Summary
In transcription initiation, all RNA polymerase molecules bound to a promoter have been conventionally supposed to proceed into elongation of transcript. However, for Escherichia coli RNA polymerase, evidence has been accumulated for a view that only its fraction can proceed into elongation and the rest is retained at a promoter in non-productive form: a pathway branching in transcription initiation. Proteins such as GreA and GreB affect these fractions at several promoters in vitro. To reveal the ubiquitous existence of the branched mechanism in E. coli, we searched for candidate genes whose transcription decreased by disruption of greA and greB using a DNA array. Among the arbitrarily selected 11 genes from over 100, the atpC, cspA and rpsA passed the test by Northern blotting. The Gre factors activated transcription initiation from their promoters in vitro, and the results demonstrated that the branched mechanism is exploited in vivo regulation. Consistently, decrease in the level of the GreA in an anaerobic stationary condition accompanied a decrease in the levels of transcripts of these genes.
Introduction
In elucidating a biological process, the mechanism tends to be supposed as a simple sequence of its essential steps, and the steps with unknown roles are often disregarded, for example, the early transcription in prokaryotes. For several decades, the mechanism has been assumed to be a sequence of three essential steps: formation of a complex between RNA polymerase and a promoter with a locally melted DNA duplex (open complex), synthesis of oligo-RNA (initiation of the chemical reaction), and escape of the RNA polymerase from the promoter associated with the progress of RNA elongation (promoter clearance). However, all prokaryotic and eukaryotic RNA polymerases so far isolated have been known to perform ‘abortive initiation’, which is an iterative synthesis and release of oligo-RNA molecules in vitro (Johnston and McClure, 1976; Carpousis and Gralla, 1980). As a minimal model based on these pioneering works, abortive transcripts were assumed to be unsuccessful precursor of the full-length transcripts, and thus abortive synthesis has been supposed to be a step of the promoter clearance in a sequential model (Munson and Reznikoff, 1981).
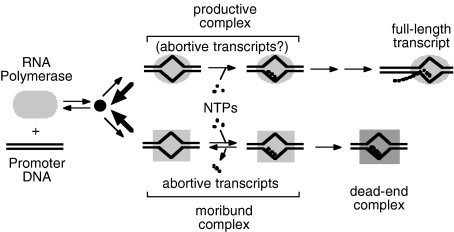
This sequential model was first tested in a kinetic study using the lacUV5 and a modified λPR (λPRAL) promoter, by comparing the times at which the full-length and abortive syntheses complete under single-round conditions. Surprisingly, abortive synthesis continues long after the full-length synthesis has completed, contradicting the idea that abortive transcripts would be an unsuccessful precursor. The long-lived transcription complex was named a moribund complex, being thus defined as a complex that produces only abortive and no full-length transcripts (Kubori and Shimamoto, 1996). The following characteristics are known for this type of complex. (i) The moribund complex is formed from the same homogeneous fraction of enzyme as the productive complex that synthesizes full-length transcript at λPRAL promoter, and dissociation of the enzyme from the promoter DNA cancels any difference between these complexes (Kubori and Shimamoto, 1996). (ii) The moribund complexes are structurally different from the productive: they slightly backtracked at the λPRAL promoter or forward-tracked at the T7A1 promoter and have more exposed conserved region 3 of σ70 (Sen et al., 1998; 2000; Susa et al., 2002). (iii) At several promoters, the moribund complex is further converted into a dead-end complex that still retains short transcript but has no elongation activity (Kubori and Shimamoto, 1996). Such complexes at λPRAL promoter are irreversibly arrested at the promoter in the form of holoenzyme (Sen et al., 2000). (iv) Initiation at the T7A1 promoter, which have long been shown no kinetic evidence for moribund complex, was found to accumulate moribund complex at a low-salt condition, thereby the initiation mechanism may be generally branching into the productive and non-productive pathways (Susa et al., 2002), and the non-productive pathway can lead to a dead-end as shown in Fig. 1. Therefore, the fates of the moribund complex are either inactivation as a dead-end complex, or reactivation by conversion into productive complex, and these conversions occur at different rates depending on the promoter, yielding variations in characteristics of promoters (Kubori and Shimamoto, 1997; Sen et al., 2001; Susa et al., 2002). (v) There are protein factors and mutant σ70 that introduce reversibility between the non-productive and productive pathways in a manner that depends on the promoter (Sen et al., 1998; 2001; Tagami and Aiba, 1998; Susa et al., 2002). This reversibility is the reason why there is little accumulation of the moribund complex at the T7A1 promoter in standard ionic conditions, converting the moribund complex into productive one rapidly consumed (Shimamoto et al., 1981; Kubori and Shimamoto, 1997; Susa et al., 2002). These results indicate the generality of the branched mechanism and explain that the seeming sequential pathway is due to reversibility between the branched pathways. This mechanism provides a concrete example of the molecular memory in the form of conformation of transcription complex on how it has been initiated (Qi et al., 1996; Berghöfer-Hochheimer et al., 2005).
Fig. 1.
Branched initiation pathway and action of Gre factors at branches. The initiation pathway is composed of two branches; one leads to a productive elongation complex and the other to a moribund complex then to a dead-end complex. RNA polymerase holoenzyme and DNA carrying a promoter are, respectively, displayed as grey ovals and double lines. ‘NTPs’ indicates ATP, GTP, CTP and UTP, and are shown as small dots and transcripts are indicated as dotted lines. Moribund complex is a major source of abortive transcripts, but the absence of abortive synthesis in the other steps has not been established. At the promoters such as λPRAL and lacUV5, irreversible branching occurs (thin arrows), and a moribund complex once formed is subsequently converted into a dead-end complex. In the presence of Gre factors, the branching becomes reversible (thick arrows) so that moribund and productive complexes can exchange with each other. In this case, the branched pathway becomes kinetically equivalent to the conventional sequential pathway if the formation of a dead-end complex is negligible. This situation also occurs in initiation at the promoter with intrinsic reversibility such as T7A1. The big black dot is the earliest branching point and could be a binary intermediate or free state depending on a promoter.
