Abstract
In a variety of yeast cellular pathways, the Ste50p protein regulates the kinase function of the mitogen extracellular signal-regulated kinase kinase (MEKK) Ste11p. Both Ste11p and Ste50p contain sterile α motif (SAM) domains; these are interchangeable, and can be replaced by other protein-interacting modules. Furthermore, the function of the Ras association (RA)-like domain of Ste50p can be mimicked by a plasma membrane recruiting signal, and direct plasma membrane targeting of Ste11p bypasses the requirement of Ste50p for Ste11p function. Thus the regulatory role of Ste50p requires both the N-terminal SAM domain to bind Ste11p and the C-terminal RA-like domain to direct kinase localization. We have identified Opy2p, an integral membrane protein that can interact with Ste50p, as a new component in the Sho1p–Ste11p/Ste50p signaling branch of the high-osmolarity glycerol (HOG) pathway. We propose that Opy2p can serve as a membrane anchor for the Ste50p/Ste11p module in the activation of the HOG pathway.
Keywords: SAM domain, RA-like domain, MEKK, HOG, Ste50p, signal transduction
Mitogen-activated protein (MAP) kinase modules are important components of eukaryotic signaling networks. The activation of the mitogen extracellular signal-regulated kinase kinase (MEKK) is a critical step in the signaling mechanism, and thus the regulatory inputs at the level of the MEKK represent important control nodes for these networks. One of the best-studied MAP kinase modules is the kinase cascade regulating mating in the yeast Saccharomyces cerevisiae (Herskowitz 1995; Banuett 1998). In this pathway, the MEKK protein is Ste11p, and the function of this kinase is regulated in part by association with the binding proteins Ste5p (Leberer et al. 1993; Choi et al. 1994; Whiteway et al. 1995; Feng et al. 1998; Elion 2001) and Ste50p (Posas et al. 1998; Ramezani Rad et al. 1998; Wu et al. 1999; Jansen et al. 2001), and by its phosphorylation by the p21-activated protein kinase (PAK) Ste20p (Leberer et al. 1992a; Wu et al. 1995; Drogen et al. 2000). Intriguingly, Ste11p also serves as a MEKK for other signaling pathways: one controlling response to hyperosmotic stress, and the other controlling pseudohyphal growth in diploids in response to specific nutrient limitations (Roberts and Fink 1994; Mosch and Fink 1997; Erdman and Snyder 2001). In these other two pathways, Ste20p and Ste50p also regulate Ste11p function (O’Rourke and Herskowitz 1998; Posas et al. 1998; Ramezani Rad et al. 1998; Wu et al. 1999; Raitt et al. 2000). The involvement of Ste20p in phosphorylating and activating Ste11p has been established (Wu et al. 1995; Drogen et al. 2000); however, the molecular role of Ste50p in all these pathways is less clear.
The involvement of Ste50p in the mating process was noted initially during a systematic analysis of yeast gene function (Rad et al. 1992). The protein sequence gave no immediate hint as to the molecular function of the protein, but did establish that the protein contained an N-terminal sequence termed a sterile α motif (SAM) domain that is shared by several other proteins and is involved in protein–protein interactions (Ponting 1995; Posas et al. 1998; Wu et al. 1999; Jansen et al. 2001). Structural analysis of the SAM domain of Ste50p (Grimshaw et al. 2004) shows that it exhibits the same fold structure as other characterized SAM domains, including the SAM domain of the Ste11p kinase (Smalla et al. 1999; Stapleton et al. 1999; Thanos et al. 1999; Kim et al. 2002; Kim and Bowie 2003; Bhattacharjya et al. 2004; Kwan et al. 2004).
Although a phenotype for the ste50 deletion was first noted in the mating pathway, loss of Ste50p function does not totally block mating. However, Ste50p was found to be absolutely required for Ste11p function in the osmotic response pathway (O’Rourke and Herskowitz 1998; Posas et al. 1998; Wu et al. 1999; Jansen et al. 2001). The osmotic response involves two distinct pathways; the role of Ste50p in this process was initially obscured by the cell’s ability to respond to osmotic stress using the Sln1p–Ssk1p–Ssk2p/Ssk22p pathway. In the absence of this two-component module, however, the activation of the high-osmolarity glycerol (HOG) pathway in response to high osmotic stress, and ability to withstand this stress, is dependent on the proper function of the Ste11p–Ste50p complex (Posas and Saito 1997; O’Rourke and Herskowitz 1998; Posas et al. 1998; Wu et al. 1999; Raitt et al. 2000; Jansen et al. 2001).
The fact that Ste50p is essential for the osmotic function of Ste11p, but only partially required for Ste11p function in mating, suggested that the Ste50p function in mating could be partially overlapping with that of Ste5p. Both Ste50p and Ste5p are capable of binding to Ste11p, and the role of Ste5p in the mating pathway is to also bind the βγ subunit of the mating response heterotrimeric G-protein and thus localize the kinase complex to the plasma membrane (Whiteway et al. 1995; Inouye et al. 1997; Pryciak and Huntress 1998). Therefore, it is possible that the role of Ste50p in the regulation of Ste11p is also to serve as a membrane-targeting signal. Here we show that the function of the C-terminal Ras association (RA)-like domain of Ste50p can be replaced by a membrane-targeting signal, and that adding a plasma membrane association signal to Ste11p bypasses the need for Ste50p in signaling pathways using the Ste11p MEKK. Furthermore, we have identified an integral membrane protein, Opy2p, as a new component of the HOG pathway that is capable of interacting through its C-terminal fragment with the RA-like domain of Ste50p.
Results
The SAM domains of Ste11p and Ste50p are functionally interchangeable, but a heterotypic configuration is required for function
We have previously shown that a protein–protein interaction through the respective SAM domains of Ste11p and Ste50p is essential for the function of Ste11p in the HOG pathway, and that both the Ste50p SAM domain and the C-terminal RA-like domain are required for Ste50p function (Wu et al. 1999). To further dissect the mechanism through which Ste50p regulates the function of Ste11p, we studied the role of both the SAM domain and the RA-like domain of Ste50p in the modulation of Ste11p function.
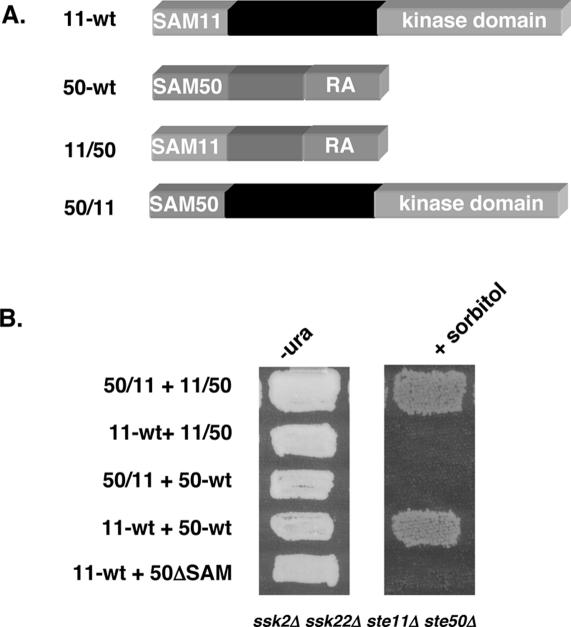
We first exchanged the SAM domains between Ste50p and Ste11p to make chimeras in which the SAM domain of Ste11p replaced that of Ste50p, creating SAM11–Ste50pΔSAM (11/50); and in which the SAM domain of Ste50p replaced that of Ste11p, creating SAM50–Ste11pΔSAM (50/11) (Fig. 1A; see Materials and Methods for details). We then measured the ability of these chimeras to function in the HOG pathway. Yeast cells (ste11Δ ste50Δ ssk2Δ ssk22Δ) are sensitive to high osmotic stress and cannot grow on hyperosmotic medium. However, cells were able to grow on hyperosmotic medium when they harbored both chimeras, indicating that the interaction between the two chimeras leads to the activation of the SAM50–Ste11pΔSAM kinase (Fig. 1B). These chimeras appear to function as well as the wild-type Ste11p and Ste50p pair, as judged by their ability to permit growth on high-osmotic media. In contrast, the SAM50–Ste11pΔSAM kinase cannot be activated by wild-type Ste50p, and neither can the wild-type Ste11p be activated by the SAM11–Ste50pΔSAM chimera. In both cases, only one type of SAM domain is present in each pair of the molecules, the Ste50p SAM domain in the former, and the Ste11p SAM in the latter. These results indicate that the SAM–SAM domain interaction between Ste11p and Ste50p must be heterotypic to permit Ste50p regulation of the Ste11p kinase, and that the relative configuration of the SAM domain to the rest of the molecule does not seem to be functionally important.
Figure 1.
The SAM domains of Ste11p and Ste50p are functionally exchangeable. (A) Schematic representation of the SAM domain-swapped chimeras of Ste11p and Ste50p. (B) Yeast cells (ste11Δ ste50Δ ssk2Δ ssk22Δ) were cotransformed with either the wild-type STE11 and STE50 or SAM domain-swapped chimeras of STE11 and STE50, and tested for the ability to grow on hyperosmotic medium. (−ura) Synthetic dextrose uracil drop-out medium; (+sorbitol) selective medium with 1.5 M sorbitol.
