoh and Yang, 1988), which presumably would lead to its rapid degradation in vivo. The half-life of ACS in vitro in the presence of saturating concentrations of S-adenosyl-Met has been reported to be similar to that of the in vivo ACS activity (Satoh and Esashi, 1986; Satoh et al., 1993). Furthermore, the inhibition of ACS activity by either the competitive inhibitor aminooxyacetic acid or the pyridoxal inhibitor aminoethoxyvinylglycine increased the half-life of ACS enzyme activity both in vitro and in vivo (Yoshii and Imaseki, 1982; Kim and Yang, 1992) and was shown in one case to increase the half-life of the ACS protein (Kim and Yang, 1992). These data support a model in which mechanism-based inactivation of ACS is a key component of the in vivo turnover of ACS. However, Spanu et al. (1990) reported that aminoethoxyvinylglycine did not affect the apparent turnover of ACS activity in tomato leaves and cultured cells, although their study was performed in the absence of cycloheximide, so the results may be confounded by de novo synthesis of the enzyme. It is possible that the eto mutations somehow affect the rate at which ACS5 undergoes mechanism-based inactivation in vivo, although this still would entail a subsequent degradation by a protease(s). Alternatively, ACS5 may be degraded independently of mechanism-based inactivation, and the eto mutants and cytokinin may affect this process.
The gene corresponding to the recessive eto1 mutation was cloned recently and reported to be similar to proteins with peptide binding domains (Cosgrove et al., 2000). ETO1 was shown to interact with wild-type ACS5 in a yeast two-hybrid assay, and this interaction was disrupted by the eto2 mutation (Cosgrove et al., 2000). The interaction of ETO1 with ACS5 could affect either the activity of the enzyme or its stability. Here, we demonstrate that eto1 acts in vivo partially through the ACS5 isoform and increases the stability of the protein strikingly. This finding suggests that eto1 mediates the turnover of ACS5 by means of an interaction with the C-terminal domain.
Protein phosphorylation has been implicated in the regulation of ACS function. The addition of a Ser/Thr protein kinase inhibitor reduced the ethylene production that occurs in response to elicitor application in tomato cell cultures, and blocking of protein phosphatase activity increased ACS activity in this same system (Spanu et al., 1994). The C-terminal domain of the tomato LE-ACS2 and LE-ACS3 enzymes has been shown to be phosphorylated by a calcium-dependent protein kinase (Tatsuki and Mori, 2001). Both LE-ACS2 and LE-ACS3 have C-terminal domains that are similar in sequence to ACS5 and ACS9, including conservation of the Ser residue that is the target of phosphorylation in the tomato proteins. Thus, it is likely that this Ser residue also is the target of a protein kinase in the Arabidopsis ACS5 and ACS9 proteins. Phosphorylation of the tomato enzymes had no effect on their in vitro activity, leading the authors to postulate that phosphorylation affected the stability of the protein. Interestingly, the eto3 mutation is predicted to result in the replacement of an uncharged Val with a negatively charged Asp residue. This substitution occurs very close to the Ser that is the likely target of phosphorylation in ACS9 (Figure 2). Thus, it is possible that the addition of the negative charge to the C terminus of ACS9 caused by the eto3 mutation mimics this phosphorylation and thus increases the stability of the enzyme.
The effect of cytokinin on ACS5 is complex. Although growth in the presence of cytokinin results in an approximately ninefold increase in the steady state level of myc-ACS5WT, we observed only an approximately twofold increase in ethylene production (Figures 7A and 7B). One explanation for this discrepancy is that the myc-ACS5WT fusion protein is not as active in the presence of cytokinin. A second possibility is that we measured the ethylene that accumulated during the course of 3 days but examined the steady state level of myc-ACS5WT only after 72 h of growth in the presence of cytokinin. It is possible that the increase in the level of myc-ACS5WT protein occurs relatively slowly, which would explain the difference between the level of ethylene that accumulated over 3 days and the level of protein observed at the 72-h time point. It also is possible that cytokinin could increase the conjugation of ACC or decrease the activity of ACC oxidase, which in either case would result in a lower level of ethylene production at equal ACS activities.