All this evidence has been obtained in vitro, and thus the most important question should be whether or not the same mechanism works in cells and contributes to the regulation of transcription. The clue to the question is the Gre factors, GreA and GreB. These factors increase reversibility between the branched pathways (Fig. 1), and consequently increase the productive initiation by enhancing conversion of the otherwise moribund complex into the productive one, in the presence of a physiological concentration of initiating nucleoside triphosphate (NTP) at the λPRAL promoter (Sen et al., 2001). The Gre factors, GreA and GreB, originally isolated as factors inducing cleavage of the nascent transcript in arrested complexes in elongation (Borukhov et al., 1992; 1993), are shown to stimulate productive initiation (Feng et al., 1994; Hsu et al., 1995). If the branched mechanism works as a regulatory mechanism in vivo, a decrease of the amount of the Gre factors should result in the reduction of productive transcription from some promoters. To test this hypothesis, we first constructed a double disruptant, ΔgreAΔgreB, of Escherichia coli and then compared the levels of mRNAs with those in the parental strain. Among approximately 200 genes less transcribed in the mutant, we arbitrarily selected 11 genes. Three genes among them, atpC (uncC), cspA and rpsA, passed a test by Northern blotting and then their transcription initiation displayed a branched pathway in a reconstituted transcription system composed of purified components. Furthermore, we confirmed that the level of GreA in the parental strain varied in different culture conditions, suggesting the role of a Gre factor as the regulator. These results provide evidence that the branched mechanism exists in vivo and can be exploited in the regulation of transcription initiation from some promoters.
Results
Phenotypes of ΔgreAΔgreB strain
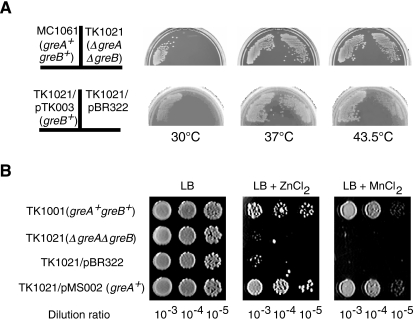
We first constructed an E. coli strain, TK1021, lacking greA and greB. This double mutant cannot grow at 30°C (Fig. 2A, top) and is hypersensitive to divalent metal ions such as Zn2+ and Mn2+ (Fig. 2B). These defects are suppressed by a plasmid harbouring either greA or greB (Fig. 2A, bottom, and Fig. 2B), or both genes (data not shown), although the suppressions by a gre gene are shown in Fig. 2. We examined the phenotype of single disruptant of greA or greB, which had been obtained on the way of constructing the double disruptant. The greA+greB– strain showed little difference from its parental strain in the temperature dependence and the sensitivity to Mn2+. In contrast, the greA–greB+ strain indicated the same phenotype as the double disruptant, but the phenotype was so slight that no further analysis was carried out.
Fig. 2.
Phenotypes of greA– and greB– strains. A. Temperature sensitivities. MC1061(greA+greB+) and TK1021(ΔgreAΔgreB) were spread, and incubated on LB plates at the indicated temperatures overnight. pTK003 is a derivative of pBR322 expressing greB.
B. Sensitivities to Zn2+ and Mn2+. Overnight cultures of TK1001(greA+greB+) and TK1021 were spotted onto LB, LB + 1.5 mM ZnCl2 and LB + 5 mM MnCl2 plates and incubated for 24 h. pMS002 is a derivative of pBR322 expressing greA.
Escherichia coli genes whose transcription is reduced in the absence of gre genes
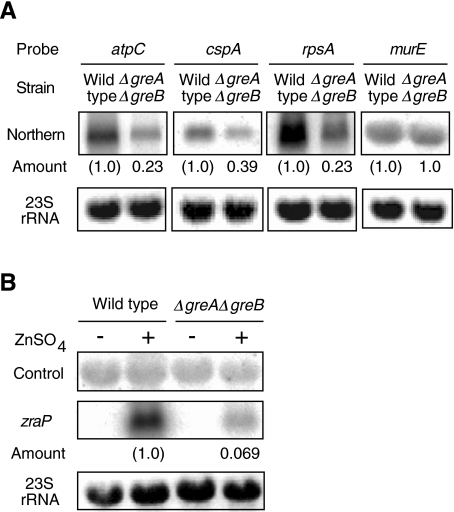
We next searched for genes whose transcription level is reduced by the disruption of greA and greB. In the primary screening using a DNA array, we could not find distinct reduction in the amounts of the cDNAs prepared from the disruptant and its parental strain. Therefore, we gave up to deduce any conclusion from the analysis, and limited its use for selecting rough candidates. Instead of quantification of the arrays by scanning individual signals, we analysed the array by subtracting a density image from another (data not shown). Among the approximately 200 genes hinted by this rough method, we selected 11 candidates showing relatively large differences for examination by Northern analysis (see Experimental procedures for selection criteria). We excluded the y-genes from the candidates because their functions have not been estimated in the literature of the lack of further suggestion on cross-talk with other regulatory circuits. Among the candidates, the levels of transcripts were lower in seven cases (atpC, cspA, rpsA, cspE, lpp, infC and ptsH). As expected from the roughness of the array analysis, three others (nagE, hlpA and pstB) actually showed increased levels, and one (hycF) unchanged in the ΔgreAΔgreB strain (data not shown). Among the seven downshifted genes, the atpC, cspA and rpsA transcripts showed the largest changes: decreased by 60–80%, while the control murE transcript remained constant (Fig. 3A). Transcription of these three genes is known to be dependent on σ70 (Porter et al., 1983; Pedersen et al., 1984; Tanabe et al., 1992), as well as the control murE (Michaud et al., 1990). The lower but significant levels of these genes in the disruptant, as well as the fact that greA and greB are not essential for growth in a standard condition, indicate that greA and greB are involved in fine-tuning their expression rather than shutting off and turning on.
Fig. 3.
Effect of disruption of greA and greB on the levels of atpC, cspA, rpsA and zraP transcripts. Northern analyses were carried out by using the 32P-labelled atpC, cspA, rpsA, murE and zraP probes. The 23S rRNA was visualized by staining with ethidium bromide before blotting, confirming the loading of the same amount of total RNA. The amounts in the ΔgreAΔgreB strain are normalized against those in the wild type.
A. The levels of the cspA, atpC and rpsA mRNA. The murE mRNA is shown as an unchanged control.
B. Effect of the greA and greB double disruption on the zraP transcription. The cells were cultured in the presence or absence of 1 mM ZnSO4 in LB media. The ‘Control’ is the bands non-specifically hybridized with the zraP probe and used as an unchanged control.
As the DNA array cannot detect transcripts of extremely low levels, we also searched for the gre-dependent genes on a functional basis. The hypersensitivity of the ΔgreAΔgreB strain to Zn2+ led us to examine a gene involved in resistance against Zn2+, zraP (yjaI), which is known to be induced by Zn2+ (Noll et al., 1998). As expected, Northern analysis showed that the induction required gre genes (Fig. 3B). This example confirms the results of the array analysis that there will be many genes whose expressions are affected by greA and greB. As transcription of zraP has been suggested to be σ54-dependent (Leonhartsberger et al., 2001), we did not study further zraP transcription in this work on σ70-dependent initiation.
