Summary
Soil bacteria are heavily exposed to environmental methylating agents such as methylchloride and may have special requirements for repair of alkylation damage on DNA. We have used functional complementation of an Escherichia coli tag alkA mutant to screen for 3-methyladenine DNA glycosylase genes in genomic libraries of the soil bacterium Bacillus cereus. Three genes were recovered: alkC, alkD and alkE. The amino acid sequence of AlkE is homologous to the E. coli AlkA sequence. AlkC and AlkD represent novel proteins without sequence similarity to any protein of known function. However, iterative and indirect sequence similarity searches revealed that AlkC and AlkD are distant homologues of each other within a new protein superfamily that is ubiquitous in the prokaryotic kingdom. Homologues of AlkC and AlkD were also identified in the amoebas Entamoeba histolytica and Dictyostelium discoideum, but no other eukaryotic counterparts of the superfamily were found. The alkC and alkD genes were expressed in E. coli and the proteins were purified to homogeneity. Both proteins were found to be specific for removal of N-alkylated bases, and showed no activity on oxidized or deaminated base lesions in DNA. B. cereus AlkC and AlkD thus define novel families of alkylbase DNA glycosylases within a new protein superfamily.
Introduction
Alkylating compounds represent one of the most abundant classes of mutagenic and genotoxic agents present in the environment. 7-methylguanine (7mG), 3-methyladenine (3mA), 3-methylguanine (3mG), O6-methylguanine (O6mG) and 1-methyladenine (1mA) are major base modifications introduced by methylating agents. While O6mG and 1mA are repaired by direct reversal of the damage, involving a DNA alkyltransferase or the iron-2-oxoglutarate dependent AlkB protein respectively (Falnes et al., 2002; Trewick et al., 2002), 3mA and other N-alkylated purines are excised from the DNA by base excision repair (BER) (Seeberg and Berdal, 1997). The first step of BER is mediated by a DNA glycosylase hydrolysing the N-glycosylic bond thus releasing the damaged base in a free form and creating an abasic (AP) site in the DNA. The AP site is incised at the 5′-side or the 3′-side by an AP endonuclease or an AP lyase respectively. The repair is completed by a DNA phosphodiesterase cleansing the ends, a DNA polymerase filling the gap of one to several nucleotides and finally a DNA ligase seals the nick (Seeberg et al., 1995; Fortini et al., 2003).
DNA glycosylases removing alkylated base residues have been identified in all organisms investigated and may be universally present in nature. As 3mA is a main substrate for these enzymes, they are generally referred to as 3mA DNA glycosylases. Escherichia coli possesses two enzymes of this type, 3mA DNA glycosylase I (Tag) which is constitutively expressed (Karran et al., 1980), and 3mA DNA glycosylase II (AlkA) which is induced by cell exposure to alkylating agents (Samson and Cairns, 1977; Evensen and Seeberg, 1982). The Tag enzyme has a rather narrow substrate specificity, limited to 3mA and 3mG (Bjelland et al., 1993), whereas AlkA is a much more versatile enzyme and removes 3mA, 3mG, 7mG, O2-methylpyrimidines, hypoxanthine, ethenoadenine and thymine residues oxidized in the methyl group (Evensen and Seeberg, 1982; Karran et al., 1982; McCarthy et al., 1984; Bjelland et al., 1994; Saparbaev and Laval, 1994; Saparbaev et al., 1995). The methyl group of 7mG protrudes into the major groove of the double-helix and does not appear to cause mutations or block DNA replication. In contrast, both 3mA and 3mG are minor groove lesions and represent blocks to DNA replication because of impaired stacking properties. These lesions therefore have severe cytotoxic effects and need to be removed prior to DNA replication (Boiteux et al., 1984; Larson et al., 1985).
The Tag and AlkA proteins share no significant sequence homology in spite of their functional similarity. The 3mA DNA glycosylases from Saccharomyces cerevisiae (Mag) and Schizosaccharomyces pombe (Mag1) both belong to the AlkA family, whereas the mammalian enzymes (Aag) are different with little or no relevant sequence homology and hence represent a third family of 3mA DNA glycosylases. This family was initially thought to be limited to mammalian cells, but genome sequencing efforts have revealed the presence of homologous proteins in certain prokaryotic species as well (Aamodt et al., 2004). Some enzymes of the endonuclease III (Nth) family of DNA glycosylases remove methylated purines from DNA and constitute a forth family of 3mA DNA glycosylases (Begley et al., 1999; O’Rourke et al., 2000).
Mutants of E. coli lacking both Tag and AlkA are extremely sensitive towards exposure to simple alkylating agents such as methyl methanesulphonate (MMS) and dimethylsulphate. Functional complementation of the tag alkA double mutant with a gene expressing 3mA DNA glycosylase activity will restore alkylation resistance. Such mutants have been instrumental for the cloning of 3mA DNA glycosylase genes from other organisms, including Micrococcus luteus (Pierre and Laval, 1986), yeast (Chen et al., 1989; Berdal et al., 1990; Memisoglu and Samson, 1996), Arabidopsis thaliana (Santerre and Britt, 1994) and mammalian cells (Chakravarti et al., 1991; O’Connor and Laval, 1990; 1991; Samson et al., 1991; Engelward et al., 1993). The same approach was utilized in this study to screen for 3mA DNA glycosylases in Bacillus cereus, which is a soil bacterium that is heavily exposed to methylating agents such as methylchloride under normal life conditions (Vaughan et al., 1993). Three different genes were recovered, termed alkC, alkD and alkE, which complemented the MMS sensitivity of the E. coli tag alkA double mutant. AlkC and AlkD represent novel genes with no homology to previously characterized DNA glycosylases. We purified both enzymes to homogeneity and found that AlkC and AlkD indeed are functional 3mA DNA glycosylases. Iterative searches of the Non-redundant Protein Sequence Database (NCBI) revealed that AlkC and AlkD are distant homologues belonging to a new superfamily of proteins.