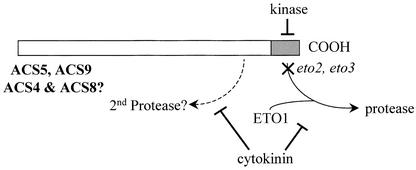
We propose the following model for the regulation of the stability of this group of ACS enzymes (Figure 10). The ETO1 protein interacts with the C-terminal domain of the ACS proteins and helps target them for proteolytic breakdown by an unidentified protease(s). Phosphorylation of the C terminus by a calcium-dependent kinase disrupts this interaction and thus increases the stability of the protein. The eto2 and eto3 mutations alter the C-terminal domain, in the latter case perhaps by mimicking phosphorylation, such that the mechanisms that target ACS for proteolysis are compromised, reducing the breakdown of the enzyme. Finally, cytokinin also reduces the turnover of the enzyme by a mechanism at least partially independent of the C-terminal domain.
Figure 10.
Model for the Regulation of ACS Protein Stability.
The C-terminal domains of ACS5 and ACS9 (and possibly also ACS4 and ACS9, because their C-terminal domains are highly similar) acts as targets for an unknown protease. The alteration of this domain caused by either the eto2 or the eto3 mutation disrupts the targeting of the protein to the proteolytic machinery. Interaction with the ETO1 protein is required for efficient turnover of the proteins. In addition, phosphorylation of the C-terminal domain may block its function as a proteolytic tag. Cytokinin acts at least partially to block the targeting of the enzyme by this domain for proteolysis. However, the observation that the eto2 mutant protein is partially stabilized by cytokinin treatment suggests that it also works through a distinct degradation pathway. See text for more details.
It is unclear how universal this mode of regulation will be among ACS enzymes. A subset of ACS proteins displays similarity to the ACS5 and ACS9 C-terminal domains, including the Arabidopsis ACS4 and ACS8 proteins and various ACS enzymes from other plant species. Thus, at least among this subset of ACS enzymes, the control of degradation directed by the C-terminal domain may be a conserved mechanism. Interestingly, Kende and Boller (1981) found a large increase in the stability of ACS activity in ripening tomatoes compared with green fruit, which suggests that the regulation of ACS turnover may play an important role in this developmental context. However, other ACS proteins have a C-terminal domain that is very diverged from the ACS5/ACS9 clade and thus may be regulated by a distinct mechanism. Numerous studies have demonstrated the importance of transcriptional control in the regulation of ACS function, and the post-transcriptional mechanism described here represents an additional level of control. Further studies should reveal other components important in the regulation of ACS protein turnover and the role that this plays in regulating ethylene production in response to various factors that increase ethylene production, such as wounding, fruit ripening, and leaf and flower senescence.
METHODS
Mapping of eto3
The eto3 mutant (ecotype Columbia [Col]) of Arabidopsis thaliana was backcrossed to the Wassilewskija (Ws) ecotype to generate a mapping population. F2 seeds were grown on Murashige and Skoog (1962) (MS) medium, and wild-type seedlings (tall phenotype) were transferred to soil and allowed to grow for 3 weeks. Genomic DNA was extracted from these plants (Edwards et al., 1991), and cleaved amplified polymorphic sequence markers polymorphic between the parental ecotypes were analyzed. The products of the cleaved amplified polymorphic sequence reaction were separated by agarose gel electrophoresis (3% agarose) and visualized by ethidium bromide staining.
Transformation of Arabidopsis with Wild-Type and eto3 ACS9
The ACS9 gene was amplified from wild-type (ecotype Col) and eto3 genomic DNA using oligonucleotide primers (gACS9 compF, 5′-GAGCTCGGACCTTGTGTCTGATTAACCCTA-3′; gACS9 compR, 5′-GGATCCCGTTACGTTATGGACTAAACCTTC-3′) with TaKaRa Ex Taq DNA polymerase as described by the manufacturer (Panvera, Madison, WI). The resulting 3.2-kb PCR product was composed of the ACS9 coding region, 960 bases of the 5′ flanking region, and the ACS9 3′ untranslated region. The PCR products were cleaved with SacI and BamHI and ligated to the plant transformation vector pCAMBIA2300 (http://www.cambia.org.au/main/r_et_vman.htm). The resulting plasmids then were transformed into wild-type Ws plants by the floral dip method (Clough and Bent, 1998). Transformants were selected on MS medium containing 50 μg/mL kanamycin.