The interaction of SAM domains can be substituted by artificially designed protein–protein interacting modules
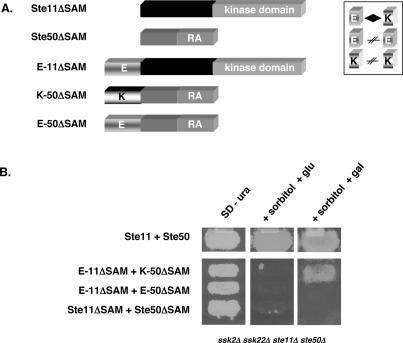
The ability to exchange the SAM domains between Ste11p and Ste50p prompted us to hypothesize that the major function of this SAM–SAM interaction is to bring the two proteins together. To test this hypothesis, we made plasmid constructs in which the SAM domains of Ste11p and Ste50p were replaced by artificially designed coiled-coiled protein–protein interaction modules, namely the E-coil (E) and K-coil (K). These coiled-coils have been shown to exhibit a specific E/K heterotypic interaction (Tripet et al. 1996, 2002). We replaced the SAM domain of Ste11p with an E-coil to create E-Ste11ΔSAMp, and replaced the SAM domain of Ste50p with a K-coil or an E-coil to create K-Ste50ΔSAMp and E-Ste50ΔSAMp. These constructs were assayed for their ability to activate the HOG pathway in response to high osmotic stress in ste11Δ ste50Δ ssk2Δ ssk22Δ cells. As shown in Figure 2, only those yeast cells cotransformed with the combination of heterotypic chimeras, K-Ste50pΔSAM and E-Ste11pΔSAM, were able to grow on high-osmotic media. In contrast, the cells with a combination of homotypic chimeras, E-Ste50pΔSAM and E- Ste11pΔSAM, were not able to grow on the same medium. These results indicated that the functionality of the heterotypic interaction between the SAM domains of Ste11p and Ste50p could be mimicked by the interaction between the E-coil and K-coil.
Figure 2.
The SAM domains of Ste11p and Ste50p can be replaced by other interaction modules. (A) Schematic diagrams of the STE11 and STE50 constructs with or without the E-coil or the K-coil. The heterotypic interaction between the E-coil and K-coil is indicated by a diamond symbol. (B) Yeast cells (ste11Δ ste50Δ ssk2Δ ssk22Δ) cotransformed with the indicated STE11 and STE50 constructs were tested for their ability to activate the HOG pathway by assaying the ability to grow on hyperosmotic medium. (SD − ura) Synthetic dextrose uracil drop-out media; (+sorbitol + glu) selective medium with 1.5 M sorbitol and glucose as carbon source; (+sorbitol + gal) selective medium with 1.5 M sorbitol and galactose as carbon source.
Fusing a nonfunctional C-terminal RA-like domain of Ste50p with a nonfunctional Ste11pΔSAM creates a chimera that complements the function of the Ste11p–Ste50p complex in the HOG pathway
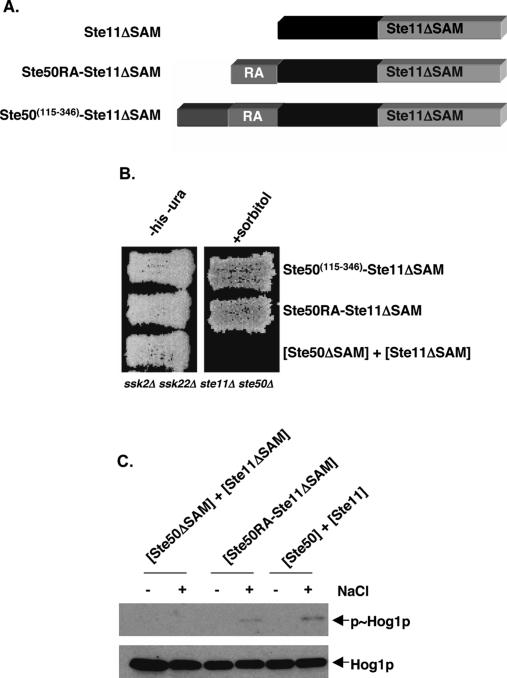
We have previously shown that both the SAM domain and the C-terminal RA-like domain of Ste50p are required for Ste50p function; these domains can function independently since the removal of the inter-region between the domains does not significantly affect the function of the protein (Wu et al. 1999). We found that the two domains, when provided in trans on separate plasmids, did not complement the function of Ste50p. Based on the results of the SAM domain swapping and the replacement with coiled-coil interaction modules, we asked if the C-terminal RA-like domain of Ste50p, provided in cis relative to the kinase domain of Ste11p, could modulate a Ste11p missing its SAM domain. We fused the Ste50p C-terminal fragments of either amino acid residues 115–346 or 219–346 to Ste11pΔSAM. This process created constructs containing the first 24 amino acid residues of Ste11p, followed by the C-terminal region of Ste50p fused to the rest of Ste11p containing amino acid residues 132–738, but lacking the region containing the SAM domain. These chimeric proteins were expressed from low-copy plasmids under the control of the STE11 promoter (Fig. 3A). We examined whether these chimeras were able to complement the function of both Ste50p and Ste11p in the HOG pathway in ste11Δ ste50Δ ssk2Δ ssk22Δ cells. As has been shown, yeast cells with either Ste50p(115–346), Ste11pΔSAM alone, or both together, were unable to grow on high-osmotic media (Posas et al. 1998; Wu et al. 1999). However, cells with either of the chimeras were resistant to hyperosmotic stress and able to grow on high-osmotic media, indicating that the chimeric constructs were able to complement the wild-type Ste11p–Ste50p function to activate the HOG pathway (Fig. 3B).
Figure 3.
Fusion of the RA domain-containing fragment of Ste50p bypasses the requirement for an interaction between the SAM domains of Ste11p and Ste50p. (A) Schematic representation of the constructs encoding fusions of the Ste50p RA domain-containing fragments and the Ste11pΔSAM fragment. (B) Yeast cells (ste11Δ ste50Δ ssk2Δ ssk22Δ) transformed with STE11 and STE50 constructs as indicated were assayed for their ability to activate the HOG pathway by testing their ability to grow on hyperosmotic medium. (−his − ura) Synthetic dextrose histidine and uracil drop-out medium; (+sorbitol) selective medium with 1.5 M sorbitol. (C) Western blot analysis of protein extracts of cells as in B—either treated (+) or untreated (−) with NaCl at 0.5 M for 5 min before the extraction of protein samples—was performed with either anti-phospho-p38 antibody to detect phosphorylated Hog1p (p∼Hog1p) or with anti-p38 antibody to detect the total amount of Hog1p.
We examined the level of expression of the chimeras to rule out the possibility that the phenotypes observed were due to changed levels of protein expression, although in the HOG pathway even overexpression of Ste11pΔSAM has no effect on pathway activity (Wu et al. 1999). Western blot analysis indicated the steady-state protein expression levels of the chimeras of Ste50p and Ste11p showed no significant difference from that of the control Ste11pΔSAM (data not shown). We also examined the possibility that the chimeras created constitutive activity of the HOG pathway by examining the activating phosphorylation level of Hog1p by Western blot analyses using specific anti-phospho-p38 antibodies. As shown in Figure 3C, the chimera of Ste50pRA–Ste11pΔSAM was able to induce the phosphorylation of Hog1p under high osmotic stress condition, whereas the Ste11pΔSAM was unable to do so under the same conditions.
Plasma membrane targeting the Ste50p SAM domain bypasses the requirement of the C-terminal domain of Ste50p
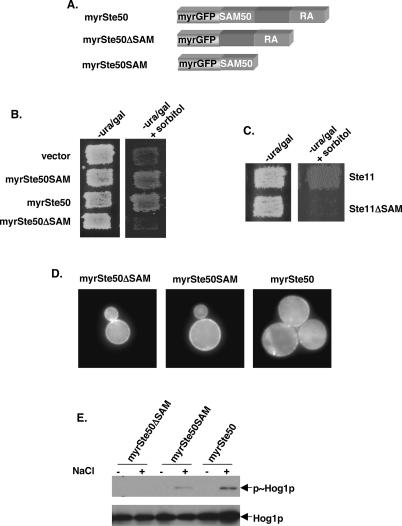
Previous studies have shown that in addition to the SAM domain at its N terminus, the C-terminal region of Ste50p is necessary for the function of Ste50p (Wu et al. 1999). The sequence alignment indicates that the C-terminal region of Ste50p has similarity to the RA domain (Ponting and Benjamin 1996). Members of the Ras superfamily are small GTPases that are typically localized to membranes through a lipid modification at their C-terminal tails (Hancock et al. 1991; Clarke 1992). If part of the function of the RA-like domain were required for localization, it is possible that independent membrane targeting could suppress the signaling defect of Ste50p lacking this region. To test this idea, we added a myristoylation signal to the N terminus of a green fluorescent protein (GFP)-tagged SAM domain of Ste50p (amino acids 1–110), creating myrGFP-Ste50pSAM. As controls, we also added the myristoylation signal to GFP only, generating myrGFP, as well as to the full-length and the C-terminal region of Ste50p to generate myrGFP-Ste50p and myrGFP-Ste50pΔSAM, respectively (Fig. 4A). We then examined the ability of these constructs to complement the function of Ste50p in the HOG pathway. Yeast cells (ste50Δ ssk2Δ skk22Δ) were transformed with the plasmid constructs mentioned above and scored for the ability to grow on hyperosmotic medium. As expected, cells with the myrGFP (vector) alone could not grow on high-osmotic media. However, the transformants with myrGFP-Ste50pSAM were able to grow on the same media, showing that the SAM domain of Ste50p with a membrane association signal is capable of complementing the requirement of wild-type Ste50p to activate the HOG pathway (Fig. 4B). In contrast, transformants with myrGFP-Ste50pΔSAM were unable to complement. As shown in Figure 4C, while a myrSte50SAM, along with a wild-type Ste11p, was able to activate the HOG pathway in ste11Δ ste50Δ ssk2Δ ssk22Δ cells, it was unable to do so when the SAM domain of Ste11p was deleted. This demonstrates that the ability of myrSte50pSAM to complement the function of Ste50p depends on its ability to interact with the SAM domain of Ste11p. We examined the localization of Ste50p constructs bearing the myristoylation signal sequence using fluorescent microscopy. As shown in Figure 4D, the GFP signal was enriched at the cell periphery, presumably the plasma membrane. To demonstrate that the Ste50pSAM bearing a myristoylation signal can activate the HOG pathway, we examined the phosphorylation of the activation loop of Hog1p. As shown in Figure 4E by Western blot analysis, Hog1p was phosphorylated upon hyperosmotic treatment in cells transformed with a myristoylated SAM domain of Ste50p, but not with a myristoylated Ste50pΔSAM. These results together suggest that plasma membrane targeting of Ste50pSAM complements the Ste50p function in the HOG pathway, and thus a predicted role of the C-terminal RA-like domain of Ste50p is to localize the Ste11p–Ste50p complex to the plasma membrane.