Results
Three open reading frames of B. cereus genome that complement the alkylation-sensitive phenotype of the E. coli strain BK2118 (tag alkA)
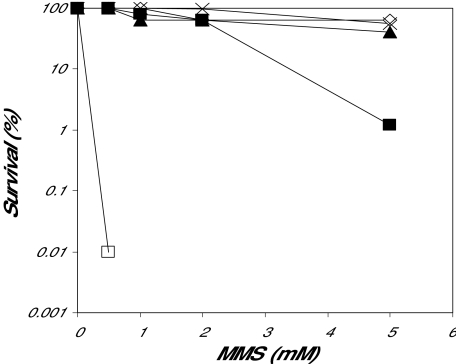
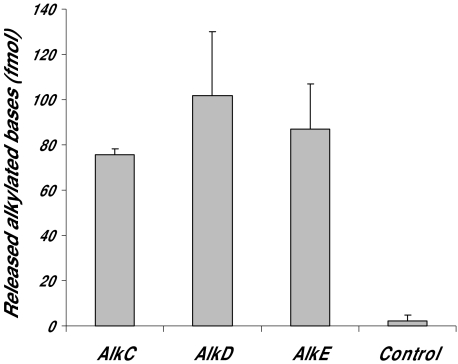
The alkylation repair-defective E. coli strain BK2118, which is lacking the AlkA and Tag 3mA DNA glycosylases, was transformed by different genome libraries made either from DNA isolated from the B. cereus strain ATCC 10987 or from commercially available B. cereus DNA (Promega, non-determined strain). Transformants surviving on media containing MMS were isolated, and plasmids were analysed by DNA sequencing and restriction cleavage. The DNA sequences were assembled into three complete open reading frames (ORFs) termed AlkC (Promega), AlkD (Promega) and AlkE (ATCC 10987). Next, three selected clones containing alkC, alkD or alkE were re-transformed into BK2118 and plated on media containing increasing amounts of MMS. Full rescue was obtained with plasmids expressing AlkC, AlkE and E. coli AlkA (control), whereas AlkD was partially complementing the MMS sensitivity of BK2118(Fig. 1). Furthermore the capability of AlkC, AlkD and AlkE to remove alkylated bases was examined in cell extracts prepared after expression of the three enzymes in BK2118 with calf thymus DNA treated with N-[3H]-methyl-N-nitrosourea as substrate. Excision of methylated bases was confirmed in all three extracts, whereas similar extracts from cells containing the pUC19 vector without insert showed no removal of methylated bases (Fig. 2). It thus appears that all three B. cereus enzymes possess alkylbase DNA glycosylase activity.
Fig. 1.
MMS survival of BK2118 (alkA tag) transformed by B. cereus clones pUC-alkC, pUC-alkD and pUC-alkE. Logarithmically growing BK2118 cells harbouring expression constructs for AlkA (◊), AlkC (▴), AlkD (▪), AlkE (×) or pUC (□) only were spread on LB plates containing 0.5, 1, 2 or 5 mM MMS and incubated at 37°C for 2 days. The percentage of surviving colonies was calculated. This survival experiment was repeated three times with similar results.
Fig. 2.
3mA DNA glycosylase activity in protein extracts of E. coli BK2118 cells expressing B. cereus AlkC, AlkD or AlkE. [3H]-methyl-N-nitrosourea treated calf thymus DNA (800 fmol modified bases) was incubated with 1 µg extracts at 37°C for 30 min, the DNA was ethanol precipitated and the supernatant subjected to scintillation counting. Control cells contained empty pUC vector.
AlkC and AlkD both belong to the same protein superfamily
The deduced amino acid sequences were compared with protein sequences in the NCBI non-redundant protein database (see Table S1 for accession numbers). The alkE gene encoded a putative protein of 287 amino acids with 26% identity and 45% similarity to the E. coli AlkA protein over an aligned region of 170 amino acids (Fig. S1). The nucleotide sequence of alkC and alkD translates into polypeptides of 256 and 237 amino acids respectively. Iterative sequence similarity searches using psi-blast in the NCBI non-redundant protein sequence database showed that homologues of both AlkC and AlkD are present in several prokaryotic organisms; however, none of these were annotated as DNA repair enzymes or other proteins with known function (Figs. S2 and S3). Further analysis of the iterative searches revealed that many of the members of the AlkC group were also present in the AlkD group and vice versa indicating that AlkC and AlkD are distant homologues belonging to a large superfamily of uncharacterized proteins. For example, alignment of homologues from Pasteurella multocida and uncultured archea GZfos12E1 with B. cereus AlkC and AlkD demonstrate the link between the two families (Fig. S4). Other examples of organisms with AlkC and AlkD homologues include: firmicutes (Bacillus subtilis), proteobacteria (Agrobacterium tumefaciens, Helicobacter hepaticus and Pseudomonas sp.), planctomycetes (Rhodopirellula baltica), proteobacteria, actinobacteria, bacteroidetes, archaeon and spirochaetes (Leptospira sp.). Cyanobacteria appear to be the only bacterial group without ORFs with sequence similarity to AlkC and AlkD. It thus appears that the AlkC/AlkD superfamily is widespread in prokaryotes. Entamoeba histolytica and Dictyostelium discoideum, which are protezoa causing amebic dysentery, seem to be the only eukaryotes yet found to harbour this protein family.