Transcription of the E. coli genes is activated in initiation but not in elongation
To determine whether the Gre factors enhance the transcription of atpC, cspA and rpsA in initiation or in elongation, we carried out in vitro transcription reconstituted by purified components on a linear DNA template containing a promoter (Fig. 4A). For atpC, a member of atp operon, the major promoter of the operon was selected because the length of atpC transcripts shown in the Northern analysis was consistent with initiation from the promoter. For rpsA, two DNA templates were prepared because there are two major promoters, P1 and P3. Transcription was labelled with a [γ-32P]-labelled initiating NTP, so that the stoichiometry of the transcripts was measured irrespective of their lengths. The amounts of full-length synthesis initiated at these promoters were all limited in the absence of GreA and GreB, and the levels increased by 2- to 17-fold in their presence (Fig. 4B). These increases were accompanied by decreases in the amounts of some abortive transcripts, suggesting that the moribund complexes are converted into the productive one. Notably no significant decrease was detected in the amounts of transcripts in size between abortive and the full-length products, indicating that the Gre factors were not acting in elongation. The full-length transcripts from the control promoter, T7A1, showed little changes by the addition of GreA or GreB, if the typical error of ± 25% is taken into account (Fig. 4B). This result indicates the absence of any contaminations activating transcription in the preparations of the Gre factors. It also confirms that the Gre factors cannot affect transcription from the promoter where the branched pathway is intrinsically reversible (Susa et al., 2002), being consistent with the introduction of reversibility by the Gre factors. The effects by GreA were equal to or slightly more than GreB (Fig. 4B; Sen et al., 2001). As GreB is generally more active in the relief of elongation arrest (Borukhov et al., 1993), this tendency may be specific to their action in initiation.
Fig. 4.
Effects of GreA or GreB on in vitro transcription from atp, cspA and rpsA promoters.
A. Promoter sequences of the template DNAs. The indicated numbers are the co-ordinates of the DNA ends relative to the transcription start sites at +1. The start sites are denoted by the single-underlined letters (Porter et al., 1983; Pedersen et al., 1984; Tanabe et al., 1992). The −10 and −35 boxes that have been previously suggested (Porter et al., 1983; Pedersen et al., 1984; Tanabe et al., 1992) are indicated by double-underlined letters.
B. Transcripts produced in the absence or presence of GreA or GreB. They are 5′-end-labelled with [γ-32P]-GTP or [γ-32P]-ATP. Open and closed triangles indicate the positions of full-length (FL) and abortive transcripts respectively. The amounts of full-length transcripts are indicated below the autoradiograms, as ratios to the amounts observed in the absence of the Gre factors.
Formation of the moribund complex at an authentic promoter ofE. coli
If the branched pathway is the mechanism of initiation activated by the Gre factors, the moribund complex should be observed at the authentic promoters. To examine this prediction, we applied the inversed pulse-chase assay (Kubori and Shimamoto, 1996) to the atp promoter chosen as a representative. This is the most sensitive method for detecting persistent abortive synthesis carried out by a moribund complex, and the result can be the sufficient condition for the existence of moribund complex. In the single-round condition ensured by a template with a bead blocking elongation at its downstream end (see Experimental procedures), transcription from the promoter was started with unlabelled NTPs at time zero, and the [γ-32P]-GTP, the initiating NTP in this case, was then added at various time points, followed by further incubation for 20 min (Fig. 5A). The amount of full-length labelled transcript is equal to the amount of binary complexes that still survive at the given time point and can still synthesize long transcripts, i.e. the productive binary complexes surviving at that time. On the other hand, the amounts of labelled abortive transcripts are proportional, but may not be stoichiometric, to the amount of surviving binary complexes that carry out abortive synthesis. Therefore, increase in the ratio of the amount of an abortive transcript to that of the full-length labelled transcript means the existence of a complex that synthesizes only abortive products, namely moribund complex. As expected, the observed ratios of the amounts of 5- or 7-mer abortive transcripts to those of full-length transcripts did increase by two- to threefold in 60 min (Fig. 5B and C). Therefore, the moribund complex is formed, namely the pathway is branching in initiation from an authentic promoter of E. coli.
Fig. 5.
Inversed pulse-chase assay for detection of persistent abortive synthesis at the atp promoter.
A. Transcripts produced from the DNA template containing the atp promoter in the pulse-chase assay.
B and C. The ratios of the amounts of 7-mer (B) and 5-mer (C) abortive transcripts to those of full-length transcripts, plotted against the time spent in the reaction with unlabelled NTPs.
The branched mechanism works in gene regulation in cells
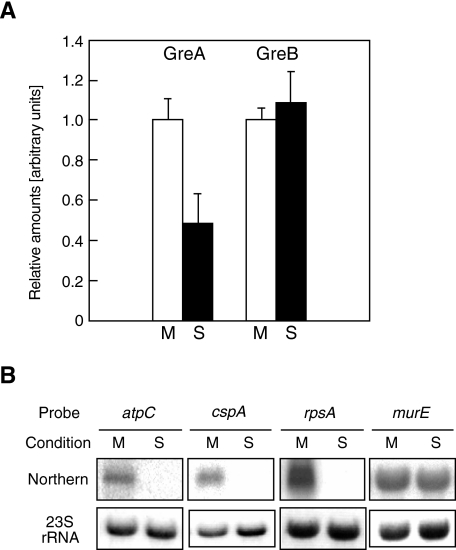
To claim that the branched pathway controlled by a Gre factor is a regulatory mechanism, we must show that the level of the Gre factors must be changed according to some environmental or physiological changes. Therefore, we measured the levels of the GreA in the greA+greB+ strain TK1001 under various growth conditions. In Western analysis, significant changes were detected under different aerobic and growth conditions. As shown in Fig. 6A, the fraction of GreA in the total protein in shaking culture at mid-log phase was almost as twice as that in standing culture at stationary phase. We also confirmed that the amounts of the atpC, cspA and rpsA transcripts were decreased in the latter condition (Fig. 6B). However, the observed decreases were much larger than that observed in the ΔgreAΔgreB strain (Fig. 3A) and than that of the level of GreA. These differences may indicate that there are more factors relating in the different growth conditions. For example, factors induced in the anaerobic and stationary condition could inhibit the basal level of the expression of the genes and/or the complete absence of the Gre factors could indirectly increase a basal level of the expression. The result obtained in vivo can be explained by many other models, even by those supposing no roles of GreA, but it does not contradict the regulatory function of GreA in initiation. Regulation of GreA has been speculated from the structure of its gene: a terminator-like stem-loop structure followed by U stretch in the 5′-untranslated region (5′-UTR) (Sparkowski and Das, 1990). This structure actually blocks transcription in vitro (data not shown), although this does not necessarily mean the function of the structure is the same in vivo.