Figure 4.
Plasma-membrane targeting of the Ste50p SAM domain complements Ste50p function in the HOG pathway. (A) Schematic representation of the constructs encoding fusions of the GFP carrying a myristoylation signal and various fragments of Ste50p. (B) Yeast cells (ste50Δ ssk2Δ ssk22Δ), transformed with either the vector control or constructs encoding various fragments of Ste50p with a myristoylation signal, were assayed for the ability to grow on hyperosmotic media. (−ura/gal) Synthetic uracil drop-out with galactose as carbon source; (+sorbitol) with 1.5 M sorbitol. (C) Yeast cells (ste11Δ ste50Δ ssk2Δ ssk22Δ) cotransformed with a myristoylated Ste50p SAM domain in combination with either the wild-type Ste11p or the Ste11pΔSAM construct were assayed for the ability to grow on hyperosmotic medium. (D) Plasma membrane localization of myristoylated GFP fusion of the SAM domain and the RA-like domain of Ste50p, as well as the full-length Ste50p in ste50Δ ssk2Δ ssk22Δ cells. (E) Western blot analysis of protein extracts of cells as in B, for detecting Hog1p phosphorylation as described in Figure 3.
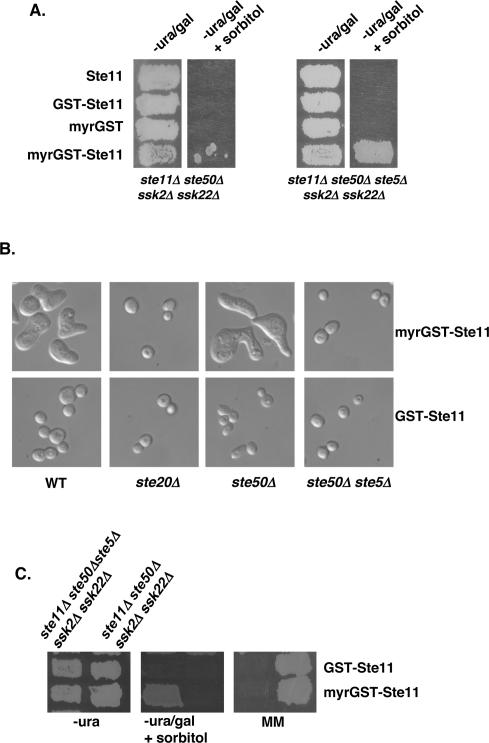
Membrane-targeting Ste11p bypasses the requirement for Ste50p
It appears that the role of Ste50p in the HOG pathway activation involves interacting with Ste11p through a SAM–SAM domain association, and localizing the heterodimer to the plasma membrane using its C-terminal RA-like domain. We asked whether direct plasma membrane targeting of Ste11p could bypass the requirement of Ste50p in the HOG pathway. We made constructs generating myristoylated GST-Ste11p (myrGST-Ste11p), and provided myrGST, nonmyristoylated GST-Ste11p, or Ste11p as controls. These constructs were expressed under control of the GAL1 promoter. We tested for the ability of these constructs to activate the HOG pathway in ste11Δ ste50Δssk2Δ ssk22Δ cells that depend on the activity of the Ste50p–Ste11p complex. As shown previously, ste11Δ ste50Δssk2Δ ssk22Δ cells overexpressing either Ste11p or GST-Ste11p could not grow on high-osmotic media (Posas et al. 1998; Wu et al. 1999). However, we found that the cells expressing myrGST-Ste11p could grow slowly on galactose-based media, but were largely unable to grow on high-osmotic media, although spontaneous papillae could form after prolonged periods of incubation (Fig. 5A, left two panels). When examined for their cellular morphology, we found that these slow-growing cells had aberrant shapes (Fig. 5B). The cultures consisted of enlarged cells with multiple projections, which resembled the morphology of cells responding to high concentrations of mating pheromone; these cells were largely arrested for proliferation, and the spontaneous formation of colonies on high-osmotic media arose from the escape of this cell cycle arrest.
Figure 5.
Plasma membrane localization of Ste11p leads to simultaneous activation of multiple signaling pathways. (A) Yeast cells of genotypes as indicated transformed with various STE11 constructs or with the control vector were assayed for their ability to grow on hyperosmotic media. (B) The morphology of yeast cells with relevant genotype (indicated at the bottom of the figure) transformed either with myristoylated GST-Ste11p (top) or with GST-Ste11p (bottom). The yeast strains used were W303-1A (wt), YEL206 (ste20Δ), YCW555 (ste50Δ ste11Δ ssk2Δ ssk22Δ), and YCW757 (ste50Δ ste5Δ ste11Δ ssk2Δ ssk22Δ). (C) Ste11p plasma membrane localization is sufficient for activation of the HOG pathway, but still needs Ste5p for mating pathway activation. Yeast cells of the indicated genotype (top) transformed with STE11 constructs as indicated on the right were assayed for their ability to grow on hyperosmotic medium (middle), and to form diploids (right). (−ura) Synthetic dextrose uracil drop-out medium; (−ura/gal) synthetic uracil drop-out with galactose as carbon source; (+sorbitol) with 1.5 M sorbitol; (MM) minimal medium.
To block the potential mating response pathway activation, we deleted STE5 in the host strain. Cells of the resulting strain transformed with myrGST-Ste11p, but not with the other plasmid constructs mentioned above, were able to grow healthily on high-osmotic media (Fig. 5A, the right two panels), indicating that the myrGST-Ste11p was able to activate the HOG pathway in the absence of Ste50p. As expected, the aberrant morphology associated with the expression of myrGST-Ste11p also disappeared in the absence of Ste5p, suggesting that the morphology was due mainly to the improper activation of the pheromone response pathway. Indeed, although the myrGST-Ste11p bypassed the requirement for Ste50p in the HOG pathway, this construct still requires the scaffold protein Ste5p for mating (Fig. 5C). The aberrant morphology caused by the overexpression of myrGST-Ste11p also required the presence of Ste20p (Fig. 5B), suggesting that while Ste11p activation involves plasma membrane targeting as a necessary step, it still requires the function of Ste20p.
Opy2p is a novel component of the HOG pathway, and appears to act upstream or at the same level of Ste11p/Ste50p
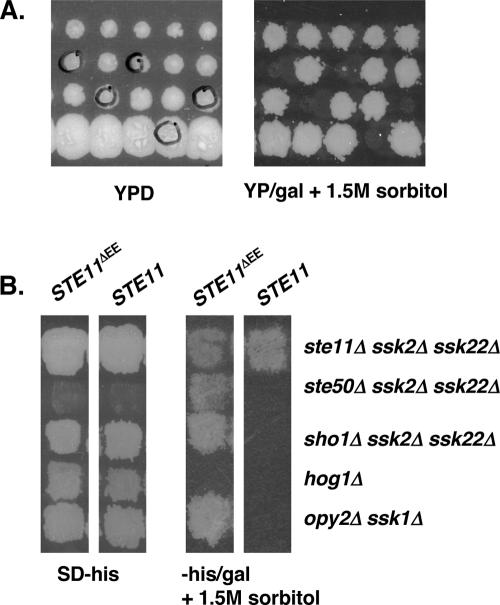
To identify the protein that provides a membrane anchor for Ste50p function in the HOG pathway, we performed a systematic genetic array (SGA) analysis (Tong et al. 2001) for ssk1Δ synthetic osmosensitivity. The analysis was designed to identify genes whose function is required for activation of the HOG pathway through the Sho1p–Ste11p/Ste50p branch. Our screen identified OPY2 as a new component of this signaling branch of the HOG pathway, since yeast cells containing the opy2Δ ssk1Δ double mutant were osmosensitive. The synthetic osmosensitive phenotype of opy2Δ ssk1Δ was confirmed by tetrad analysis as shown in Figure 6A, indicating that Opy2p is a component of the Sho1p–Ste11p/Ste50p arm of the HOG pathway. This was corroborated with genetic analysis that cells of either the opy2Δ sho1Δ or the opy2Δ ste11Δ ste50Δ mutant strains, unlike the opy2Δ ssk1Δ strain, were resistant to hyperosmotic stress (data not shown).