Removal of alkylated bases by AlkC and AlkD
To investigate the enzymatic properties of AlkC and AlkD proteins in more detail, the coding sequences were subcloned in the expression vector pT7-SCII and the proteins were produced in E. coli strain BL21. Both AlkC and AlkD were purified to near physical homogeneity by a three-step procedure including AffiGel Blue, MonoQ and DNA cellulose chromatography. AlkC and AlkD migrate on SDS-PAGE as proteins of 28 kDa and 25 kDa respectively, which is in good agreement with the molecular weights calculated from the amino acid sequence (29.9 and 28.2 kDa respectively).
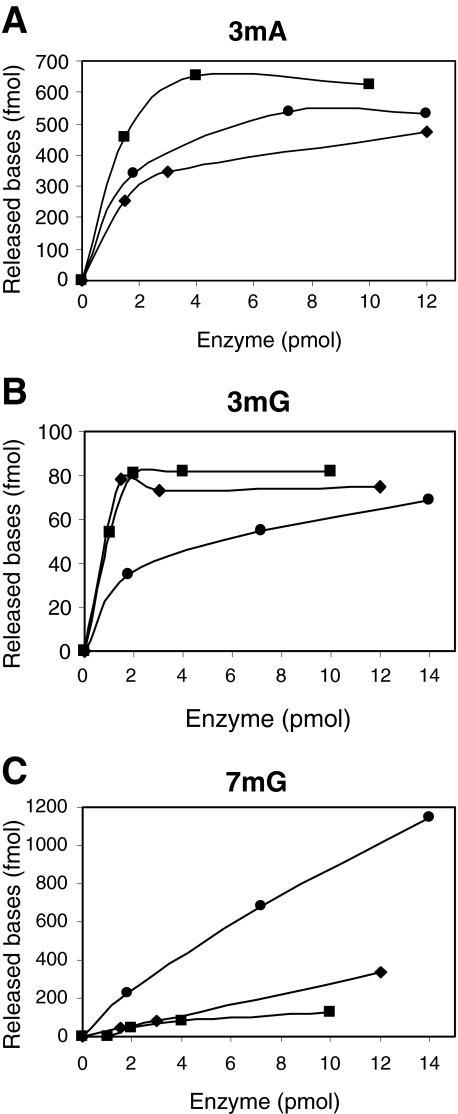
We examined the abilities of the purified AlkC and AlkD enzymes to remove alkylated bases by using DNA treated with N-[3H]-methyl-N-nitrosourea as substrate and separation of the radiolabelled excision products by high-performance liquid chromatography (HPLC) (Fig. 3). The amounts of methylpurines formed in such DNA are 65% 7mG, 10% 3mA and 0.7% 3mG (Bjelland et al., 1993). From these measurements it appears that AlkD has a high activity towards 7mG (Fig. 3C), but removes 3mG more slowly as compared with E. coli AlkA (Fig. 3B). 3mA is excised at a comparable rate for AlkD and E. coli AlkA (Fig. 3A). AlkC is more efficient in removing 3mA as compared with E. coli AlkA (Fig. 3A), whereas excision of 3mG proceeds at a similar rate (Fig. 3B). Further, AlkC shows only limited removal of 7mG (Fig. 3C), and appears to be essentially 3-methylpurine specific. AlkC therefore compares with the Tag enzyme from E. coli in its specificity for 3-methylpurines, except that the efficiency of 3mG removal is much higher than for Tag. AlkC and AlkD thus appear to functionally complement each other by efficiently removing the major N-alkylated purine products in alkylated DNA. Furthermore, inefficient removal of the cytotoxic 3mG lesion by AlkD (Fig. 3B) could explain why expression of AlkD in alkA tag E. coli mutant cells does not restore the alkylation resistance completely (Fig. 1).
Fig. 3.
Reverse phase HPLC of methylated bases released by AlkA, AlkC and AlkD from [3H]-methyl-N-nitrosourea treated calf thymus DNA. [3H]-MNU-DNA was incubated with increasing amounts of AlkA (◊), AlkC (▪) and AlkD (•) at 37°C for 30 min. The DNA was precipitated with ethanol and the supernatant was analysed by HPLC. Radioactivity in fractions corresponding to 3mA (A), 3mG (B) and 7mG (C) was measured in a liquid scintillation counter. The experiments in this figure were repeated three times with similar results.