Fig. 6.
Changes in the amount of Gre factors and in the levels of atpC, cspA and rpsA transcriptions. TK1001(greA+greB+) was cultured with shaking and harvested at mid-log (M), or cultured without shaking and harvested at stationary phase (S).
A. The amounts of GreA and GreB measured by Western analyses using their antibodies. The average amount at mid-log phase in shaking culture is set as 1.0 and the relative amount at stationary phase in standing culture is shown. The standard deviations were obtained from three or more independent runs.
B. Northern analyses of atpC, cspA, rpsA and murE mRNA. Total RNAs were applied to each lane, and the mRNAs are detected by the indicated probes. The 23S rRNAs are shown as in Fig. 2.
In contrast to GreA, no significant changes in the amount of the GreB were so far observed (Fig. 6A). A possible speculation is that the GreB may contribute to the maintenance of the basal levels of atpC, cspA and rpsA, or may have no effects on their expressions, being paralleled by the observed lack of the phenotype for the greB– single mutant. In this context, it is interesting that disruption of the greB has little effect on the relief of the elongation pause at the early pause region of phage λ late gene (Marr and Roberts, 2000). Near the greB promoter, there are no definite structures suggesting its regulation. However, absence of evidence is not evidence for absence. In conclusion, transcription initiation of at least several genes is likely to be under regulation at least partly by using the branched mechanism in response to the level of the GreA.
Discussion
ΔgreAΔgreB strain shows pleiotropic phenotype
We found that our MC1061ΔgreAΔgreB strain, TK1021, grew at 43.5°C but not at 30°C, while the previously reported strains with disrupted greA and greB, MC4100ΔgreAΔ(ompR-greB) named AD8571 (Orlova et al., 1995) and MG1655ΔgreAΔgreB named N5306 (Trautinger and Lloyd, 2002), are sensitive to temperatures of 42–43°C. Therefore, we transferred our greA disruption into the MC4100Δ(ompR-greB). We found that the strain was sensitive to temperature of 43.5°C (data not shown), and concluded that the differences between these mutants may well be due to their different genetic backgrounds.
Our finding of hypersensitivity of our ΔgreAΔgreB strain to Mn2+ and Zn2+ may have more physiological significance, because these ions are essential to E. coli but their excess is generally toxic. The replacement of an Mg2+ at the catalytic site of the RNA polymerase with Mn2+ may induce misincorporation of NTP (Niyogi and Feldman, 1981). As the Gre factors are suggested to stabilize an Mg2+ at the site (Leptenko et al., 2003; Opalka et al., 2003; Sosunova et al., 2003), it is possible that they prevent the replacement of Mg2+ and thus contribute to the fidelity of transcription. This model is consistent with the observed prevention of the misincorporation by GreA (Erie et al., 1993), although other interpretations are also possible. The concentration of Zn2+ in cells is strictly regulated by the balance between its uptake and excretion (Outten and O’Halloran, 2001). In E. coli, the ZraP (YJAI) protein, of unknown function, is required for Zn2+ homeostasis: the upshift of ZraP makes cells more tolerant to Zn2+ (Noll et al., 1998). Therefore, insufficient induction of zraP transcription in the ΔgreAΔgreB strain provides a possible explanation for the hypersensitivity to Zn2+.
In our DNA array and Northern analysis, the downshifted genes cover various functional categories, energy production (atpC), translation (rpsA, infC), cold-shock response (cspA, cspE), membrane component (lpp) and sugar metabolism (ptsH), implying that the effect of the gre genes is pleiotropic in E. coli. Recently, an important role of the Gre factors is indirectly suggested; DksA was found to be a critical factor explaining the difference between the large effects of ppGpp in vivo and its moderate effects in vitro (Paul et al., 2004). According to the structural similarity of DksA to GreA (Perederina et al., 2004), these factors seem to share the same binding site on RNA polymerase, the smaller channel. As a consequence of the common binding site, a Gre factor will compete with the DksA, and possibly other unknown factors, for the binding site. Therefore, the changes in the levels of the Gre factors may indirectly affect the alternative regulations by DksA or the other factors sharing the same site on the RNA polymerase, and vice versa. In Thermus thermophilus, Gfh1, a GreA homologue lacking the cleavage-stimulation activity, competes with GreA for a binding site on the polymerase (Hogan et al., 2002), supporting this hypothesis. Therefore, a decrease of a Gre factor could produce not only factor-free RNA polymerase but also the enzyme bound to DksA and possibly other unknown factors, and the enzyme in these forms can contributes to the variation in responses to different conditions. These direct and indirect effects of the Gre factors on transcription regulation may explain the reasons why the transcripts of the three genes decrease more drastically in the stationary aerobic condition than in the disruptant, and why the phenotype of the ΔgreAΔgreB strain is pleiotropic.
How do Gre factors relieve the initiation arrest?
Arrest of RNA polymerase occurs in initiation and elongation stages, and Gre factors relieve both arrests. In the elongation arrest, backtracking of the RNA polymerase disengages catalytic centre from the RNA 3′-end and inactivates the complex, and the Gre factors allow such enzyme to re-elongate by inducing intrinsic transcript cleavage to expose a newly generated 3′-end to the catalytic centre (see Borukhov et al., 2001; 2005). Thus, the cleavage-stimulation activity of the Gre factors is essential for their relief of polymerase from the arrest in elongation.
In initiation, on the other hand, RNA cleavage by the Gre factors may not be necessary for relief of the arrest. The success of the inversed pulse-chase experiment indicates the existence of a binary complex committed to abortive synthesis, binary moribund complex. In addition, a kinetic study indicated that GreA and B have to be bound to RNA polymerase at the binary complex stage to relieve initiation arrest at the λPRAL promoter and that a later binding abolishes the activity (Sen et al., 2001). These results provide the evidence that the binary complex is the target for the Gre factors to bind. The most straightforward speculation would be that the Gre factors relief as an simple allosteric effector without RNA cleavage, but the results do not deny the involvement of RNA cleavage in the relief by GreA. In this context, an interesting example is the effect of cAMP receptor protein (CRP) on the binary complex formed at the malT promoter. The binary complex in the absence of CRP satisfies the definition of moribund complex, synthesizing only abortive transcripts. The coexistence of CRP with the binary complex is sufficient to relieve the initiation arrest and its coexistence with a ternary complex, binary complex plus transcript, is not required (Tagami and Aiba, 1998). Because no RNA cleaving activity has been so far observed for the protein and its presence at the ternary complex stage is not required, this experiment provides the example of the relief from initiation arrest by an allosteric effector. As the position of branching point can be more than one and can depend on a promoter, the existence of abortive synthesis in the productive branch is still an open question.