Ethylene Measurements
Seed sterilization and ethylene measurements were conducted as described previously (Vogel et al., 1998). Arabidopsis seedlings were grown on MS medium containing 5 μM benzyladenine or a DMSO vehicle control in 22-mL gas chromatography (GC) vials, and the vials then were capped and incubated for 3 days at 23°C in the dark. The accumulated ethylene was measured by gas chromatography as described by Vogel et al. (1998).
Real-Time Reverse Transcriptase–Mediated PCR
Total RNA was prepared from 3-day-old etiolated wild-type and eto3 seedlings using the TRIZOL reagent as described by the manufacturer (Gibco BRL, Rockville, MD). cDNA was synthesized from 5 μg of total RNA using oligo(dT)12-18–primed reverse transcription with the SuperScript first-strand synthesis system (Gibco BRL). The following sets of primers were used for reverse transcriptase–mediated PCR to amplify the specific DNA fragments of ACS9 and actin: ACS9 rt F1 (5′-GTACGTAGAGTCAACAGATAGTAGAAGAGTGATT-3′) and ACS9 rt R1 (5′-GATTTGCTTTGTCTTAACTTGGGGC-3′) for ACS9 and ACTIN8 rt F (5′-TCCAGCAATGTGGATCTCTAAGGCA-3′) and ACTIN8 rt R (5′-TCCCGTCATGGAAACGATGTCT-3′) for actin. The SYBR Green reverse transcriptase–mediated PCR amplifications were performed in a 25-μL reaction volume containing 0.15× SYBR Green I nucleic acid gel stain (×10,000; Molecular Probes, Eugene, OR), 0.3 μM each of forward and reverse primers, 4 mM MgCl2, 0.2 mM deoxynucleotide triphosphates, 1× Pfx amplification buffer, 1 μL of cDNA, and 0.5 units of Platinum Pfx DNA polymerase (Invitrogen, Carlsbad, CA) using the SmartCycler system (Cephid, Sunnydale, CA). The temperature profile of the PCR for both ACS9 and actin was 94°C for 105 s, followed by 40 cycles of 94°C for 15 s and 64°C for 15 s, and finally 72°C for 15 s. A melt curve analysis was performed immediately after PCR by monitoring the fluorescence as the temperature was increased slowly from 60 to 95°C. An aliquot of the PCR product was run on a 3% agarose gel to confirm that each primer pair amplified a single band of the expected molecular mass. The threshold cycle was defined as the cycle at which a statistically significant increase in the fluorescence value above the threshold value was first detected (Dhar et al., 2002).
Expression and Purification of Recombinant Wild-Type and eto2 ACS5
The coding region of ACS5 was amplified from wild-type (ecotype Col) cDNA using oligonucleotide primers ACS5 impactS (5′-CATATG-AAACAGCTTTCGACAAAAG-3′) and ACS5 impactA (5′-CTCGAG-TCGTTCATCAGGTACACGATC-3′) and from eto2 cDNA using oligonucleotide primers ACS5 impactS (5′-CATATGAAACAGCTTTCG-ACAAAAG-3′) and eto2 impactA2 (5′-CTCGAGGGTACACGATCGGTCCATGAAA-3′). The PCR products were double digested with NdeI-XhoI restriction enzymes and ligated to the expression vector pTYB2 (New England Biolabs). Cells harboring the pTYB2-ACS5 recombinant plasmids were grown in Luria-Bertani medium containing 100 μg/mL ampicillin at 37°C until the OD600 reached 0.5 to 0.8. Isopropylthio-β-galactoside then was added to a final concentration of 0.5 mM, and the culture was transferred to 15°C and incubated overnight. The cells were harvested by centrifugation at 5000g for 10 min at 4°C, and the cell pellet was stored at −80°C.