Several 3mA DNA glycosylases have been reported to be active against a broad range of lesions including deaminated and oxidized bases. The mammalian Aag and E. coli AlkA DNA glycosylases excise pre-mutagenic lesions such as deaminated adenine (hypoxanthine) (Saparbaev and Laval, 1994) and cyclic etheno adducts (1, N6-ethenoadenine and 1, N2-ethenocytosine (Dosanjh et al., 1994a). Furthermore, mammalian Aag was reported to remove oxidized guanine, 7, 8-dihydro-8-oxoguanine (8oxoG) (Bessho et al., 1993) whereas E. coli AlkA are removing methyl-oxidized thymines (5-formyluracil and 5-hydroxymethyluracil) (Bjelland et al., 1994). The specificity of AlkC and AlkD towards hypoxanthine, 1,N6-ethenoadenine, 8oxoG and 5-formyluracil was examined on oligonucleotides containing a single lesion. Neither AlkC nor AlkD showed any detectable affinity for these DNA base lesions (data not shown). In addition, AlkC and AlkD showed no activity towards other important base lesions such as methyl-formamidopyrimidine and adenine mismatch (A:G). Finally, it was shown that AlkC and AlkD were not associated with an AP lyase activity when assayed with a double-stranded 32P-labelled oligonucleotide containing a single AP site. From these data it seems evident that the AlkC and AlkD are involved exclusively in the repair of alkylation damage in B. cereus.
Discussion
In this work genomic libraries of B. cereus were screened by functional complementation of the alkylation sensitivity of the E. coli tag alkA mutant to identify 3mA DNA glycosylases. By this approach two novel ORFs, termed AlkC and AlkD, were identified encoding 3mA DNA glycosylases. Amino acid sequence analysis of AlkC and AlkD revealed no sequence homology to known DNA repair enzymes or other proteins with known function. Furthermore, similarity searches of the NCBI non-redundant database with the psi-blast program showed that the AlkC and AlkD families are ubiquitous in prokaryotic organisms. Moreover, searches initiated with AlkC or AlkD revealed several common ORFs, indicating that AlkC and AlkD belong to the same superfamily and have a common ancestral origin. Biochemical characterization was performed with purified AlkC and AlkD and compared with E. coli AlkA. Both AlkC and AlkD remove the major cytotoxic alkylation product 3mA efficiently, whereas the minor cytotoxic 3mG adduct is less efficiently removed by AlkD as compared with AlkC and E. coli AlkA. Several 3mA DNA glycosylases, including mammalian Aag and E. coli AlkA, remove pre-mutagenic base lesions such as deaminated adenine (hypoxanthine) and cyclic etheno adducts; however, AlkC and AlkD showed no activity towards these lesions. It thus appears that AlkC and AlkD are specific for removal of alkylated bases.
The activity of AlkD towards 7mG is substantially different from other alkylation repair activities so far described. The enzyme specificity for 7mG is surprising in the view of the notion that 7mG is supposed to be an innocuous lesion. It could be that 7mG removal is important to prevent possible interference caused by 7mG in protein/DNA interactions or to avoid the formation of secondary derivatives of 7mG. Alkylation of guanine at the N7 position will destabilize the N-glycosylic bond and promote spontaneous release of base residues resulting in the formation of cytotoxic and pre-mutagenic AP sites. Glycosylase removal of the base is likely to be more advantageous than spontaneous release because this will result in rapid completion of the BER pathway in a controlled manner (Seeberg and Berdal, 1997). 7mG can also be converted by imidazole ring opening to a formamidopyrimidine residue, which is a strong cytotoxic lesion (Boiteu et al., 1984), and removal of 7mG will limit such conversion.
The substrate specificity of AlkC is similar to E. coli Tag which showed no significant affinity for 7mG and efficient excision of 3mA (Bjelland et al., 1993). However, in contrast to Tag, AlkC removes 3mG with high efficiency. This may be of advantage to an organism being exposed to high levels of alkylation where the formation of 3mG may be substantial, even though the relative rate of formation is low. It is clear from studies of E. coli that the Tag enzyme is essential for the first protection against sudden exposure to alkylation before the adaptive response is turned on (Evensen and Seeberg, 1982; Sedgwick and Lindahl, 2002). B. cereus also has an adaptive response to alkylation (Morohoshi and Munakata, 1995) and the alkC gene may serve a similar function in B. cereus as tag in E. coli.
Bacillus sp. are aerobic, endospore-forming, Gram positive rods widely distributed in soil, air and water and may be heavily exposed to alkylating agents such as methylchloride. Our data could support that Bacillus have special requirements for repair of alkylated DNA. Several Bacillus sp., including B. cereus, Bacillus anthracis and Bacillus thuringiensis, appear to contain as much as five different 3mA DNA glycosylases (Table S2). For example, the B. cereus strains ATCC 14579, E33L and G9241 contains in addition to AlkC and AlkD, two ORFs with homology to AlkA and one putative Aag glycosylase. Morohoshi and Munakata (1995) have shown that the overall level of 3mA DNA glycosylase activity in B. cereus is enhanced by treatment with low doses of alkylating agents, suggesting a DNA damaging inducible response similar to the adaptive response in E. coli. Genome sequence analysis showed that the alkA and ada operons of the adaptive response are conserved in B. cereus (Morohoshi and Munakata, 1995), indicating that the putative AlkA homologues of the alkA operons could be inducible. The complex life cycle of Bacillus may also require more pathways for maintaining genome stability. In the spores of B. subtilis a specific repair process for reversal of photoproducts has been identified (Fajardo-Cavazos et al., 1993) and repair of alkylated bases may also be necessary in spores.