Figure 6.
Opy2p is a new component of the HOG pathway. (A) Synthetically osmosensitive phenotype of opy2Δ ssk1Δ cells. Tetrad analysis of meiotic products from genetic cross between yeast strains YCW1323 (MATα ssk1Δ∷NatR ura3 his3 leu2 lys2) and YCW1377 (MATa opy2Δ∷KanR ura3 his3 leu2 met15). The spores circled are resistant to both Geneticin and nourseothricin (KanR and NatR) and are also sensitive to high osmolarity. (B) Opy2p acts upstream or at the same level of Ste11p/Ste50p. Yeast cells bearing indicated mutations were transformed with either wild-type STE11 or activated STE11ΔEE that functions independently of Ste50p in the HOG pathway (Wu et al. 1999) to test their ability to suppress the osmosensitivity of the original strains as indicated by the ability to grow on high-osmolarity media. (SD − his) Synthetic dextrose histidine drop-out medium; (−his/gal) synthetic histidine drop-out medium with galactose as carbon source.
To map the point of action of Opy2p, we performed an epistatic analysis. Osmotically sensitive opy2Δ ssk1Δ cells were transformed with either wild-type STE11 or the activated STE11 allele, STE11ΔEE (Wu et al. 1999), which functions independently of Ste50p in the HOG pathway. These transformants were tested for their ability to grow on high-osmolarity medium. As shown in Figure 6B, cells bearing STE11ΔEE, but not wild-type STE11, were able to grow during osmotic stress. In contrast, STE11ΔEE was unable to allow hog1Δ cells to grow on the same high-osmolarity medium. These results suggest that Opy2p acts upstream of or at the same level as Ste11p/Ste50p in the HOG pathway.
Although both Sho1p and Opy2p act upstream of Ste11p, and are required for the activation of the HOG pathway, overexpression of one could not suppress the deletion of the other, that is, overexpression of Sho1p does not suppress the HOG pathway activation defect in opy2Δ ssk2Δ ssk22Δ cells; neither does the overexpression of Opy2p in sho1Δ ssk2Δ ssk22Δ cells (data not shown). This observation suggests a likely possibility that Sho1p acts together with Opy2p, an integral plasma membrane protein (see below), to activate the HOG pathway.
Although first named after the observation that when overexpressed, it interferes with mating-pheromone-induced cell cycle arrest (Edwards et al. 1997), deletion of OPY2 caused no detectable effect on the pheromone response pathway as measured by cell cycle arrest and mating efficiency (data not shown).
Opy2p is an integral membrane protein and localized to the plasma membrane
Sequence analysis indicates that Opy2p is a potential single-transmembrane domain protein. To examine the subcellular localization of Opy2p, we constructed and expressed the fusion Opy2p-GFP under the control of the GAL1 promoter (the signal of endogenous level of Opy2p was too low to detect), which was able to complement the function of Opy2p (data not shown). As shown in Figure 7A, the fluorescent signal of Opy2p-GFP was enriched at the cell periphery, presumably the plasma membrane; other locations also enriched with the fluorescent signal appeared to be vacuoles, and these localizations are reduced in endocytosis-defective end3Δ cells.
Figure 7.
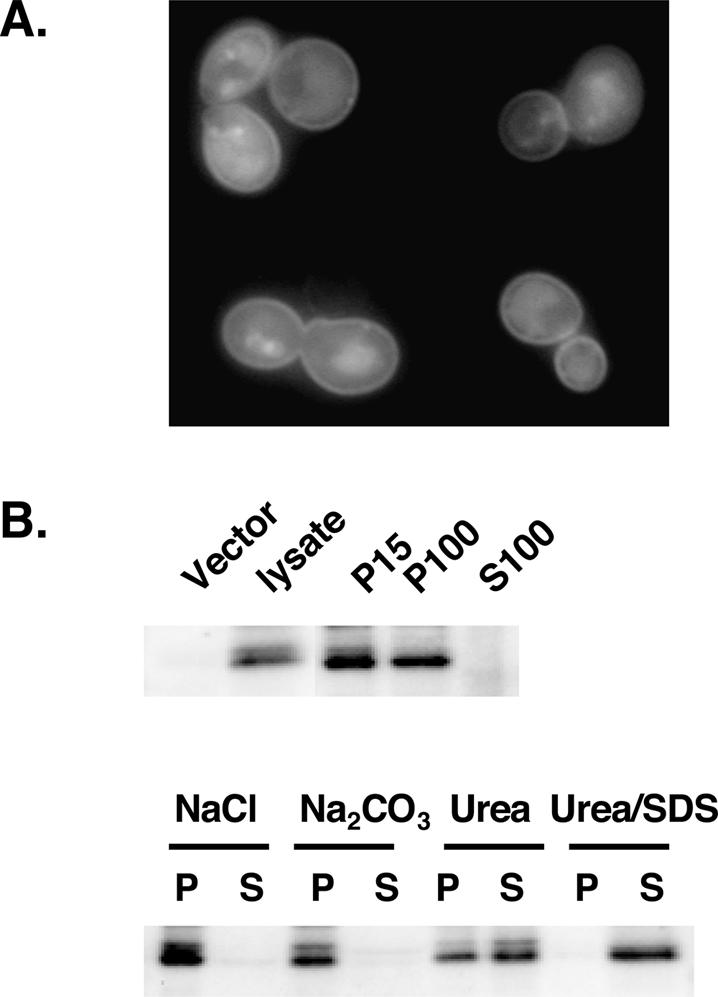
Opy2p is an integral membrane protein localized at the plasma membrane. (A) Plasma membrane localization of C-terminally GFP-tagged Opy2p in mid-log-phase end3Δ cells by fluorescent microscopy. (B) Subcellular localization of Opy2p. (Top) Cell extracts prepared from cells expressing GFP-tagged Opy2p were separated by centrifugation into pellet (P) and supernatant (S) fractions, with 15 and 100 indicating 15,000 × g and 100,000 × g, respectively. (Vector) A lysate containing no GFP-tagged Opy2p. (Bottom) Analysis of P15 was performed by treating the fraction with NaCl (0.5 M), Na2CO3 (200 mM at pH 11), urea (5 M), or SDS (5%) plus urea (5 M). Western blotting was performed using an antibody against GFP.
Subcellular fractionation confirmed that Opy2p is an integral membrane protein. Fractions of supernatant and pellet were prepared by differential centrifugation from cell lysates containing Opy2p-GFP, and probed with an anti-GFP antibody. The Opy2p-GFP was detected in the 15,000 × g (P15) pellet fraction, which contained the plasma membrane and associated proteins, and also partially in the 100,000 × g (P100) pellet fraction, but was absent in the 100,000 × g supernatant (S100) (Fig. 7B, top panel). Treatment of the P15 fraction with conditions that solubilize peripheral membrane proteins, such as high salt or sodium carbonate, failed to release Opy2p into the supernatant fraction, and urea treatment, a harsher condition that can even liberate some integral membrane proteins, could only partially liberate Opy2p into the supernatant fraction. However, treatments that solubilize membranes completely release Opy2p into the supernatant (Fig. 7B, lower panel); these results confirm that Opy2p is an integral membrane protein.
Opy2p interacts with Ste50p
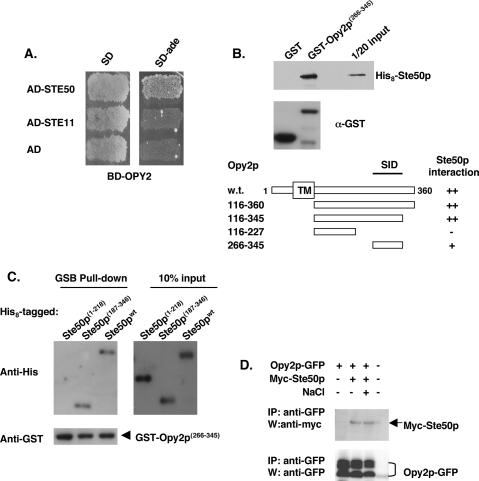
We tested if Opy2p and Ste50p physically interact, using yeast two-hybrid analysis. As shown in Figure 8A, BD-Opy2p(238–354)-containing cells cotransformed with AD-Ste50p, but not with AD-Ste11Np or with empty AD-vector, were able to grow on medium lacking adenine, indicating that BD-Opy2p specifically interacts with AD-Ste50p. To map the interaction domains of both Opy2p and Ste50p, we performed in vitro binding assays in which GST-tagged Opy2p full-length protein or deletions and His8-tagged Ste50p full-length protein or fragments were expressed in Escherichia coli. To map the region of Opy2p required for Ste50p interaction, either GST-Opy2p fragments or GST alone were mixed and incubated with His8-Ste50p, recovered by glutathione Sepharose beads, and the presence of His8-Ste50p was analyzed by Western blotting. As shown in Figure 8B, we identified that the fragment of Opy2p corresponding to amino acids 266–345 was able to pull down His8-Ste50p, establishing that Opy2p is able to interact with Ste50p in vitro. This C-terminal fragment was designated the Ste50p interaction domain (SID). Similarly, using the SID of Opy2p as a GST fusion, we performed resin-binding assays with His8-tagged Ste50p fragments. As shown in Figure 8C, both the full-length protein and the C-terminal region of Ste50p(187–346) were capable of binding GST-Opy2p(266–345), whereas the N terminus of Ste50p(1–218) was not, establishing that the C-terminal RA-like domain of Ste50p interacts with the SID of Opy2p.