Purification of the recombinant proteins was conducted as described by the manufacturer (New England Biolabs) with the following modifications. The frozen cell pellet was resuspended in 50 mL of lysis buffer (20 mM Hepes, pH 8.0, 500 mM NaCl, 1 mM EDTA, and protease inhibitors [Complete, EDTA-free; Roche Molecular Biochemicals, Mannheim, Germany]) and sonicated to disrupt the cells. The soluble ACS5 fusion protein was affinity purified using chitin resin as described by the manufacturer (New England Biolabs), and the ACS5 proteins were released from the chitin column by intein-mediated self-cleavage in the presence of 50 mM DTT, yielding ACS5 protein lacking any fused domain. The protein fractions from each purification step were analyzed by SDS-PAGE to monitor the purification. Protein concentrations were determined using the Bradford assay as described by the manufacturer (Bio-Rad).
ACS Assay
ACS activity was assayed using purified recombinant ACS5 as described previously (Peck and Kende, 1995) with the following modifications. Five microliters of the purified protein was placed into 22-mL GC vials containing 2 mL of buffer A (250 mM phosphate buffer, pH 8.0, 10 μM pyridoxal phosphate, 1 mM EDTA, 5 mM DTT, and protease inhibitors [Complete, EDTA-free; Roche Molecular Biochemicals]), and 100 μL of 5 mM S-adenosyl-Met was added. The mixture was incubated for 30 min at room temperature. The 1-aminocyclopropane-1-carboxylic acid formed was converted to ethylene by the addition of 100 μL of 20 mM HgCl, followed by 100 μL of a 1:1 mix of saturated NaOH:bleach (Lizada and Yang, 1979). The tubes were capped immediately after addition of the NaOH:bleach and incubated on ice for 10 min. Ten milliliters of headspace was removed with a syringe and injected into a new vial, and the ethylene was measured as described (Vogel et al., 1998). All reactions were performed in triplicate and compared with controls to which S-adenosyl-Met was not added.
Dexamethasone-Inducible myc-ACS5WT and myc-ACS5eto2 Expression
The coding regions of wild-type and eto2 ACS5 were fused to a 6 × myc cassette, and the resulting fusion protein was cloned into the binary GVG vector pTA7002 (Aoyama and Chua, 1997). Wild-type plants (ecotype Ws) were transformed with the plasmids by the floral dip method (Clough and Bent, 1998), and transformants were selected on MS medium containing hygromycin. T2 seedlings were grown on MS medium containing 10 μM dexamethasone (DEX) for 3 days to screen for lines that expressed the myc-tagged proteins at low levels in an inducible manner.
Immunoblot Analysis
Forty seedlings were ground in a 1.5-mL tube in 80 μL of 1 × SDS loading buffer (62.5 mM Tris, pH 6.8, 2% SDS, 5% β-mercaptoethanol, 10% glycerol, and 0.02% bromphenol blue), boiled for 3 min, and then centrifuged for 3 min at 16,000g. Twenty microliters of each extract was applied to a 10% SDS-PAGE gel and electrophoresed, and the proteins were electroblotted to a supported nitrocellulose membrane (Micron Separations, Westborough, MA) (Ausubel et al., 1994). To confirm the equal efficiency of protein extraction and loading, an identical parallel gel was stained with Coomassie Brilliant Blue R250. The C-myc epitope tag was detected using a monoclonal anti (c-myc)–peroxidase antibody as described by the manufacturer (Roche Molecular Biochemicals). Immunoblots were digitized and signal intensity was quantified using a Fluorochem densitometer (Alpha Innotech Corp., San Leandro, CA).