DNA glycosylases is classified based on biochemical features and similarity in amino acid sequence and three-dimensional structure. One superfamily of DNA glycosylases, which is characterized by the helix hairpin helix (HhH) motif, comprises several families with different substrate specificities. For example, E. coli AlkA, Nth1 and MutY remove alkylated bases, oxidized pyrimidines and adenine mismatches respectively. Therefore, we may speculate if the AlkC/AlkD superfamily possesses DNA glycosylase activities with specificities towards a broader spectrum of base lesions including oxidized and deaminated bases.
Experimental procedures
Bacterial strains, plasmid vectors, gene libraries and growth conditions
DNA libraries were constructed using DNA isolated from B. cereus ATCC 10987, obtained from the American Type Culture Collection or by using DNA obtained from Promega. The Promega DNA was originally marketed as being from the yeast S. pombe; however, further analysis of this DNA by 16S RNA DNA sequencing and hybridization analysis revealed that the DNA purchased indeed was from B. cereus. The alkC and alkD genes were isolated from the Promega DNA and the alkE gene from ATCC 10987. Cloning vectors pUC18 and pUC19 were used for construction of libraries and for subcloning.
Escherichia coli strains DH5α and BL21 were used as recombinant hosts. The DNA glycosylase-deficient strain E. coli BK2118 (tag, alkA) described by Clark et al. (1984) was used for the complementation screening. Clones complementing the alkylation-sensitive phenotype of BK2118 were selected on Luria–Bertani (LB) agar containing 1, 3 or 5 mM MMS. From isolated colonies plasmids were isolated and checked for complementation by a second round of transformation and testing for MMS resistance. All bacteria were grown in LB broth or on LB agar at 37°C. Ampicillin was used at a concentration of 50 µg ml−1, where appropriate.
DNA sequencing and sequence analysis
Sequence analysis was performed using the Geneting program (Lillestrom, Norway) and the GCG Sequence Analysis Software (Devereux et al., 1984). Homology searches were carried out using salsa (Rognes and Seeberg, 1998), paralign (Rognes, 2001), blast and psi-blast (Altschul et al., 1997). Multiple sequence alignments were created using clustal w (Thompson et al., 1994), t-coffee (Notredam et al., 2000) and muscle (Edgar, 2004). Alignment graphics were produced using genedoc (Nicholas and Nicholas, 1997) and clustal x (Thompson et al., 1997). Accession numbers for AlkC, AlkD and AlkE for EMBL, UniProt and GenBank are given in Table S1.
Alkylation survival of BK2118 (tag, alkA) and transformed derivatives
Escherichia coli BK2118 transformed by expression constructs for the different alkylbase DNA glycosylases were grown in LB to an OD of 1.0–1.2, incubated on ice for 2–3 h, diluted in M9 buffer and spread on LB plates containing MMS at the concentrations indicated. Plates were incubated at 37°C for 2 days and the number of surviving cells was counted. The AlkA plasmid was pBK161 (Kaasen et al., 1986).
Expression and purification of AlkC and AlkD
The alkC containing fragment (1347 bp; alkC ORF 771 bp) was excised from the pUC-alkC plasmid by cleavage with EcoRI and PstI, and reinserted at the corresponding restriction sites of the expression vector pT7-SCII (Stratagene) to yield pT7-alkC. The AlkD coding region (714 bp) was PCR amplified with primers gcggatcccATGCATCCATTTGTAA AAGCA (BamHI hinge in lower case and start codon in bold) and cccaagcttAAGTCCGTCATCGCTAC (HindIII hinge in lower case) from the pUC19 construct and inserted into pT7-SCII to yield pT7-alkD. The NdeI–BamHI fragment of the polylinker of pT7-alkD was removed to shorten the distance between the ribosomal binding site and the start codon. The correct sequence of both constructs was verified by DNA sequencing.
Escherichia coli strain BL21 harbouring pT7-AlkD plasmid was grown in LB medium (10 l) to an OD600 of 0.7. The culture was induced with IPTG (0.1 mM) for 2 h at 37°C and cell extract was prepared by a combination of plasmolysis and lysozyme treatment as previously described (Seeberg, 1978). To monitor AlkD purification, 3mA DNA glycosylase activity was measured by the method of Riazuddin and Lindahl (1978) as modified (Bjelland and Seeberg, 1987). Cell extract was applied to an Affigel Blue (Bio-Rad) column (2 × 8 cm) equilibrated with buffer A (0.1 M Tris HCl, pH 8.0, 1 mM EDTA, 20% glycerol, 10 mM β-mercaptoethanol). After washing, active fractions were eluted with buffer A containing 1 M KCl. Fractions with alkylbase activity were pooled, dialysed against buffer A and applied to a MonoQ column (HR 5/5; Pharmacia). The column was eluted by a 0–2.0 M NaCl linear gradient in buffer A and peak fractions eluting between 0.2 and 0.3 M NaCl were pooled. Active fractions were diluted 1:4 in buffer A and applied to a calf thymus DNA cellulose column (HR 5/5; Pharmacia). The column was eluted with a linear gradient of 0–1.0 M KCl in buffer A and purified AlkD eluted at 0.25 M KCl.