Figure 8.
Opy2p interacts with Ste50p. (A) Opy2p interacts with Ste50p in a yeast two-hybrid assay. (B) Mapping the SID of Opy2p. Bacterially expressed GST fusion proteins as indicated were purified onto glutathione-Sepharose beads; the beads were then blocked with 2% BSA, incubated with bacterial extract containing His8-tagged Ste50p, washed, and suspended in SDS-PAGE sample buffer. The presence of His8-tagged Ste50p was detected by an anti-His antibody, and visualized using the ECL detection system. The levels of the GST fusion proteins used in the binding assay were monitored by the anti-GST antibody. (Top) An amount of ∼5% input of the His 8-tagged Ste50p used in the assay is shown. (Bottom) Schematic representation of using various fragments of Opy2p, analyzed in the same way as described above, to map the SID of Opy2p. (C) Opy2p interacts with the C-terminal RA-like domain of Ste50p. Resin-binding assay using bacterially expressed GST-Opy2p SID and various His8-tagged Ste50p and fragments as indicated under the same conditions as described in B. (D) Opy2p interacts with Ste50p in vivo. GFP-tagged Opy2p was immunoprecipitated from cell extracts containing Myc-tagged Ste50p, and the presence of Myc-Ste50p in the immunocomplex was analyzed by Western blotting with an antibody against the Myc epitope (shown in the top panel). The relative amount of Opy2p in the immunoprecipitate was revealed by Western blotting with an antibody against GFP. (NaCl) Treatment of cells with 0.5 M NaCl for 5 min before harvest for the preparation of cells extracts.
To examine whether Opy2p and Ste50p interact in vivo, coimmunoprecipitation (co-IP) analysis was performed using both tagged Opy2p and Ste50p. GFP-tagged Opy2p was immunoprecipitated from cell lysates containing both Opy2p-GFP and Myc-tagged Ste50p, both of which were expressed under the control of a GAL1 promoter for higher level of expression for immunodetection, as endogenous levels of both genes are too low to be readily detectable. The presence of Myc-Ste50p in the immunocomplex was analyzed, as shown in Figure 8D, and Opy2p was able to pull down Ste50p, indicating that these two proteins interact in vivo. This association appeared to be constitutive, as treatment with hyperosmotic shock (0.5 M NaCl) did not seem to affect the interaction.
Discussion
Ste50p as a bifunctional adaptor protein
Our data show that the two functional domains of Ste50p, the N-terminal SAM domain and the C-terminal RA-like domain, act together as a necessary step to modulate the activation of Ste11p MEKK. The primary role of the interactions of the SAM domains is to bring together Ste50p and Ste11p, and this function can be replaced by synthetic interacting protein modules (Figs. 1, 2). The role of the RA-like domain of Ste50p is plasma membrane localization, because the function of this region can be mimicked by a plasma-membrane-targeting signal (Fig. 4). The observation that ste11Δ ste50Δssk2Δ ssk22Δ cells bearing Ste11p and Ste50p chimeras with homotypic SAM domain combinations are unable to activate the HOG pathway suggests that homotypic interactions of either the Ste11p or Ste50p SAM domains is not sufficient to elicit the biological readout being measured, and thus the reported homotypic interactions of the Ste11p SAM domain in vitro (Bhattacharjya et al. 2004; Kwan et al. 2004) may not be physiologically relevant. This is, however, consistent with the result (Grimshaw et al. 2004) that the Ste50p SAM domain is monomeric in solution.
The combination of the mutant alleles Ste50pΔSAM and Ste11pΔSAM is unable to activate the HOG pathway in ste11Δ ste50Δssk2Δ ssk22Δ cells when these two alleles are provided in the trans configuration on separate plasmids, but the alleles are functional when they are fused together (Fig. 3). This behavior demonstrates that one of the roles of the Ste50p SAM domain is to connect the C-terminal RA-like domain to Ste11p through the interaction with the SAM domains of Ste11p; this interaction becomes dispensable if the RA-like domain of Ste50p is connected to the kinase in the cis configuration by fusion.
Although the construct of the Ste11pΔSAM containing the “built-in” Ste50p RA-like domain functions almost as effectively as the wild-type Ste11p–Ste50p pair, the architecture of Ste50p as a discrete molecule with two domains has been conserved among all homologs of Ste50p found so far in fungi, including Schizosaccharomyces pombe and Candida albicans. It is conceivable that adaptors of this kind offer more flexibility to regulate other partners and to be regulated by yet other signals. Such flexibility may become especially important when the components of interest, like Ste11p and Ste50p, are involved in multiple signaling pathways, and where regulating signals, often combinatorial in nature, are required to coordinate or differentially distribute signals among different pathways.
Plasma membrane localization and MEKK activation
The activation of both mammalian Raf and MEKK3 involves associating the protein kinases to the plasma membrane through interactions with small GTPases; this association plays a critical role in the activation of pathways in response to growth factors as well as environmental stresses (Leevers et al. 1994; Stokoe et al. 1994; Uhlik et al. 2003). The osmosensing scaffold for MEKK3 (OSM) of mammalian cells also mediates MEKK3 plasma membrane localization and is critical for the activation of p38 MAP kinase, the mammalian counterpart of the yeast Hog1p. Intriguingly, OSM has been proposed to be a Ste50p analog in terms of pathway organization and of MEKK activation (Uhlik et al. 2003).
Our data that localization of Ste11p to the plasma membrane through myristoylation bypasses the requirement for Ste50p in the activation of the HOG pathway demonstrates that plasma membrane localization of Ste11p is a critical step for its activation (Fig. 5A). This finding supports the notion that the major role of Ste50p is to bring Ste11p to the plasma membrane. Cells expressing a myristoylated Ste11p show an aberrant morphology that is very similar to cells expressing myristoylated Ste5p, which specifically activates the mating pheromone response pathway (Pryciak and Huntress 1998), and this aberrant morphology is dependent on the function of Ste5p and Ste20p (Fig. 5B). Cells expressing a myristoylated Ste50p, however, show no morphological aberrancies (data not shown). These observations indicate that while membrane localization of Ste11p is required for activation in general, the specific localization and relationship to associating proteins may be under tight differential regulation through Ste5p and/or Ste50p in response to different stimuli. Specifically, Ste5p appears to be used for mating, and Ste50p used for other pathways that require Ste11p; a direct unregulated membrane localization of Ste11p leads to the concomitant activation of multiple signaling pathways using Ste11p. Our results therefore suggest that differential plasma membrane targeting of the Ste11p MEKK contributes substantially to pathway specificity.
Opy2p, a new component of the HOG pathway, may act as a membrane anchor for the Ste50p/Ste11p complex
We used SGA to screen for synthetic osmosensitivity with ssk1Δ, and identified Opy2p as a new component of the HOG pathway. Since opy2Δ was synthetically osmosensitive only with known components from the Sln1p–Ssk1p branch, and not with components from the Sho1p–Ste11p/Ste50p branch (data not shown), we conclude that Opy2p functions in the Sho1p–Ste11p/Ste50p branch of the HOG pathway. Based on the result of an epistatic analysis with the constitutively active allele STE11ΔEE, we propose that the point of action for Opy2p is upstream of or at the same level as the Ste50p/Ste11p complex. Deletion of OPY2 has no detectable effect in the mating pheromone response and the filamentous growth pathways (Cullen et al. 2004; data not shown), indicating that Opy2p functions preferentially in the HOG pathway. Sequence analysis of Opy2p predicts a potential transmembrane domain corresponding to amino acids 93–115; and subcellular localization confirms its association with the plasma membrane (Fig. 7). This, along with the results of the epistatic analysis and the fact that Opy2p interacts with Ste50p, suggests that Opy2p could function as the membrane “anchor” that is necessary for Ste50p/Ste11p activation in response to high osmolarity stress.
Sho1p is a plasma membrane protein that acts as an osmosensor and is essential for HOG pathway activation. Ste11p has been shown to associate with Sho1p through Pbs2p (Posas and Saito 1997), and an interaction between Ste11p and Sho1p has been demonstrated to contribute specifically to cross-talk to the mating pathway (Zarrinpar et al. 2004). However, there has been no direct interaction detected between Ste50p and Sho1p (Posas et al. 1998). The genetic result that Sho1p requires Opy2p to activate the HOG pathway puts Opy2p downstream of or at the same level as Sho1p. Our observation that both Sho1p and Opy2p are necessary for the activation of the HOG pathway, and overexpression of one does not suppress the defect in activating the pathway caused by the deletion of the other, suggests a model in which these two integral plasma membrane proteins act together to activate the HOG pathway in response to hyperosmotic stress.