Significance of branched mechanisms
The moribund complex was recently tried to be dismissed as an artefactual inactivation in vitro (Vo et al., 2003), but the possibility they examined has been already denied in the first article (Kubori and Shimamoto, 1996) by the conversion from productive into moribund complex and by its reverse conversion in the presence of the Gre factors (Sen et al., 2001). The finding of moribund complex at authentic promoters in this work may be a more convincing answer to the question. Does abortive synthesis occur in the productive pathway? Vo et al. (2003) claimed the existence of abortive synthesis in the productive branch at the T5N25 promoter, based on the assumption that elongation rates are the same in two branches. However, the rate of non-productive elongation is much slower than that of the productive one at least at the λPRAL promoter, judging from the time-course of elongation retained by moribund complex continues long after the completion of pen question, and its test requires a promoter that has been proved to produce no moribund complex.
The branched mechanism may contribute to the high fidelity of transcription by aborting transcripts with misincorporation. Several promoters are known to produce abortive transcripts with anomalous mobilities caused by misincorporation, including slippage (Metzger et al., 1993). At the λPRAL promoter, all the transcripts including misincorporation are found in abortive but not the full-length (32 nucleotides) transcripts, and are always elongated 1–2 nucleotides beyond the site of misincorporation, allowing a kind of proofreading (Kubori and Shimamoto, 1997). We are inclined to explain that misincorporation switches the conformation of transcription complex from productive into moribund, and elongation of the transcript and its dissociation occurs simultaneously. Such a mechanism is impossible in a classic sequential model.
The branched mechanism in initiation is not limited to the branching into moribund complex. Recently, Travers and Muskhelishvili (2005) proposed the branching of productive initiation into a supercoiling-sensitive pathway involving FIS and a ppGpp-sensitive pathway involving DksA, which supposes complexes interacting differently with the spacer region between −35 and −10 hexemer sequences. The regulatory mechanism of the stuttering synthesis must involve another branching mechanism because an event in initiation changes the fate of the transcript at a later step (Qi et al., 1996). Caution should be given to the rate-limiting step of a branched mechanism with more than one parallel pathway leading to the product. In such a mechanism, deceleration of a rate-limiting step can make a new rate-limiting step in another pathway, which never happens if the product is formed in a single pathway, allowing contradicting properties of the rate-limiting steps.
Another caution must be paid when a non-productive branched pathway is handled as a hypothetical step in a sequential pathway, for example, inclusion of the moribund pathway in the promoter escape because transcription initiation can be regulated differently in the classic sequential and the branched mechanisms with productive and non-productive pathways. In the former, the level of initiation can be regulated only at the rate-limiting step, while in the latter the level is determined only by the fraction of enzyme molecules in the productive pathway independently of the position of the rate-limiting step. When some physiological changes shift the rate-limiting step from one process to another, the branched mechanism can provide a unique and robust regulation insensitive to such a change. The second possible benefit of the mechanism is that blocking at the promoter prevents the wasteful queue of RNA polymerase molecules, and that moribund complex can reserve potentially active RNA polymerase for emergency. For the branched mechanism involving moribund complex and GreA, this idea is consistent with the observation that expression of greA is greatly stimulated by σE-dependent transcription (C.A. Gross, pers. comm.). The third possible benefit is maintenance of the effect of a regulator. If conversion between two pathways is essentially irreversible, the moribund or productive conformation is maintained long after the dissociation of a factor that has triggered a new conformation. Therefore, the mechanism can work as a molecular memory, and a mechanism involving a memory is described as a branched mechanism. These distinct properties of the branched mechanism provide variety in gene regulation that will be required for the survival in various conditions.
Experimental procedures
Materials
All strains were grown either in LB medium or in M9 medium supplemented with 0.2% (w/v) glycerol, 10 µg ml−1 thiamine, 1 mM MgSO4, 0.2% (w/v) casamino acids and 10 µg ml−1l-tryptophan. Antibiotics were used at the following concentrations: ampicillin, 50 µg ml−1; kanamycin, 25 µg ml−1; chloramphenicol, 20 µg ml−1; and tetracycline, 25 µg ml−1. The E. coli strains and plasmids newly constructed in this study are listed in Table 1.
Table 1.
Newly constructed E. coli strains and plasmids used in this study.
| Strain/plasmid | Genotype/property |
|---|---|
| Strain | |
| TK1001 | MC1061 zgj-203::Tn10 |
| TK1007 | MC1061 greB::cat |
| TK1009 | MC1061 greA::kan, zgj-203::Tn10 |
| TK1021 | MC1061 greA::kan, greB::cat, zgj-203::Tn10 |
| Plasmid | |
| pTK002 | Derivative of pKH5002 containing greA gene |
| pTK003 | Derivative of pBR322 containing greB gene |
| pTK007 | Derivative of pBR322 containing greB gene disrupted with CmR cassette |
| pTK008 | Derivative of pKH5002 containing greB gene disrupted with CmR cassette |
| pTK010 | Derivative of pKH5002 containing greA gene disrupted with KanR cassette |
| pMS002 | Derivative of pBR322 containing greA gene |
The 3.3 kb BamHI–EcoRI fragment of Kohara's library phage ♯521 (Kohara et al., 1987) that harbours greA was inserted between the corresponding sites of a suicide plasmid pKH5002 (Ohmori et al., 1995). The greA gene of the resulting plasmid, pTK002, was disrupted by replacing the BglI–BglI segment of 454 bp, which corresponds to 64 nucleotides of 5′-UTR and residues 1–130 of the GreA protein, with a 1.4 kb HaeII–HaeII KanR cassette prepared from pACYC177 (New Engl and Biolabs), by fill-in and following ligation using a BglII linker (New Engl and Biolabs). The disrupted greA gene of the resulting plasmid, pTK010, was used to replace the chromosomal greA gene in MC1061, employing conjugation as previously described (Ohmori et al., 1995).