AlkC was also expressed in E. coli BL21 and purified by a protocol similar to that used for AlkD. Extract made from 10 l culture was applied to an Affigel Blue column equilibrated with buffer A and eluted with 2 M KCl. Active fractions were pooled, dialysed against buffer A and applied to a MonoQ column. AlkC was collected in the flow-through and applied to a DNA cellulose column. The column was eluted with a linear gradient of 0–1.2 M KCl in buffer A and peak fractions eluted at 0.3 M KCl. The DNA cellulose chromatography was repeated to remove minor impurities.
Notably, E. coli AlkA and Tag showed no affinity to Affigel blue. Consequently, contaminations of endogenous 3mA DNA glycosylases were excluded during purification of AlkC and AlkD.
HPLC analysis of alkylated base derivatives
Reverse phase HPLC of methylated bases released by the purified glycosylases was performed as described by Bjelland et al. (1993). Briefly, 2.5 µg DNA (15 000 dpm µg−1) of calf thymus DNA alkylated with N-[3H]-methyl-N-nitrosourea (1.5 Ci mmol−1; NET-408, Du Pont NEN) was incubated with different amounts of enzymes as indicated for 30 min at 37°C. The DNA was precipitated with ethanol, the supernatant concentrated by lyophilization and mixed with unlabelled alkylated bases as markers. The samples were analysed by HPLC (Spheri-5 RP-18, 220 × 4.6 mm, Brownlee Laboratories) using a linear gradient of 100–75% (v/v) 0.1 M triethylammoniumacetate buffer pH 7.3 or pH 5.4 in methanol for elution (1 ml min−1). Fractions of 0.5 ml were collected and the radioactivity was measured in a liquid scintillation counter. At pH 7.3, 3mG was well separated from 3mA and 7mG whereas pH 5.4 gave good separation of 7mG from the two 3-methyl purines. The reference compounds 3mA, 3mG and 7mG were from Fluka.
DNA substrates and enzyme assays
The AP site-, 8oxoG- and faPy-containing DNA was prepared as described by Alseth et al. (1999) and 5-formyluracil and 5-hydroxymethyluracil substrates as described by Bjelland et al. (1994). The hypoxanthine-containing DNA substrate was a 25-mer oligonucleotide with hypoxanthine at position 13 (Alseth et al., 1999). The A/G mismatch substrate was identical to the hypoxanthine-containing oligonucleotide except for the substitution of an adenine for hypoxanthine. All enzyme activities were assayed as described (Alseth et al., 1999).
Acknowledgments
This research was supported by the Norwegian Research Council and The Norwegian Cancer Society. This work is dedicated to the late Erling Seeberg, who initiated this research project.
Supplementary material
The following supplementary material is available for this article online:
Fig. S1. Sequence alignment of B. cereus AlkE and E. coli AlkA.
Fig. S2. Multiple sequence alignment of B. cereus AlkC and selected homologues.
Fig. S3. Bacillus cereus AlkD and selected homologues.
Fig. S4. Sequence alignment of B. cereus AlkC (top) and AlkD (bottom) and selected ‘stepping stone’ proteins that link them together.
Table S1. Accession numbers of AlkC, AlkD and AlkE.
Table S2. 3mA DNA glycosylases in different strains of B. anthracis, B. thuringiensis and B. cereus indicated by accession numbers, and classified by sequence homology to the AlkA, AlkC, AlkD and Aag family.
This material is available as part of the online article from http://www.blackwell-synergy.com
References
- Aamodt RM, Falnes PO, Johansen RF, Seeberg E, Bjoras M. The Bacillus subtilis counterpart of the mammalian 3-methyladenine DNA glycosylase has hypoxanthine and 1,N6-ethenoadenine as preferred substrates. J Biol Chem. 2004;279:13601–13606. doi: 10.1074/jbc.M314277200. [DOI] [PubMed] [Google Scholar]
- Alseth I, Eide L, Pirovano M, Rognes T, Seeberg E, Bjoras M. The S. cerevisiae homologues of endonuclease III from E. coli, Ntg1 and Ntg2, are both required for efficient repair of spontaneous and induced oxidative DNA damage in yeast. Mol Cell Biol. 1999;19:3779–3787. doi: 10.1128/mcb.19.5.3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begley TJ, Haas BJ, Noel J, Shekhtman A, Williams WA, Cunningham RP. A new member of the endonuclease III family of DNA repair enzymes that removes methylated purines from DNA. Curr Biol. 1999;9:653–656. doi: 10.1016/s0960-9822(99)80288-7. [DOI] [PubMed] [Google Scholar]
- Berdal KG, Bjoras M, Bjelland S, Seeberg E. Cloning and expression in Escherichia coli of a gene for an alkylbase DNA glycosylase from Saccharomyces cerevisiae; a homologue to the bacterial alkA gene. EMBO J. 1990;9:4563–4568. doi: 10.1002/j.1460-2075.1990.tb07909.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessho T, Roy R, Yamamoto K, Kasai H, Nishimura S, Tano K, Mitra S. Repair of 8-hydroxyguanine in DNA by mammalian N-methylpurine-DNA glycosylase. Proc Natl Acad Sci USA. 1993;90:8901–8904. doi: 10.1073/pnas.90.19.8901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjelland S, Seeberg E. Purification and characterization of 3-methyladenine DNA glycosylase I from Escherichia coli. Nucleic Acids Res. 1987;15:2787–2801. doi: 10.1093/nar/15.7.2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjelland S, Bjoras M, Seeberg E. Excision of 3-methylguanine from alkylated DNA by 3-methyladenine DNA glycosylase I of Escherichia coli. Nucleic Acids Res. 1993;21:2045–2049. doi: 10.1093/nar/21.9.2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjelland S, Birkeland NK, Benneche T, Volden G, Seeberg E. DNA glycosylase activities for thymine residues oxidized in the methyl group are functions of the AlkA enzyme in Escherichia coli. J Biol Chem. 1994;269:30489–30495. [PubMed] [Google Scholar]
