Abstract
The inactivation of either subunit of the Ku70-Ku80 heterodimer, which functions in nonhomologous end-joining and telomere maintenance, generates severe defects such as sensitivity to DNA damage, telomere shortening, and increased gross chromosomal rearrangements (GCRs) that are frequently observed in many cancers. To understand the mechanism of Ku as a genome gatekeeper, we overexpressed the yKu70-yKu80 heterodimer and monitored the formation of GCRs. Ku overexpression suppressed the formation of either spontaneously generated GCRs or those induced by treatments with different DNA damaging agents. Interestingly, this suppression depended on Ku’s interaction with DNA damage checkpoints and not through nonhomologous end-joining. We also demonstrate that the inactivation of telomerase inhibitor, Pif1 along with Ku overexpression or the overexpression of Pif1 in either yku70 or yku80 strains arrested the cell cycle at S phase in a DNA damage checkpoint-dependent fashion. Lastly, Ku overexpression causes cell growth delay, which depends on intact Rad27. In summary, the results presented here suggest that Ku functions as a genomic gatekeeper through its crosstalk with DNA damage checkpoints.
Keywords: cancer, DNA repair
Many genetic disorders, particularly cancer, increase genome instability throughout their progression (1, 2). Gross chromosomal rearrangements (GCRs) including translocations, deletions of chromosome arms, interstitial deletions, inversions, amplifications, and aneuploidy is a class of genome instability found in most cancers (1, 2). Genomic instabilities in cancer cells can be caused by mutator mutations and as a result facilitate further accumulations of genetic changes (1).
The mechanisms for suppression and progression of GCRs have been studied by using different quantitative assays developed in Saccharomyces cerevisiae as a model organism (3–17). The cell cycle checkpoint pathways appear to redundantly suppress GCR formation (7). Cell cycle checkpoints are surveillance mechanisms designed to ensure correct transmission of genetic information during cell division (18, 19). S. cerevisiae and other organisms contain a number of checkpoints that respond to DNA damage as well as aberrant DNA structures that occur when DNA replication is blocked (20–24). Genetic defects in the various DNA damage and S phase checkpoints have previously been demonstrated to result in different degrees of increased spontaneous GCR rates, increased chromosome loss, and increased recombination (6, 7).
Recently, we have identified additional pathways that are important for the formation of GCRs (11, 25, 26). Interestingly, proteins identified as being required for the formation of GCRs normally function to protect the genome. For example, the Ku70-Ku80 heterodimer functions in genome stability by participating in the nonhomologous end-joining (NHEJ) and telomere maintenance pathways but is clearly also involved in the formation of GCRs (11, 27,30–31). How proteins normally functioning in genomic protection change their roles to participate in the formation of GCRs is still not clearly understood.
The Ku70-Ku80 heterodimer was originally identified as an autoantigen recognized by sera from patients with rheumatic disorders (32). Biochemical analyses of the Ku70-Ku80 heterodimer protein demonstrated that it bound in a sequence-nonspecific fashion to virtually all double-stranded DNA ends including 5′- or 3′-protruding ends, blunt ends (32), and duplex DNA ending in stem–loop structures (33). The DNA binding activity of the Ku70-Ku80 heterodimer is thought to function by holding broken double-stranded DNAs for repair (29). At telomeres, the terminal structures of linear chromosomes, the Ku70-Ku80 heterodimer presumably also binds at the end of DNA to protect double-stranded DNA from end-to-end fusion (28, 34). Inactivation experiments in different species demonstrated that the Ku70-Ku80 heterodimer is important for the protection of DNA ends both during DNA repair and telomere maintenance (27–31, 35–40).
Because of the multiple roles of Ku, it is difficult to understand how the Ku70-Ku80 heterodimer protects the genome from rearrangements. In the present study, we demonstrate that overexpression of both yeast Ku70 and Ku80 proteins reduces GCR formation through a genetic interaction with DNA damage checkpoints.
Results
Recently, we demonstrated that the haploinsufficiency of human Ku80 could result in profound telomere loss, accompanied by an increase of chromosomal aberrations such as translocations, chromosomal end-to-end fusions including ring chromosome formation, and aneuploidy (28). To understand the mechanism of GCR suppression by the Ku70-Ku80 heterodimer, we chose a more genetically tractable system, S. cerevisiae. Because yKu70-yKu80 is required for GCR formation in the absence of proper DNA repair (11, 41), we could not study the role of yKu70-yKu80 for GCR suppression by mutating yKU70 or yKU80. Therefore, we decided to investigate GCR formation in presence of excess yKu70-yKu80.
Overexpression of yKu70-yKu80 Reduces GCRs Induced by DNA Damage or Generated Spontaneously.