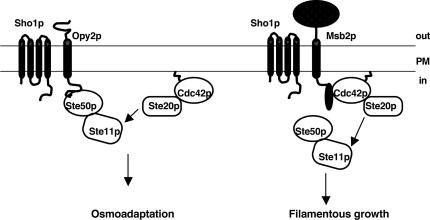
As a general factor, Sho1p is involved in both the HOG and the filamentous growth pathways (O’Rourke and Herskowitz 1998; Cullen et al. 2000, 2004), and has been shown to specifically activate the filamentous growth pathway in combination with the specificity factor Msb2p, a signaling mucin member localized at the plasma membrane and functioning at the head of the pathway (Cullen et al. 2004). Although Msb2p was also proposed to have a role in osmotolerance (O’Rourke and Herskowitz 2002), other results suggest its function is strain background dependent and conclude that the role of Msb2p is specific to the filamentous growth pathway (Cullen et al. 2004). Intriguingly, our data show that Opy2p, which is similar in membrane topology and analogous in function to Msb2p, also works together with Sho1p for HOG pathway activation. These observations suggest a model whereby the integral membrane protein Opy2p functions as a specificity factor or a co-osmosensor with Sho1p for HOG pathway activation by interacting with Ste50p. This association aids in localizing the Ste50p/Ste11p complex formed through SAM–SAM domain interaction to the plasma membrane to be activated by the upstream factors including Ste20p (Fig. 9). This model suggests that differential plasma membrane localization/activation of MEKK occurs through a combination of general as well as specificity factor(s) at the level of cell surface receptors/sensors to achieve localized activation of the shared protein kinase, and thus contributes significantly to the pathway specificity.
Figure 9.
Model of Opy2p as a component of the HOG pathway. Opy2p has a single transmembrane domain, shown as a cylinder, and its cytosolic tail interacts with Ste50p, which in turn interacts with Ste11p through SAM–SAM domain association. Through these protein–protein interactions, Opy2p acts together with Sho1p to present Ste11p to the plasma membrane, where it is phosphorylated by Ste20p under the control of Cdc42p, itself isoprenylated and attached to the plasma membrane. Under these conditions, the Ste50p/Ste11p complex that interacts with Opy2p is activated, and this may contribute to the HOG pathway specificity in a way that parallels that of Msb2p providing specificity for the filamentous growth pathway. Whether Sho1p functions the same way in the activation of the two pathways remains unknown. The model for Msb2p function is adapted from Cullen et al. (2004). See the reference for details.
Materials and methods
Materials
Restriction endonucleases and DNA-modifying enzymes were obtained from New England Biolabs and Amersham Biosciences. High-fidelity Expand thermostable DNA polymerase, anti-His monoclonal antibody, and tablet protease inhibitors were purchased from Roche Molecular Biochemicals. Acid-washed glass beads (450–600 μm), synthetic α-mating factor, protease inhibitors, and bovine serum albumin (BSA) were purchased from Sigma. Geneticin was purchased from Life Technologies, and nourseothricin (clonNAT) from Werner BioAgents. Plasmids pGEX-4T-3 and pGEX-2TK, glutathione-Sepharose beads, glutathione, and protein A/G Sepharose beads were obtained from Amersham Biosciences. The antibody against GST was described previously (Wu et al. 1999). Anti-Myc(9E10) monoclonal antibody, anti-Ste11p, and horseradish peroxidase conjugated secondary antibodies were purchased from Santa Cruz Biotechnology; rabbit polyclonal antibodies, anti-p38, and anti-phospho-p38 were from Cell Signaling Technology. Nitrocellulose membranes were purchased from Bio-Rad. The enhanced chemiluminescence (ECL) assay system was purchased from Amersham Biosciences.
Yeast strains and manipulations
Yeast media, culture conditions, and manipulations of yeast strains were as described (Rose et al. 1990). Yeast transformations with circular or linearized plasmid DNA were carried out after treatment of yeast cells with lithium acetate (Rose et al. 1990). The yeast GST-ORF library was purchased from Invitrogen, and the yeast deletion strain collection was purchased from ATCC. The yeast strains used in this study are listed in Table 1.
Table 1.
Yeast strains used in this study
Screen for ssk1Δ synthetic osmosensitivity
A systematic genetic array analysis was used to identify genes essential for growth on high-osmolarity media in the absence of ssk1. First, the ssk1Δ∷kanMX MATa strain from the deletion strain collection (ATCC) was switched to ssk1Δ∷natMX by transformation with a kanMX-targeted natMX (Goldstein and McCusker 1999) PCR product. NatR transformants were mated to Y3656 (gift from C. Boone, University of Toronto, Toronto, Ontario, Canada) to obtain a MATα strain (YCW1390) with the genotype needed for the SGA analysis by subsequent tetrad dissection.
The SGA analysis was carried out essentially as described (Tong et al. 2001) to create all possible viable double deletion strains of ssk1 with the haploid set of deletion mutants. In addition to the described procedure (Tong et al. 2001), we performed a second selection on media containing ClonNAT, G418, and canavanine before replicating the whole set of derived double-deletion strains onto rich media containing 4% galactose, ClonNAT, G418, and 1.5 M sorbitol. These plates were incubated for 2 d at 30°C, and replicated again onto the same media to clean up background before scoring growth of double deletions on sorbitol high-osmolarity media after a further incubation for 2 d at 30°C. The high-osmolarity-sensitive deletion strains hog1Δ and pbs2Δ were used as internal controls.
Construction of plasmids
For SAM domain swapping among Ste11p and Ste50p, plasmid pCW555 was constructed by cloning a PCR-amplified STE50 fragment corresponding to amino acid residues 27–149 into the ClaI and EcoRI sites of pCW204 (Wu et al. 1999), resulting in a chimera Ste11p bearing the Ste50p SAM domain. Plasmid pCW556 was constructed by cloning into the BamHI site of pCW362 an STE11 PCR fragment corresponding to amino acid residues 22–119, creating an Ste50p chimera bearing the SAM domain of Ste11p. The plasmid pCW362 was constructed by exchanging the STE50 fragment encoding amino acids 115–346 as a BamHI–EcoRI fragment from pCW207 with that of pCW267 (Wu et al. 2003). Plasmids pCW592 and pCW594, which are STE11ΔSAM constructs containing the sequences of STE50 encoding the amino acids 115–346 and 219–346 of the C terminus of Ste50p in the place of its SAM domain, respectively, were constructed by cloning the ClaI- and EcoRI-digested STE50 PCR products into the ClaI- and EcoRI-digested pCW204. To replace the SAM domain of Ste50p with the coiled-coil interaction modules, the E-coil and K-coil (kindly provided by M. O’Connor, National Research Council, Montreal, Quebec, Canada) were cloned into the EcoRI site of pGREG586 (Jansen et al. 2005) to create pCW386 and pGJ1273, respectively; the resulting plasmids were digested with SalI and recombined in vivo with a PCR-amplified STE50 fragment encoding amino acids 115–346 to generate plasmids pCW608 and pCW609, respectively. To generate either the K-coil- or the E-coil-tagged STE11 or STE11ΔSAM, the K-coil and E-coil (as EcoRI fragments from pCW386 and pGJ1273) were cloned into the EcoRI sites of pCW199 and pCW204 (Wu et al. 1999) to generate the K-coil-tagged STE11 and STE11ΔSAM constructs pCW547 and pCW549 and the E-coil-tagged STE11 and STE11ΔSAM constructs pCW548 and pCW550, respectively.
To construct myristoylated Ste50p and its various fragments, the corresponding ORF and fragments were cloned into vector plasmid pGREG596, which expresses GFP N-terminally tagged with the myristoylation signal sequence of the N-terminal 10 amino acids of Gpa1p, the α-subunit of the heterotrimeric G-protein of S. cerevisiae (Jansen et al. 2005); the resulting constructs express myrGFP-Ste50p, myrGFP-Ste50pSAM (amino acids 1–110), and myrGFP-Ste50pΔSAM (amino acids115–346) under the control of a galactose promoter GAL1. Similarly, the construct myrGST-Ste11p expressing myristoylated GST-Ste11p was created by cloning STE11 into plasmid pGREG556 (Jansen et al. 2005). The plasmid expressing Myc-tagged Ste50p under the control of the GAL1 promoter was created by cloning STE50 into pGREG523, and the plasmid expressing the C-terminally GFP-tagged Opy2p was constructed by cloning OPY2 into pGREG600 (Jansen et al. 2005).
The Escherichia coli expression constructs used in this study generated either His-tagged proteins or GST fusion proteins. The His-tagged Ste50p constructs were made using a derivative of the pET15 vector to generate pCW429 expressing the full-length Ste50p, pCW520 expressing the 187–346-amino-acid fragment of Ste50p, and pCW521 expressing the 1–218-amino-acid fragment of Ste50p. The GST-Opy2p fusions were constructed by cloning the ORF of OPY2 and fragments thereof into the pGEX-4T-3 to give rise to pCW674, pCW676, pCW678, pCW680, and pCW681, which express as GST fusion of the full-length, amino acids 116–345, 116–227, 266–345, and 116–360 of Opy2p, respectively.
The construct of BD-Opy2 was made by cloning the fragment corresponding to amino acids 238–354 into the DNA-binding domain vector pMBD (Jansen et al. 2005). The constructs of the full-length Ste50p and Ste11p-NT (amino acids 1–415) fused to the activation domains were created by cloning Ste50p into vector pAD2 (Jansen et al. 2005) and Ste11p-NT into pGAD424 (BD Biosciences Clontech), respectively.