- Boiteux S, Huisman O, Laval J. 3-Methyladenine residues in DNA induce the SOS function sfiA in Escherichia coli. EMBO J. 1984;3:2569–2573. doi: 10.1002/j.1460-2075.1984.tb02175.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarti D, Ibeanu GC, Tano K, Mitra S. Cloning and expression in Escherichia coli of a human cDNA encoding the DNA repair protein N-methylpurine-DNA glycosylase. J Biol Chem. 1991;266:15710–15715. [PubMed] [Google Scholar]
- Chen J, Derfler B, Maskati A, Samson L. Cloning of eukaryotic DNA glycosylase repair gene by the suppression of a DNA repair defect in Escherichia coli. Proc Natl Acad Sci USA. 1989;86:7961–7965. doi: 10.1073/pnas.86.20.7961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke ND, Kvaal M, Seeberg E. Cloning of Escherichia coli genes encoding 3-methyladenine DNA glycosylases I and II. Mol Gen Genet. 1984;197:368–372. doi: 10.1007/BF00329931. [DOI] [PubMed] [Google Scholar]
- Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the Vax. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dosanjh MK, Chenna A, Kim E, Fraenkel-Conrat H, Samson L, Singer B. All four known cyclic adducts formed in DNA by the vinyl chloride metabolite chloroacetaldehyde are released by a human DNA glycosylase. Proc Natl Acad Sci USA. 1994a;91:1024–1028. doi: 10.1073/pnas.91.3.1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dosanjh MK, Roy R, Mitra S, Singer B. 1,N6-ethenoadenine is preferred over 3-methyladenine as substrate by a cloned human N-methylpurine-DNA glycosylase (3-methyladenine-DNA glycosylase) Biochemistry. 1994b;33:1624–1628. doi: 10.1021/bi00173a002. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelward BP, Boosalis M, Chen BJ, Deng Z, Siciliano MJ, Samson L. Cloning and characterization of a mouse 3-methyladenine/7-methylguanine/3-methylguanine DNA glycosylase cDNA whose gene maps to chromosome 11. Carcinogenesis. 1993;14:175–181. doi: 10.1093/carcin/14.2.175. [DOI] [PubMed] [Google Scholar]
- Evensen G, Seeberg E. Adaptation to alkylation resistance involves the induction of a DNA glycosylase. Nature. 1982;296:773–775. doi: 10.1038/296773a0. [DOI] [PubMed] [Google Scholar]
- Fajardo-Cavazos P, Salazar C, Nicholson WL. Molecular cloning and characterization of the Bacillus subtilis spore photoproduct lyase (spl) gene, which is involved in repair of UV radiation-induced DNA damage during spore germination. J Bacteriol. 1993;175:1735–1744. doi: 10.1128/jb.175.6.1735-1744.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falnes PO, Johansen R, Seeberg E. AlkB mediated oxidative demethylation of reverses DNA damage in Escherichia coli. Nature. 2002;419:178–182. doi: 10.1038/nature01048. [DOI] [PubMed] [Google Scholar]
- Fortini P, Pascucci B, Parlanti E, Errico MD, Simonelli V, Dogliotti E. The base excision repair: mechanisms and its relevance for cancer susceptibility. Biochimie. 2003;85:1053–1071. doi: 10.1016/j.biochi.2003.11.003. [DOI] [PubMed] [Google Scholar]
- Kaasen I, Evensen G, Seeberg E. Amplified expression of the tag+ and alkA+ genes in Escherichia coli: identification of gene products and effects on alkylation resistance. J Bacteriol. 1986;168:642–647. doi: 10.1128/jb.168.2.642-647.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karran P, Lindahl T, Øfsteng I, Evensen GB, Seeberg E. Escherichia coli mutants deficient in 3-methyladenine-DNA glycosylase. J Mol Biol. 1980;140:101–127. doi: 10.1016/0022-2836(80)90358-7. [DOI] [PubMed] [Google Scholar]
- Karran P, Hjelmgren T, Lindahl T. Induction of a DNA glycosylase for N-methylated purines is part of the adaptive response to alkylating agents. Nature. 1982;296:770–773. doi: 10.1038/296770a0. [DOI] [PubMed] [Google Scholar]
- Larson K, Sahm J, Shenkar R, Strauss B. Methylation-induced blocks to in vivo DNA replication. Mutat Res. 1985;150:77–84. doi: 10.1016/0027-5107(85)90103-4. [DOI] [PubMed] [Google Scholar]
- McCarthy TV, Karran P, Lindahl T. Inducible repair of O-alkylated DNA pyrimidines in Escherichia coli. EMBO J. 1984;3:545–550. doi: 10.1002/j.1460-2075.1984.tb01844.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memisoglu A, Samson L. Cloning and characterization of a cDNA encoding a 3-methyladenine DNA glycosylase from the fission yeast Schizosaccharomyces pombe. Gene. 1996;177:229–235. doi: 10.1016/0378-1119(96)00308-3. [DOI] [PubMed] [Google Scholar]
- Morohoshi F, Munakata N. Diverse capacities for the adaptive response to DNA alkylation in Bacillus species and strains. Mutat Res. 1995;337:97–110. doi: 10.1016/0921-8777(95)00013-a. [DOI] [PubMed] [Google Scholar]
- Nicholas KB, Nicholas HB., Jr 1997 GeneDoc version 2.6.002 URL http://www.psc.edu/biomed/genedoc.