The plasmid pTK003 was constructed by replacing the EcoRI–BamHI segment of pBR322 with a greB-containing 3.2 kb BamHI–EcoRI fragment of Kohara's lambda ♯620. A CmR cassette of 1.3 kb was prepared by PCR from pACYC184 (New England Biolabs) using primers 5′-TC CCCCGGGAGTATACACTCCGCTAGCG-3′ and 5′-CCATCG ATCCTACATCTGTATTAACGAA-3′. This cassette was used to replace the 338 bp segment between the SmaI and ClaI sites in the greB gene on pTK003, which corresponds to residues 21–131 of GreB. The disrupted greB locus in the resulting plasmid, pTK007, was transferred into the suicide plasmid pKH5002 to produce pTK008. The disrupted greB of pTK008 was then used to replace the chromosomal greB in MC1061 by conjugation, to construct the ΔgreB strain TK1007. The expected allelic exchanges by double cross-over recombination in the ΔgreA and ΔgreB strains were confirmed by Southern and PCR analyses (data not shown).
ΔgreAΔgreB double mutant strains were constructed by transduction, using a selection marker of Tn10 that had been inserted near the greA locus. The Tn10 marker was 90% linked to greA. The resulting TK1009 (greA::kan zgj203::Tn10) was then used as a donor in transduction of the disrupted greA allele into the ΔgreB strain TK1007. The double mutant strain, TK1021, was selected by TetR and CmR.
RNA analyses
Total RNA was prepared from the E. coli strains grown in a condition as previously reported (Aiba et al., 1981). For the DNA array, the RNA (40–70 µg) was treated with 10 units of RQ1 DNase (Promega) to remove residual DNA, and the RNA was purified by phenol extraction followed by ethanol precipitation, and dissolved in 50 µl of diethyl pyrocarbonate-treated distilled water. The preparations of 32P-labelled cDNA and its hybridization with Panorama E. coli Gene Array (Sigma Genosys) were carried out according to the maker's manual.
For the Northern analysis, 10–20 µg of total RNA was subjected to electrophoresis on a 1% agarose gel containing 1× MOPS buffer and 1.3% formaldehyde. RNA was then transferred from the gel onto Hybond-N+ nylon membrane (Amersham), treated with 50 mM NaOH for 5 min, and hybridized with a 32P-labelled DNA probe for more than 12 h in the presence of 5× SSC containing 5× Denhardt's solution, 0.5% SDS and 17.5 µg ml−1 salmon sperm DNA. The membrane was washed twice at 42°C in 2× SSC containing 0.1% SDS and once with 0.1× SSC containing 0.1% SDS, followed by autoradiography. The signal intensities were normalized to the intensities of an internal control, either murE (for cspA) or cspA (for atpC and rpsA), which migrates a position different from the target bands. In the case of zraP, the signal intensities were normalized to the low-mobility bands observed with similar intensities in each lane (‘Control’ bands in Fig. 3B). The sequences of the DNA probes used for hybridization are complementary to the C-terminal coding regions of the target genes: 5′-TCGCTTTTTTGGTCAACT CG-3′ for atpC, 5′-TACAGGCTGGTTACGTTACC-3′ for cspA, 5′-ACTCGCCTTTAGCTGCTTTG-3′ for rpsA, 5′-TGCAATCA CCCCCAGCAGAC-3′ for murE and 5′-TTACCAGTGGC CCATACCCATATGACCGCC-3′ for zraP.
In vitro transcription assays
The template DNAs used in Fig. 4 were prepared by PCR, using Kohara's library phages ♯560 and ♯217, pJJG02 (Jiang et al., 1996) and pAR1435 (Dunn and Studier, 1983), respectively, for the atp, rpsA, cspA and T7A1 promoters (Fig. 4A). RNA polymerase (0.10 pmol for T7A1 and 1.0 pmol for the others) and DNA template (0.15 pmol for T7A1 and 0.50 pmol for the rest) were mixed and GreA or GreB was added to a final concentration of 50 or 5 µM respectively. The mixture was pre-incubated for 10 min at 37°C in 8 µl of T-buffer [50 mM Tris-HCl (pH 7.9), 100 mM KCl, 10 mM MgCl2, 1 mM DTT, 150 µg ml−1 partially hydrolysed casein] and then transcription was started by adding substrates: 20 µM [γ-32P]-ATP (5 Ci mmol−1) and 100 µM each of the other three NTPs in the case of T7A1, or 20 µM [γ-32P]-ATP (100 Ci mmol−1) or GTP (24 Ci mmol−1) as well as 500 µM each of the other three NTPs in the other cases. Transcription was continued for 20 min and the reaction was quenched with phenol. The transcripts were analysed by electrophoresis on a 20% polyacrylamide sequencing gel.
Inversed pulse-chase experiments
A downstream-biotinylated 169 bp DNA harbouring the atp promoter was immobilized on avidin-coated beads (Fujioka et al., 1991). Being attached at the downstream end relative to transcription, the bead traps RNA polymerase molecules that have reached the end and thus ensures single-round transcription. RNA polymerase (1.0 pmol) and the immobilized DNA (1.5 pmol) were pre-incubated at 37°C in 8 µl of T-buffer for 10 min. A substrate mixture was then added to give finally 5 µM GTP and 100 µM each of ATP, CTP and UTP. After various reaction times, 3.3 µCi [γ-32P]-GTP was added and the reaction was continued for a further 20 min. The reaction was then stopped by addition of phenol. The transcripts were subjected to 20% sequencing gel electrophoresis.
Western analysis
The E. coli TK1001 strain was cultured at 37°C overnight in 50 ml of LB medium containing tetracycline in either a 500 ml flask with vigorous shaking (shaking culture) or a 50 ml tube without shaking (standing culture). Cells were harvested at mid-log phase for shaking culture or stationary phase for standing culture. They were suspended in the buffer containing 10 mM Tris-HCl (pH 7.5), 5% glycerol, 0.1 mM EDTA, 0.5% SDS and 1 mM Pefabloc (Roche) and heated at 70°C for 10 min. Cell debris was removed by centrifugation, and the protein concentration in the supernatant was measured with the BCA Protein Assay Kit (Pierce). Total protein of 2–10 µg was subjected to SDS-PAGE using 5–20% gradient or 15% gel, and then blotted onto an Immobilon-P membrane (Millipore) according to the manufacturer's protocol. The GreA was immunostained with HRP-1000 Kit (Konica) and GreB with ECL advance (Amersham). The rabbit anti-GreA was raised against purified GreA and the anti-GreB was provided by Dr V. James Hernandez.
Acknowledgments
We thank Dr Richard S. Hayward for a critical reading of the manuscript, and Dr V. James Hernandez for anti-GreB. The metal sensitivity of the strain was first suggested by Dr Kazuhisa Sekimizu. This work was supported in part by grants from Ministry of Education, Culture, Sports, Science and Technology (to N.S.) and Itoh scholarship foundation (to M.S.).