For immunodetection of recombinant ACS5, 10 ng of purified proteins was analyzed by SDS-PAGE and transferred to nitrocellulose as described above. The ACS5 proteins were detected using a rabbit polyclonal antiserum against a full-length ACS5 protein made in Escherichia coli as described above followed by incubation with alkaline phosphatase–linked goat anti-rabbit Ig (Chemicon International, Temecula, CA).
Analysis of Protein Stability
Transgenic seedlings harboring DEX-inducible myc-ACSWT and myc-ACSeto2 were grown on filter paper on MS medium containing the amounts of DEX indicated in the figures for 3 days at 22°C in the dark. The seedlings were washed twice with liquid MS medium and then transferred to liquid MS medium containing 100 μM cycloheximide in the presence of 5 μM benzyladenine or a DMSO vehicle control. After incubation in the dark for the times indicated in the figures, total proteins were extracted and used for immunoblot analysis as described above.
Analysis of eto1 cin5 Double Mutants and eto1/myc-ACS5
The eto1-1 cin5-1 double mutant was obtained by crossing homozygous eto1 and cin5 lines. The cin5 allele used contains a T-DNA inserted (encoding kanamycin resistance) within the ACS5 promoter and is phenotypically a null allele (Vogel et al., 1998). The F1 population was allowed to self, and kanamycin-resistant F2 seedlings displaying a constitutive triple response were selected. PCR was used to identify two lines that were homozygous for the T-DNA insertion in ACS5.
eto1/myc-ACSWT plants were obtained by crossing a homozygous eto1 mutant to the same myc-ACSWT transgenic line that was used for all of the other experiments described in this article. The F2 population was selected on MS plates containing hygromycin and allowed to self. Two lines in the F3 generation that displayed a constitutive triple-response phenotype and hygromycin resistance were identified and used for further analysis.
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes.
Acknowledgments
We thank Nam-Hai Chua for providing the DEX-inducible system and Claire Hutchison for critical reading of the manuscript. This work was supported by U.S. Department of Agriculture Grants 2000-01540 and 97-01425 to J.J.K. and by a Korea Science and Engineering Foundation postdoctoral fellowship to H.S.C.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.006882.
References
- Abel, S., Nguyen, M.D., Chow, W., and Theologis, A. (1995). ACS4, a primary indoleacetic acid-responsive gene encoding 1-ami-nocyclopropane-1-carboxylate synthase in Arabidopsis thaliana. J. Biol. Chem. 270, 19093–19099. [DOI] [PubMed] [Google Scholar]
- Abeles, F.B., Morgan, P.W., and Saltveit, M.E., Jr. (1992). Ethylene in Plant Biology. (San Diego, CA: Academic Press).
- Acaster, M.A., and Kende, H. (1983). Properties and partial purification of 1-aminocyclopropane-1-carboxylate synthase. Plant Physiol. 72, 139–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso, J., and Ecker, J. (2001). The ethylene pathway: A paradigm for plant hormone signaling and interaction. Sci. STKE 70, RE1. [DOI] [PubMed] [Google Scholar]
- Aoyama, T., and Chua, N.-H. (1997). A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J. 11, 605–612. [DOI] [PubMed] [Google Scholar]
- Ausubel, F.M., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., Smith, J.A., and Struhl, K. (1994). Current Protocols in Molecular Biology. (New York: John Wiley & Sons).