Preparation of GST fusion or His-tagged proteins from E. coli, and resin-binding assays
The GST fusions were expressed in E. coli strain BL21(DE3) (Invitrogen), extracted, bound to glutathione Sepharose beads, and washed as previously described (Wu et al. 1995). The His-tagged proteins were expressed and released from E. coli strain BL21(DE3). For the resin-binding assay with the GST fusion proteins and the His-tagged protein from E. coli, the GST fusion proteins were immobilized onto glutathione-Sepharose beads (30 μL), further incubated for 1 h at 4°C with 2% BSA in lysis buffer containing 50 mM HEPES (pH 7.0), 150 mM NaCl, 5 mM EDTA, 2 mM DTT, and 0.1% Triton X-100, and then incubated with E. coli extracts containing His-tagged proteins (0.1–0.5 μg) for 1 h at 4°C followed by three washes, 1 mL each time with lysis buffer; the presence of His-tagged proteins was analyzed by Western blotting, detected by anti-His antibody and visualized by the ECL system.
Co-IP and subcellular localization analyses
Cell lysates were prepared from mid-log-phase yeast cells essentially as described (Wu et al. 1999). Co-IP of epitope-tagged proteins from the lysates was either performed using monoclonal anti-GFP antibody (3E6) generated in our group (commercially available from Molecular Probes) immobilized onto activated Sepharose beads (Amersham Biosciences), or using agarose beads immobilized with anti-Myc antibody (9E10) (Santa Cruz). Elution of the epitope-tagged proteins from the washed beads was performed using 0.2 M glycine (pH 3.0). Induction of GAL1-promoter-controlled expression was performed by shifting cells grown in mid-log phase in raffinose to galactose media for 4 h. Treatment of yeast cells with hyperosmotic stress was performed by adding NaCl to a final concentration of 0.5 M for 5 min before harvesting the cells.
For subcellular localization analysis, cell lysates were prepared by breaking mid-log-phase cells with glass beads in breaking buffer containing: 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 5 mM EDTA, and 0.3 M sorbitol and protease inhibitors, followed by centrifugation at 1000 × g for 10 min at 4°C. The lysates were then centrifuged at the required centrifugal force to generate the corresponding pellet and supernatant fractions.
Yeast mating, two-hybrid, and other assays
Plate mating tests and quantitative mating assays were performed as described (Leberer et al. 1992b). The yeast two-hybrid assay was performed essentially as described (Jansen et al. 2005). Halo assays to test cell growth inhibition in response to α-mating factor, assays for the ability of cells to grow on hyperosmotic media to test the function of the HOG pathway, and yeast extract preparation and Western blot analyses were performed as described previously (Wu et al. 1999).
Photomicroscopy
Cells were grown as indicated, and fixed with formaldehyde at a final concentration of 3.7% with 150 mM NaCl. Cells were viewed with a microscope equipped with Nomarski optics, and microscopic photographs were acquired with a 100× objective using a Micro Max camera (Princeton Instruments Inc.) with Northern Eclipse imaging software (Empix Imaging Inc.). The fluorescent microscopy of the GFP signals was performed as described (Wu et al. 1997). The microscopic photographs were processed using Adobe Photoshop for Macintosh.
Acknowledgments
G.J. was a recipient of a Deutsche Forschungsgemeinschaft post-doctoral fellowship and an NRC/NSERC post-doctoral fellowship. This work was supported by the NRC/GHI and grants from the CIHR to M.W. and D.Y.T. The National Research Council of Canada publication number for this work is 47497.
Footnotes
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1375706
References
- Banuett F. Signalling in the yeasts: An informational cascade with links to the filamentous fungi. Microbiol. Mol. Biol. Rev. 1998;62:249–274. doi: 10.1128/mmbr.62.2.249-274.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharjya S., Xu P., Gingras R., Shaykhutdinov R., Wu C., Whiteway M., Ni F. Solution structure of the dimeric SAM domain of MAPKKK Ste11 and its interactions with the adaptor protein Ste50 from the budding yeast: Implications for Ste11 activation and signal transmission through the Ste50–Ste11 complex. J. Mol. Biol. 2004;344:1071–1087. doi: 10.1016/j.jmb.2004.09.018. [DOI] [PubMed] [Google Scholar]
- Choi K.-Y, Satterberg B., Lyons D.M., Elion E.A. Ste5 tethers multiple protein kinases in the MAP kinase cascade required for mating in S. cerevisiae. Cell. 1994;78:499–512. doi: 10.1016/0092-8674(94)90427-8. [DOI] [PubMed] [Google Scholar]
- Clarke S. Protein isoprenylation and methylation at carboxyl-terminal cysteine residues. Annu. Rev. Biochem. 1992;61:355–386. doi: 10.1146/annurev.bi.61.070192.002035. [DOI] [PubMed] [Google Scholar]
- Cullen P.J., Schultz J., Horecka J., Stevenson B.J., Jigami Y., Sprague G.F., Jr. Defects in protein glycosylation cause SHO1-dependent activation of a STE12 signaling pathway in yeast. Genetics. 2000;155:1005–1018. doi: 10.1093/genetics/155.3.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen P.J., Sabbagh W., Jr., Graham E., Irick M.M., van Olden E.K., Neal C., Delrow J., Bardwell L., Sprague G.F., Jr. A signaling mucin at the head of the Cdc42- and MAPK-dependent filamentous growth pathway in yeast. Genes & Dev. 2004;18:1695–1708. doi: 10.1101/gad.1178604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drogen F., O’Rourke S.M., Stucke V.M., Jaquenoud M., Neiman A.M., Peter M. Phosphorylation of the MEKK Ste11p by the PAK-like kinase Ste20p is required for MAP kinase signaling in vivo. Curr. Biol. 2000;10:630–639. doi: 10.1016/s0960-9822(00)00511-x. [DOI] [PubMed] [Google Scholar]
- Edwards M.C., Liegeois N., Horecka J., DePinho R.A., Sprague G.F., Jr., Tyers M., Elledge S.J. Human CPR (cell cycle progression restoration) genes impart a Far- phenotype on yeast cells. Genetics. 1997;147:1063–1076. doi: 10.1093/genetics/147.3.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elion E.A. The Ste5p scaffold. J. Cell Sci. 2001;114:3967–3978. doi: 10.1242/jcs.114.22.3967. [DOI] [PubMed] [Google Scholar]
- Erdman S., Snyder M. A filamentous growth response mediated by the yeast mating pathway. Genetics. 2001;159:919–928. doi: 10.1093/genetics/159.3.919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y., Song L.Y., Kincaid E., Mahanty S.K., Elion E.A. Functional binding between Gβ and the LIM domain of Ste5 is required to activate the MEKK Ste11. Curr. Biol. 1998;8:267–278. doi: 10.1016/s0960-9822(98)70108-3. [DOI] [PubMed] [Google Scholar]
- Goldstein A.L., McCusker J.H. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast. 1999;15:1541–1553. doi: 10.1002/(SICI)1097-0061(199910)15:14<1541::AID-YEA476>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Grimshaw S.J., Mott H.R., Stott K.M., Nielsen P.R., Evetts K.A., Hopkins L.J., Nietlispach D., Owen D. Structure of the sterile α motif (SAM) domain of the Saccharomyces cerevisiae mitogen-activated protein kinase pathway-modulating protein STE50 and analysis of its interaction with the STE11 SAM. J. Biol. Chem. 2004;279:2192–2201. doi: 10.1074/jbc.M305605200. [DOI] [PubMed] [Google Scholar]
- Hancock J.F., Cadwallader K., Paterson H., Marshall C.J. A CAAX or a CAAL motif and a second signal are sufficient for plasma membrane targeting of ras proteins. EMBO J. 1991;10:4033–4039. doi: 10.1002/j.1460-2075.1991.tb04979.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herskowitz I. MAP kinase pathways in yeast: For mating and more. Cell. 1995;80:187–197. doi: 10.1016/0092-8674(95)90402-6. [DOI] [PubMed] [Google Scholar]
- Inouye C., Dhillon N., Thorner J. Ste5 RING-H2 domain: Role in Ste4-promoted oligomerization for yeast pheromone signaling. Science. 1997;278:103–106. doi: 10.1126/science.278.5335.103. [DOI] [PubMed] [Google Scholar]
- Jansen G., Buhring F., Hollenberg C.P., Ramezani Rad M. Mutations in the SAM domain of STE50 differentially influence the MAPK-mediated pathways for mating, filamentous growth and osmotolerance in Saccharomyces cerevisiae. Mol. Genet. Genomics. 2001;265:102–117. doi: 10.1007/s004380000394. [DOI] [PubMed] [Google Scholar]
- Jansen G., Wu C., Schade B., Thomas D.Y., Whiteway M. Drag&Drop cloning in yeast. Gene. 2005;344:43–51. doi: 10.1016/j.gene.2004.10.016. [DOI] [PubMed] [Google Scholar]
- Kim C.A., Bowie J.U. SAM domains: Uniform structure, diversity of function. Trends Biochem. Sci. 2003;28:625–628. doi: 10.1016/j.tibs.2003.11.001. [DOI] [PubMed] [Google Scholar]