References
- Aiba H, Adhya S, de Crombrugghe B. Evidence for two functional gal promoters in intact Escherichia coli cells. J Biol Chem. 1981;256:11905–11910. [PubMed] [Google Scholar]
- Berghöfer-Hochheimer Y, Lu CZ, Gross CA. Altering the interaction between σ70 and RNA polymerase generates complexes with distinct transcription-elongation properties. Proc Natl Acad Sci USA. 2005;102:1157–1162. doi: 10.1073/pnas.0408973102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borukhov S, Polyakov A, Nikiforov V, Goldfarb A. GreA protein: a transcription elongation factor from Escherichia coli. Proc Natl Acad Sci USA. 1992;89:8899–8902. doi: 10.1073/pnas.89.19.8899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borukhov S, Sagitov V, Goldfarb A. Transcript cleavage factors from E. coli. Cell. 1993;72:459–466. doi: 10.1016/0092-8674(93)90121-6. [DOI] [PubMed] [Google Scholar]
- Borukhov S, Laptenko O, Lee J. Escherichia coli transcript cleavage factors GreA and GreB: functions and mechanisms of action. Methods Enzymol. 2001;342:64–76. doi: 10.1016/s0076-6879(01)42536-5. [DOI] [PubMed] [Google Scholar]
- Borukhov S, Lee J, Laptenko O. Bacterial transcription elongation factors: new insights into molecular mechanism of action. Mol Microbiol. 2005;55:1315–1324. doi: 10.1111/j.1365-2958.2004.04481.x. [DOI] [PubMed] [Google Scholar]
- Carpousis AJ, Gralla JD. Cycling of ribonucleic acid polymerase to produce oligonucleotides during initiation in vitro at the lacUV5 promoter. Biochemistry. 1980;19:3245–3253. doi: 10.1021/bi00555a023. [DOI] [PubMed] [Google Scholar]
- Dunn JJ, Studier FW. Complete nucleotide sequence of bacteriophage T7 DNA and the locations of T7 genetic elements. J Mol Biol. 1983;166:477–535. doi: 10.1016/s0022-2836(83)80282-4. [DOI] [PubMed] [Google Scholar]
- Erie DA, Hajiseyedjavadi O, Young MC, von Hippel PH. Multiple RNA polymerase conformations and GreA: control of the fidelity of transcription. Science. 1993;262:867–873. doi: 10.1126/science.8235608. [DOI] [PubMed] [Google Scholar]
- Feng GH, Lee DN, Wang D, Chan CL, Landick R. GreA-induced transcript cleavage in transcription complexes containing Escherichia coli RNA polymerase is controlled by multiple factors, including nascent transcript location and structure. J Biol Chem. 1994;269:22282–22294. [PubMed] [Google Scholar]
- Fujioka M, Hirata T, Shimamoto N. Requirement for the β,γ-pyrophosphate bond of ATP in a stage between transcription initiation and elongation by Escherichia coli RNA polymerase. Biochemistry. 1991;30:1801–1807. doi: 10.1021/bi00221a011. [DOI] [PubMed] [Google Scholar]
- Hogan BP, Hartsch T, Erie DA. Transcript cleavage by Thermus thermophilus RNA polymerase. Effects of GreA and anti-GreA factors. J Biol Chem. 2002;277:967–975. doi: 10.1074/jbc.M108737200. [DOI] [PubMed] [Google Scholar]
- Hsu LM, Vo NV, Chamberlin MJ. Escherichia coli transcript cleavage factors GreA and GreB stimulate promoter escape and gene expression in vivo and in vitro. Proc Natl Acad Sci USA. 1995;92:11588–11592. doi: 10.1073/pnas.92.25.11588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Fang L, Inouye M. The role of the 5′-end untranslated region of the mRNA for CspA, the major cold-shock protein of Escherichia coli, in cold-shock adaptation. J Bacteriol. 1996;178:4919–4925. doi: 10.1128/jb.178.16.4919-4925.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston DE, McClure WR. Abortive initiation of in vitro RNA synthesis on bacteriophage λDNA. In: Losick R, Chamberlin M, editors. RNA Polymerase. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1976. pp. 413–428. [Google Scholar]
- Kohara Y, Akiyama K, Isono K. The physical map of the whole E. coli chromosome: application of a new strategy for rapid analysis and sorting of a large genomic library. Cell. 1987;50:495–508. doi: 10.1016/0092-8674(87)90503-4. [DOI] [PubMed] [Google Scholar]
- Kubori T, Shimamoto N. A branched pathway in the early stage of transcription by Escherichia coli RNA polymerase. J Mol Biol. 1996;256:449–457. doi: 10.1006/jmbi.1996.0100. [DOI] [PubMed] [Google Scholar]
- Kubori T, Shimamoto N. Physical interference between Escherichia coli RNA polymerase molecules transcribing in tandem enhances abortive synthesis and misincorporation. Nucleic Acids Res. 1997;25:2640–2647. doi: 10.1093/nar/25.13.2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonhartsberger S, Huber A, Lottspeich F, Bock A. The hydH/G genes from Escherichia coli code for a zinc and lead responsive two-component regulatory system. J Mol Biol. 2001;307:93–105. doi: 10.1006/jmbi.2000.4451. [DOI] [PubMed] [Google Scholar]
- Leptenko O, Lee J, Lomakin I, Borukhov S. Transcript cleavage factors GreA and GreB act as transient catalytic components of RNA polymerase. EMBO J. 2003;22:6322–6334. doi: 10.1093/emboj/cdg610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marr MT, Roberts JW. Function of transcription cleavage factors GreA and GreB at a regulatory pause site. Mol Cell. 2000;6:1275–1285. doi: 10.1016/s1097-2765(00)00126-x. [DOI] [PubMed] [Google Scholar]
- Metzger W, Schickor P, Meier T, Werel W, Heumann H. Nucleation of RNA chain formation by Escherichia coli DNA-dependent RNA polymerase. J Mol Biol. 1993;232:35–49. doi: 10.1006/jmbi.1993.1368. [DOI] [PubMed] [Google Scholar]
- Michaud C, Parquet C, Flouret B, Blanot D, van Heijenoort J. Revised interpretation of the sequence containing the murE gene encoding the UDP-N-acetylmuramyl-tripeptide synthetase of Escherichia coli. Biochem J. 1990;269:277–278. doi: 10.1042/bj2690277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munson LM, Reznikoff WS. Abortive initiation and long ribonucleic acid synthesis. Biochemistry. 1981;20:2081–2085. doi: 10.1021/bi00511a003. [DOI] [PubMed] [Google Scholar]