- Notredame C, Higgins D, Heringa J. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J Mol Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- O'Connor TR, Laval F. Isolation and structure of a cDNA expressing a mammalian 3-methyladenine-DNA glycosylase. EMBO J. 1990;9:3337–3342. doi: 10.1002/j.1460-2075.1990.tb07534.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor TR, Laval J. Human cDNA expressing a functional DNA glycosylase excising 3-methyladenine and 7-methylguanine. Biochem Biophys Res Commun. 1991;176:1170–1177. doi: 10.1016/0006-291x(91)90408-y. [DOI] [PubMed] [Google Scholar]
- O'Rourke EJ, Chevalier C, Boiteux S, Labigne A, Ielpi L, Radicella JP. A novel 3-methyladenine DNA glycosylase from Helicobacter pylori defines a new class within the endonuclease III family of base excision repair glycosylases. J Biol Chem. 2000;275:20077–20083. doi: 10.1074/jbc.M001071200. [DOI] [PubMed] [Google Scholar]
- Pierre J, Laval J. Cloning of Micrococcus luteus 3-methyladenine-DNA glycosylase genes in Escherichia coli. Gene. 1986;43:139–146. doi: 10.1016/0378-1119(86)90017-x. [DOI] [PubMed] [Google Scholar]
- Riazuddin S, Lindahl T. Properties of 3-methyladenine-DNA glycosylase from Escherichia coli. Biochemistry. 1978;17:2110–2118. doi: 10.1021/bi00604a014. [DOI] [PubMed] [Google Scholar]
- Rognes T. ParAlign: a parallel sequence alignment algorithm for rapid and sensitive database searches. Nucleic Acids Res. 2001;29:1647–1652. doi: 10.1093/nar/29.7.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rognes T, Seeberg E. SALSA: improved protein database searching by a new algorithm for assembly of sequence fragments into gapped alignments. Bioinfomatics. 1998;14:839–845. doi: 10.1093/bioinformatics/14.10.839. [DOI] [PubMed] [Google Scholar]
- Samson L, Cairns J. A new pathway for DNA repair in Escherichia coli. Nature. 1977;19:281–283. doi: 10.1038/267281a0. [DOI] [PubMed] [Google Scholar]
- Samson L, Derfler B, Call K. Cloning and characterization of a 3-methyladenine DNA glycosylase cDNA from human cells whose gene maps to chromosome 16. Proc Natl Acad Sci USA. 1991;88:9127–9131. doi: 10.1073/pnas.88.20.9127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santerre A, Britt AB. Cloning of a 3-methyladenine-DNA glycosylase from Arabidopsis thaliana. Proc Natl Acad Sci USA. 1994;91:2240–2244. doi: 10.1073/pnas.91.6.2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saparbaev M, Laval J. Excision of hypoxanthine from DNA containing dIMP residues by the Escherichia coli, yeast, rat, and human alkylpurine DNA glycosylases. Proc Natl Acad Sci USA. 1994;91:5873–5877. doi: 10.1073/pnas.91.13.5873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saparbaev M, Kleibl K, Laval J. Escherichia coli, Saccharomyces cerevisiae, rat and human 3-methyladenine DNA glycosylases repair 1,N6-ethenoadenine when present in DNA. Nucleic Acids Res. 1995;23:3750–3755. doi: 10.1093/nar/23.18.3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedgwick B, Lindahl T. Recent progress on the Ada response for inducible repair of DNA alkylation damage. Oncogene. 2002;21:8886–8894. doi: 10.1038/sj.onc.1205998. [DOI] [PubMed] [Google Scholar]
- Seeberg E. Reconstitution of an Escherichia coli repair endonuclease activity from the separated uvrA+ and uvrB+/uvrC+ gene products. Proc Natl Acad Sci USA. 1978;75:2569–2573. doi: 10.1073/pnas.75.6.2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeberg E, Berdal KG. Repair of alkylation damage to DNA. In: Hickson ID, editor. Base Excision Repair of DNA Damage. Georgetown, TX: Landes Bioscience; 1997. pp. 151–168. [Google Scholar]
- Seeberg E, Eide L, Bjoras M. The base excision repair pathway. Trends Biochem Sci. 1995;20:391–397. doi: 10.1016/s0968-0004(00)89086-6. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;24:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trewick SC, Henshaw TF, Hausinger RP, Lindahl T, Sedgwick B. Oxidative demethylation by Escherichia coli AlkB reverts DNA base damage. Nature. 2002;419:174–178. doi: 10.1038/nature00908. [DOI] [PubMed] [Google Scholar]
- Vaughan P, Lindahl T, Sedgwick B. Induction of the adaptive response of Escherichia coli to alkylation damage by the environmental mutagen, methyl chloride. Mutat Res. 1993;293:249–257. doi: 10.1016/0921-8777(93)90076-s. [DOI] [PubMed] [Google Scholar]