- Kim C.A., Gingery M., Pilpa R.M., Bowie J.U. The SAM domain of polyhomeotic forms a helical polymer. Nat. Struct. Biol. 2002;9:453–457. doi: 10.1038/nsb802. [DOI] [PubMed] [Google Scholar]
- Kwan J.J., Warner N., Pawson T., Donaldson L.W. The solution structure of the S. cerevisiae Ste11 MAPKKK SAM domain and its partnership with Ste50. J. Mol. Biol. 2004;342:681–693. doi: 10.1016/j.jmb.2004.06.064. [DOI] [PubMed] [Google Scholar]
- Leberer E., Dignard D., Harcus D., Thomas D.Y., Whiteway M. The protein kinase homologue Ste20p is required to link the yeast pheromone response G-protein βγ subunits to downstream signalling components. EMBO J. 1992a;11:4815–4824. doi: 10.1002/j.1460-2075.1992.tb05587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leberer E., Dignard D., Hougan L., Thomas D.Y., Whiteway M. Dominant-negative mutants of a yeast G-protein β subunit identify two functional regions involved in pheromone signalling. EMBO J. 1992b;11:4805–4813. doi: 10.1002/j.1460-2075.1992.tb05586.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leberer E., Dignard D., Harcus D., Hougan L., Whiteway M., Thomas D.Y. Cloning of Saccharomyces cerevisiae STE5 as a suppressor of a Ste20 protein kinase mutant: Structural and functional similarity of Ste5 to Far1. Mol. Gen. Genet. 1993;241:241–254. doi: 10.1007/BF00284675. [DOI] [PubMed] [Google Scholar]
- Leevers S.J., Paterson H.F., Marshall C.J. Requirement for Ras in Raf activation is overcome by targeting Raf to the plasma membrane. Nature. 1994;369:411–414. doi: 10.1038/369411a0. [DOI] [PubMed] [Google Scholar]
- Mosch H.U., Fink G.R. Dissection of filamentous growth by transposon mutagenesis in Saccharomyces cerevisiae. Genetics. 1997;145:671–684. doi: 10.1093/genetics/145.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Rourke S.M., Herskowitz I. The Hog1 MAPK prevents cross talk between the HOG and pheromone response MAPK pathways in Saccharomyces cerevisiae. Genes & Dev. 1998;12:2874–2886. doi: 10.1101/gad.12.18.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Rourke S.M., Herskowitz I. A third osmosensing branch in Saccharomyces cerevisiae requires the Msb2 protein and functions in parallel with the Sho1 branch. Mol. Cell. Biol. 2002;22:4739–4749. doi: 10.1128/MCB.22.13.4739-4749.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting C.P. SAM: A novel motif in yeast sterile and Drosophila polyhomeotic proteins. Protein Sci. 1995;4:1928–1930. doi: 10.1002/pro.5560040927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting C.P., Benjamin D.R. A novel family of Ras-binding domains. Trends Biochem. Sci. 1996;21:422–425. doi: 10.1016/s0968-0004(96)30038-8. [DOI] [PubMed] [Google Scholar]
- Posas F., Saito H. Osmotic activation of the HOG MAPK pathway via Ste11p MAPKKK: Scaffold role of Pbs2p MAPKK. Science. 1997;276:1702–1705. doi: 10.1126/science.276.5319.1702. [DOI] [PubMed] [Google Scholar]
- Posas F., Witten E.A., Saito H. Requirement of STE50 for osmostress-induced activation of the STE11 mitogen-activated protein kinase kinase kinase in the high-osmolarity glycerol response pathway. Mol. Cell. Biol. 1998;18:5788–5796. doi: 10.1128/mcb.18.10.5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryciak P.M., Huntress F.A. Membrane recruitment of the kinase cascade scaffold protein Ste5 by the Gβγ complex underlies activation of the yeast pheromone response pathway. Genes & Dev. 1998;12:2684–2697. doi: 10.1101/gad.12.17.2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rad M.R., Xu G., Hollenberg C.P. STE50, a novel gene required for activation of conjugation at an early step in mating in Saccharomyces cerevisiae. Mol. Gen. Genet. 1992;236:145–154. doi: 10.1007/BF00279653. [DOI] [PubMed] [Google Scholar]
- Raitt D.C., Posas F., Saito H. Yeast Cdc42 GTPase and Ste20 PAK-like kinase regulate Sho1-dependent activation of the Hog1 MAPK pathway. EMBO J. 2000;19:4623–4631. doi: 10.1093/emboj/19.17.4623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramezani Rad M., Jansen G., Buhring F., Hollenberg C.P. Ste50p is involved in regulating filamentous growth in the yeast Saccharomyces cerevisiae and associates with Ste11p. Mol. Gen. Genet. 1998;259:29–38. doi: 10.1007/s004380050785. [DOI] [PubMed] [Google Scholar]
- Roberts R.L., Fink G.R. Elements of a single MAP kinase cascade in Saccharomyces cerevisiae mediate two developmental programs in the same cell type: Mating and invasive growth. Genes & Dev. 1994;8:2974–2985. doi: 10.1101/gad.8.24.2974. [DOI] [PubMed] [Google Scholar]
- Rose M.D., Winston F., Hieter P.1990. . Methods in yeast genetics. A laboratory manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY [Google Scholar]
- Smalla M., Schmieder P., Kelly M., Ter Laak A., Krause G., Ball L., Wahl M., Bork P., Oschkinat H. Solution structure of the receptor tyrosine kinase EphB2 SAM domain and identification of two distinct homotypic interaction sites. Protein Sci. 1999;8:1954–1961. doi: 10.1110/ps.8.10.1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapleton D., Balan I., Pawson T., Sicheri F. The crystal structure of an Eph receptor SAM domain reveals a mechanism for modular dimerization. Nat. Struct. Biol. 1999;6:44–49. doi: 10.1038/4917. [DOI] [PubMed] [Google Scholar]
- Stokoe D., MacDonald S.G., Cadwallader K., Symons M., Hancock J.F. Activation of Raf as a result of recruitment to the plasma membrane. Science. 1994;264:1463–1467. doi: 10.1126/science.7811320. [DOI] [PubMed] [Google Scholar]
- Thanos C.D., Faham S., Goodwill K.E., Cascio D., Phillips M., Bowie J.U. Monomeric structure of the human EphB2 sterile α motif domain. J. Biol. Chem. 1999;274:37301–37306. doi: 10.1074/jbc.274.52.37301. [DOI] [PubMed] [Google Scholar]
- Tong A.H., Evangelista M., Parsons A.B., Xu H., Bader G.D., Page N., Robinson M., Raghibizadeh S., Hogue C.W., Bussey H., et al. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 2001;294:2364–2368. doi: 10.1126/science.1065810. [DOI] [PubMed] [Google Scholar]
- Tripet B., Yu L., Bautista D.L., Wong W.Y., Irvin R.T., Hodges R.S. Engineering a de novo-designed coiled-coil heterodimerization domain off the rapid detection, purification and characterization of recombinantly expressed peptides and proteins. Protein Eng. 1996;9:1029–1042. doi: 10.1093/protein/9.11.1029. [DOI] [PubMed] [Google Scholar]
- Tripet B., De Crescenzo G., Grothe S., O’Connor-McCourt M., Hodges R.S. Kinetic analysis of the interactions between troponin C and the C-terminal troponin I regulatory region and validation of a new peptide delivery/capture system used for surface plasmon resonance. J. Mol. Biol. 2002;323:345–362. doi: 10.1016/s0022-2836(02)00883-5. [DOI] [PubMed] [Google Scholar]
- Uhlik M.T., Abell A.N., Johnson N.L., Sun W., Cuevas B.D., Lobel-Rice K.E., Horne E.A., Dell’Acqua M.L., Johnson G.L. Rac-MEKK3-MKK3 scaffolding for p38 MAPK activation during hyperosmotic shock. Nat. Cell Biol. 2003;5:1104–1110. doi: 10.1038/ncb1071. [DOI] [PubMed] [Google Scholar]
- Whiteway M.S., Wu C., Leeuw T., Clark K., Fourest A., Thomas D.Y., Leberer E. Association of the yeast pheromone response G protein βγ subunits with the MAP kinase scaffold Ste5p. Science. 1995;269:1572–1575. doi: 10.1126/science.7667635. [DOI] [PubMed] [Google Scholar]
- Wu C., Whiteway M., Thomas D.Y., Leberer E. Molecular characterization of Ste20p, a potential mitogen-activated protein or extracellular signal-regulated kinase kinase (MEK) kinase kinase from Saccharomyces cerevisiae. J. Biol. Chem. 1995;270:15984–15992. doi: 10.1074/jbc.270.27.15984. [DOI] [PubMed] [Google Scholar]
- Wu C., Lytvyn V., Thomas D.Y., Leberer E. The phosphorylation site for Ste20p-like protein kinases is essential for the function of myosin-I in yeast. J. Biol. Chem. 1997;272:30623–30626. doi: 10.1074/jbc.272.49.30623. [DOI] [PubMed] [Google Scholar]
- Wu C., Leberer E., Thomas D.Y., Whiteway M. Functional characterization of the interaction of Ste50p with Ste11p MAPKKK in Saccharomyces cerevisiae. Mol. Biol. Cell. 1999;10:2425–2440. doi: 10.1091/mbc.10.7.2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C., Arcand M., Jansen G., Zhong M., Iouk T., Thomas D.Y., Meloche S., Whiteway M. Phosphorylation of the MAPKKK regulator Ste50p in Saccharomyces cerevisiae: A casein kinase I phosphorylation site is required for proper mating function. Eukaryot. Cell. 2003;2:949–961. doi: 10.1128/EC.2.5.949-961.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarrinpar A., Bhattacharyya R.P., Nittler M.P., Lim W.A. Sho1 and Pbs2 act as coscaffolds linking components in the yeast high osmolarity MAP kinase pathway. Mol. Cell. 2004;14:825–832. doi: 10.1016/j.molcel.2004.06.011. [DOI] [PubMed] [Google Scholar]