- Bleecker, A.B., and Kende, H. (2000). Ethylene: A gaseous signal molecule in plants. Annu. Rev. Cell Dev. Biol. 16, 1–18. [DOI] [PubMed] [Google Scholar]
- Chappell, J., Hahlbrock, K., and Boller, T. (1984). Rapid induction of ethylene biosynthesis in cultured parsley cells by fungal elicitor and its relationship to the induction of phenylalanine ammonia lyase. Planta 161, 475–480. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Cosgrove, D.J., Gilroy, S., Kao, T.-h., Ma, H., and Schultz, J.C. (2000). Plant signaling 2000: Cross talk among geneticists, physiologists, and ecologists. Plant Physiol. 124, 499–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crocker, W., and Knight, L.I. (1908). Effect of illuminating gas and ethylene upon flowering carnation. Bot. Gaz. 46, 259–276. [Google Scholar]
- Dhar, A.K., Roux, M.M., and Klimpel, K.R. (2002). Quantitative assay for measuring the Taura syndrome virus and yellow head virus load in shrimp by real-time RT-PCR using SYBR Green chemistry. J. Virol. Methods 104, 69–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards, K., Johnstone, C., and Thompson, C. (1991). A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res. 6, 1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felix, G., Grosskopf, D.G., Regenass, M., Basse, C., and Boller, T. (1991). Elicitor-induced ethylene biosynthesis in tomato cells: Characterization and use as a bioassay for elicitor action. Plant Physiol. 97, 19–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funke, G.L., DeCoeyer, F., DeDecker, A., and Maton, J. (1938). The influence of the emanation of apples on several life phenomena of plants. Biol. Jaarb. 5, 335–381. [Google Scholar]
- Guzman, P., and Ecker, J.R. (1990). Exploiting the triple response of Arabidopsis to identify ethylene-related mutants. Plant Cell 2, 513–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kende, H. (1989). Enzymes of ethylene biosynthesis. Plant Physiol. 91, 1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kende, H. (1993). Ethylene biosynthesis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 44, 283–307. [Google Scholar]
- Kende, H., and Boller, T. (1981). Wound ethylene and 1-aminocyclopropane-1-carboxylate synthase in ripening tomato fruit. Planta 151, 476–481. [DOI] [PubMed] [Google Scholar]
- Kieber, J.J., Rothenburg, M., Roman, G., Feldmann, K.A., and Ecker, J.R. (1993). CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the Raf family of protein kinases. Cell 72, 427–441. [DOI] [PubMed] [Google Scholar]
- Kim, W.T., and Yang, S.F. (1992). Turnover of 1-aminocyclopropane-1-carboxylic acid synthase protein in wounded tomato fruit tissue. Plant Physiol. 100, 1126–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight, L.I., Rose, R.C., and Crocker, W. (1910). Effects of various gases and vapors upon etiolated seedlings of the sweet pea. Science 31, 635–636. [Google Scholar]
- Li, N., and Mattoo, A.K. (1994). Deletion of the carboxyl-terminal region of 1-aminocyclopropane-1-carboxylic acid synthase, a key protein in the biosynthesis of ethylene, results in catalytically hyperactive monomeric enzyme. J. Biol. Chem. 269, 6908–6917. [PubMed] [Google Scholar]
- Liang, X., Oono, Y., Shen, N.F., Köhler, C., Li, K., Scolnik, P.A., and Theologis, A. (1995). Characterization of two members (ACS1 and ACS3) of the 1-aminocyclopropane-1-carboxylate synthase gene family of Arabidopsis thaliana. Gene 167, 17–24. [DOI] [PubMed] [Google Scholar]
- Lizada, M.C.C., and Yang, S.F. (1979). A simple and sensitive assay for 1-aminocyclopropane-1-carboxylic acid. Anal. Biochem. 100, 140–145. [DOI] [PubMed] [Google Scholar]
- Mattoo, A.K., and Suttle, J.C. (1991). The Plant Hormone Ethylene. (Boca Raton, FL: CRC Press).