- Niyogi SK, Feldman RP. Effect of several metal ions on misincorporation during transcription. Nucleic Acids Res. 1981;9:2615–2627. doi: 10.1093/nar/9.11.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noll M, Petrukhin K, Lutsenko S. Identification of a novel transcription regulator from Proteus mirabilis, PMTR, revealed a possible role of YJAI protein in balancing zinc in Escherichia coli. J Biol Chem. 1998;273:21393–21401. doi: 10.1074/jbc.273.33.21393. [DOI] [PubMed] [Google Scholar]
- Ohmori H, Saito M, Yasuda T, Nagata T, Fujii T, Wachi M, Nagai K. The pcsA gene is identical to dinD in Escherichia coli. J Bacteriol. 1995;177:156–165. doi: 10.1128/jb.177.1.156-165.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opalka N, Chlenov M, Chacon P, Rice WJ, Wriggers W, Darst SA. Structure and function of the transcription elongation factor GreB bound to bacterial RNA polymerase. Cell. 2003;114:335–345. doi: 10.1016/s0092-8674(03)00600-7. [DOI] [PubMed] [Google Scholar]
- Orlova M, Newlands J, Das A, Goldfarb A, Borukhov S. Intrinsic transcript cleavage activity of RNA polymerase. Proc Natl Acad Sci USA. 1995;92:4596–4600. doi: 10.1073/pnas.92.10.4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Outten CE, O'Halloran TV. Femtomolar sensitivity of metalloregulatory proteins controlling zinc homeostasis. Science. 2001;292:2488–2492. doi: 10.1126/science.1060331. [DOI] [PubMed] [Google Scholar]
- Paul BJ, Barker MM, Ross W, Schneider DA, Webb C, Foster JW, Gourse RL. DksA: a critical component of the transcription initiation machinery that potentiates the regulation of rRNA promoters by ppGpp and the initiating NTP. Cell. 2004;118:311–322. doi: 10.1016/j.cell.2004.07.009. [DOI] [PubMed] [Google Scholar]
- Pedersen S, Skouv J, Kajitani M, Ishihama A. Transcriptional organization of the rpsA operon of Escherichia coli. Mol Gen Genet. 1984;196:135–140. doi: 10.1007/BF00334105. [DOI] [PubMed] [Google Scholar]
- Perederina A, Svetlov V, Vassylyeva MN, Tahirov TH, Yokoyama S, Artsimovitch I, Vassylyev DG. Regulation through the secondary channel – structural framework for ppGpp-DksA synergism during transcription. Cell. 2004;118:297–309. doi: 10.1016/j.cell.2004.06.030. [DOI] [PubMed] [Google Scholar]
- Porter AC, Brusilow WS, Simoni RD. Promoter for the unc operon of Escherichia coli. J Bacteriol. 1983;155:1271–1278. doi: 10.1128/jb.155.3.1271-1278.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi F, Liu C, Heath LS, Turnbough CL., Jr In vitro assay for reiterative transcription during transcriptional initiation by Escherichia coli RNA polymerase. Methods Enzymol. 1996;273:71–85. doi: 10.1016/s0076-6879(96)73007-0. [DOI] [PubMed] [Google Scholar]
- Sen R, Nagai H, Hernandez VJ, Shimamoto N. Reduction in abortive transcription from the λPR promoter by mutations in region 3 of the σ70 subunit of Escherichia coli RNA polymerase. J Biol Chem. 1998;273:9872–9877. doi: 10.1074/jbc.273.16.9872. [DOI] [PubMed] [Google Scholar]
- Sen R, Nagai H, Shimamoto N. Polymerase arrest at the λPR promoter during transcription initiation. J Biol Chem. 2000;275:10899–10904. doi: 10.1074/jbc.275.15.10899. [DOI] [PubMed] [Google Scholar]
- Sen R, Nagai H, Shimamoto N. Conformational switching of Escherichia coli RNA polymerase–promoter binary complex is facilitated by elongation factor GreA and GreB. Genes Cells. 2001;6:389–401. doi: 10.1046/j.1365-2443.2001.00436.x. [DOI] [PubMed] [Google Scholar]
- Shimamoto N, Wu FY, Wu CW. Mechanism of ribonucleic acid chain initiation. Molecular pulse-labeling study of ribonucleic acid syntheses on T7 deoxyribonucleic acid template. Biochemistry. 1981;20:4745–4755. doi: 10.1021/bi00519a034. [DOI] [PubMed] [Google Scholar]
- Sosunova E, Sosunov V, Kozlov M, Nikiforov V, Goldfarb A, Mustaev A. Donation of catalytic residues to RNA polymerase active center by transcription factor Gre. Proc Natl Acad Sci USA. 2003;100:15469–15474. doi: 10.1073/pnas.2536698100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparkowski J, Das A. The nucleotide sequence of greA, a suppressor gene that restores growth of an Escherichia coli RNA polymerase mutant at high temperature. Nucleic Acids Res. 1990;18:6443. doi: 10.1093/nar/18.21.6443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susa M, Sen R, Shimamoto N. Generality of the branched pathway in transcription initiation by Escherichia coli RNA polymerase. J Biol Chem. 2002;277:15407–15412. doi: 10.1074/jbc.M112481200. [DOI] [PubMed] [Google Scholar]
- Tagami H, Aiba H. A common role of CRP in transcription activation: CRP acts transiently to stimulate events leading to open complex formation at a diverse set of promoters. EMBO J. 1998;17:1759–1767. doi: 10.1093/emboj/17.6.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanabe H, Goldstein J, Yang M, Inouye M. Identification of the promoter region of the Escherichia coli major cold shock gene, cspA. J Bacteriol. 1992;174:3867–3873. doi: 10.1128/jb.174.12.3867-3873.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trautinger BW, Lloyd RG. Modulation of DNA repair by mutations flanking the DNA channel through RNA polymerase. EMBO J. 2002;21:6944–6953. doi: 10.1093/emboj/cdf654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travers A, Muskhelishvili G. DNA supercoiling – a grobal transcriptional regulator for entrobacterial growth? Nat Rev Microbiol. 2005;3:157–169. doi: 10.1038/nrmicro1088. [DOI] [PubMed] [Google Scholar]
- Vo NV, Hsu LM, Kane CM, Chamberlin MJ. In vitro studies of transcript initiation by Escherichia coli RNA polymerase. 2. Formation and characterization of two distinct classes of initial transcribing complexes. Biochemistry. 2003;42:3787–3797. doi: 10.1021/bi0269613. [DOI] [PubMed] [Google Scholar]