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473–497. [Google Scholar]
- Neljubov, D. (1901). Uber die horizontale Nutation der Stengel von Pisum sativum und einiger Anderer. Pflanzen Beih. Bot. Zentralb. 10, 128–139. [Google Scholar]
- Peck, S.C., and Kende, H. (1995). Sequential induction of the ethylene biosynthetic enzymes by indole-3-acetic acid in etiolated peas. Plant Mol. Biol. 28, 293–301. [DOI] [PubMed] [Google Scholar]
- Rodrigues-Pousada, R.A., De Rycke, R., Dedonder, A., van Caeneghem, W., Engler, G., van Montagu, M., and Van der Straeten, D. (1993). The Arabidopsis 1-aminocyclopropane-1-car-boxylate synthase gene 1 is expressed during early development. Plant Cell 5, 897–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato, T., and Yang, S.F. (1988). Inactivation of 1-aminocyclopropane-1-carboxylic acid synthase by l-vinylglycine as related to the mechanism-based inactivation of the enzyme by S-adenosyl-l-methionine. Plant Physiol. 91, 1036–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh, S., and Esashi, Y. (1986). Inactivation of 1-aminocyclopropane-1-carboxylic acid synthase of etiolated mung bean hypocotyl segments by its substrate, S-adenosyl-l-methionine. Plant Cell Physiol. 27, 285–291. [Google Scholar]
- Satoh, S., Mori, H., and Imaseki, H. (1993). Monomeric and dimeric forms and the mechanism-based inactivation of 1-aminocyclopropane-1-carboxylate synthase. Plant Cell Physiol. 34, 752–760. [Google Scholar]
- Satoh, S., and Yang, S.F. (1988). S-Adenosylmethionine-dependent inactivation and radiolabeling of 1-aminocyclopropane-1-carboxylate synthase isolated from tomato fruits. Plant Physiol. 88, 109–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller, G.E., and Kieber, J.J. (September 30, 2002). Ethylene. In The Arabidopsis Book, C.R. Somerville and E.M. Meyerowitz, eds (Rockville, MD: American Society of Plant Biologists), doi/10.1199/tab.0071, http://www.aspb.org/publications/arabidopsis.
- Spanu, P., Felix, G., and Boller, T. (1990). Inactivation of stress induced 1-aminocyclopropane carboxylate synthase in vivo differs from substrate-dependent inactivation in vitro. Plant Physiol. 93, 1482–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spanu, P., Grosskopf, D.G., Felix, G., and Boller, T. (1994). The apparent turnover of 1-aminocyclopropane-1-carboxylate synthase in tomato cells is regulated by protein phosphorylation and dephosphorylation. Plant Physiol. 106, 529–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarun, A.S., and Theologis, A. (1998). Complementation analysis of mutants of 1-aminocyclopropane-1-carboxylate synthase reveals the enzyme is a dimer with shared active sites. J. Biol. Chem. 273, 12509–12514. [DOI] [PubMed] [Google Scholar]
- Tatsuki, M., and Mori, H. (2001). Phosphorylation of tomato 1-aminocyclopropane-1-carboxylic acid synthase, LE-ACS2, at the C-terminal region. J. Biol. Chem. 276, 28051–28057. [DOI] [PubMed] [Google Scholar]
- Vahala, J., Schlagnhaufer, C.D., and Pell, E.J. (1998). Induction of an ACC synthase cDNA by ozone in light-grown Arabidopsis thaliana leaves. Physiol. Plant. 103, 45–50. [Google Scholar]
- Vogel, J.P., Woeste, K.W., Theologis, A., and Kieber, J.J. (1998). Recessive and dominant mutations in the ethylene biosynthetic gene ACS5 of Arabidopsis confer cytokinin insensitivity and ethylene overproduction, respectively. Proc. Natl. Acad. Sci. USA 95, 4766–4771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, K.L.-C., Li, H., and Ecker, J.R. (2002). Ethylene biosynthesis and signaling networks. Plant Cell 14 (suppl.), S131.–S151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woeste, K., Ye, C., and Kieber, J.J. (1999). Two Arabidopsis mutants that overproduce ethylene are affected in the post-transcriptional regulation of ACC synthase. Plant Physiol. 119, 521–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, S.F., and Hoffman, N.E. (1984). Ethylene biosynthesis and its regulation in higher plants. Annu. Rev. Plant Physiol. 35, 155–189. [Google Scholar]
- Yoshii, H., and Imaseki, H. (1982). Regulation of auxin-induced ethylene biosynthesis: Repression of inductive formation of 1-aminocyclopropane-1-carboxylate synthase by ethylene. Plant Cell Physiol. 23, 639–649. [Google Scholar]
- Zarembinski, T.I., and Theologis, A. (1994). Ethylene biosynthesis and action: A case of conservation. Plant Mol. Biol. 26, 1579–1597. [DOI] [PubMed] [Google Scholar]