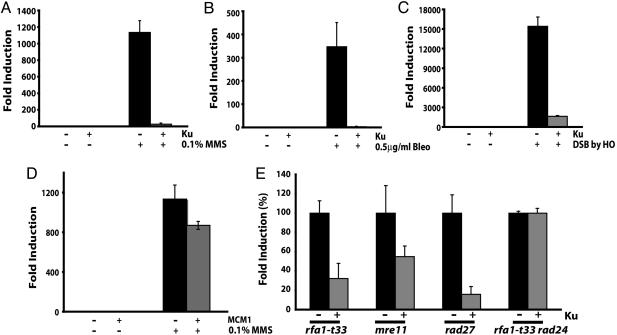
We previously showed that treatment of a wild-type strain with methyl methane sulfonate (MMS) increased the frequency of GCRs, especially the de novo telomere addition type of GCRs (7). To determine whether yKu70-yKu80 heterodimer functions to suppress GCRs, plasmids encoding both subunits of Ku were introduced into a wild-type yeast strain that carried the GCR assay system. The overexpression of Ku was induced by galactose and could be detected by Western blot with antibodies, which recognized Ku70 and Ku80 ( Fig. 5A, which is published as supporting information on the PNAS web site). Induced and uninduced cells were then treated with either MMS, an alkylating agent (Fig. 1A); bleomycin, a radiomimetic agent (Fig. 1B); or co-induction of the HO endonuclease, which makes a site-specific double-strand break (DSB) (Fig. 1C). The exposure of control cells to any one of these three conditions resulted in increased numbers of GCRs that were two to four orders of magnitude more frequent than the number of GCRs observed for untreated cells (Fig. 1). In striking contrast, the Ku-overexpressing strain was extremely resistant to GCRs induced by DNA damage. However, the decrease of GCR frequency was not observed when another DNA binding protein, MCM1 that binds to the replication origin and also functions as a transcription factor (42), was overexpressed in the same manner (Fig. 1D). For each strain, a limited number of breakpoints corresponding to the GCR were characterized by DNA sequencing but no obvious change in the spectra of rearrangement breakpoints was observed (Table 1, which is published as supporting information on the PNAS web site). Ku overexpression did not alter the viability in response to MMS in wild type or in the rad52 background ( Fig.5 B and C), suggesting that Ku overexpression did not change the capability of DSB repair, especially NHEJ. In summary, the overexpression of yKu70-yKu80 heterodimer dramatically suppressed genome rearrangements induced by a variety of DNA damage, and this suppression was independent from yKu70-yKu80’s role in DSB repair.
Fig. 1.
yKu70-yKu80 overexpression suppresses GCR formation induced by different types of DNA damage or generated spontaneously. The increased GCR frequency was suppressed by the overexpression of the yKu70-yKu80 heterodimer in the presence of 0.1% MMS (A), 0.5 μg/ml bleomycin (Bleo) (B), and a single DSB introduced by an HO endonuclease (C). (D) Mcm1, a DNA binding protein, did not suppress the GCRs induced by MMS treatment. (E) GCR rates in different GCR mutator strains were reduced by Ku overexpression. Each GCR rate of a GCR mutator strain with control plasmids was recalculated to 100%, and the GCR rate with Ku overexpression was converted to a percentage compared with that of strains carrying control plasmids. 100% GCR rate presented here for rfa1-t33, mre11, rad27, and rfa1-t33 rad24 are 5.8 × 10−7, 5.9 × 10−7, 2.2 × 10−6, and 4.9 × 10−7, respectively. “+” and “−” represent the presence and absence of indicated proteins or the treatment and no treatment of indicated DNA damaging agents, respectively.
If overexpression of yKu70-yKu80 suppresses GCR formation by DNA damage, a logical extension of this observation would be that the overexpression of yKu70-yKu80 might suppress spontaneous GCR formation in GCR mutator strains. More than 70 GCR mutator genes have been identified, and a mutation in each of these GCR mutator genes generates preferentially one or two different types of GCRs (3–9, 11, 12, 15–17, 41, 43). We chose four GCR mutator genes, the mutation of which increases the GCR rate up to 1, 000-fold compared with wild type. RFA1, which encodes a single-strand DNA binding protein that is important during DNA replication and recombination (43, 44); MRE11, which encodes a protein participating in the MRX (Mre11-Rad50-Xrs2) complex and functions in DNA recombination, S phase cell cycle checkpoint, DNA repair, and telomere maintenance (45–47); and RAD27, which encodes a flap endonuclease necessary for the processing and maturation of Okazaki fragments during DNA replication as well as for long patch base excision repair (48). PIF1, which encodes a helicase, functions as a telomerase inhibitor, and its inactivation increases the GCR formation rate and telomere length (10, 11).
When the GCR rates of controls without Ku overexpression in rfa1-t33, mre11, and rad27 were set to 100%, the overexpression of yKu70-yKu80 heterodimer reduced the GCR rate to 30%, 49%, and 16%, respectively (Fig. 1E). Therefore, the overexpression of yKu70-yKu80 suppressed spontaneously generating GCR formation as well as GCRs induced by exogenous DNA damage.
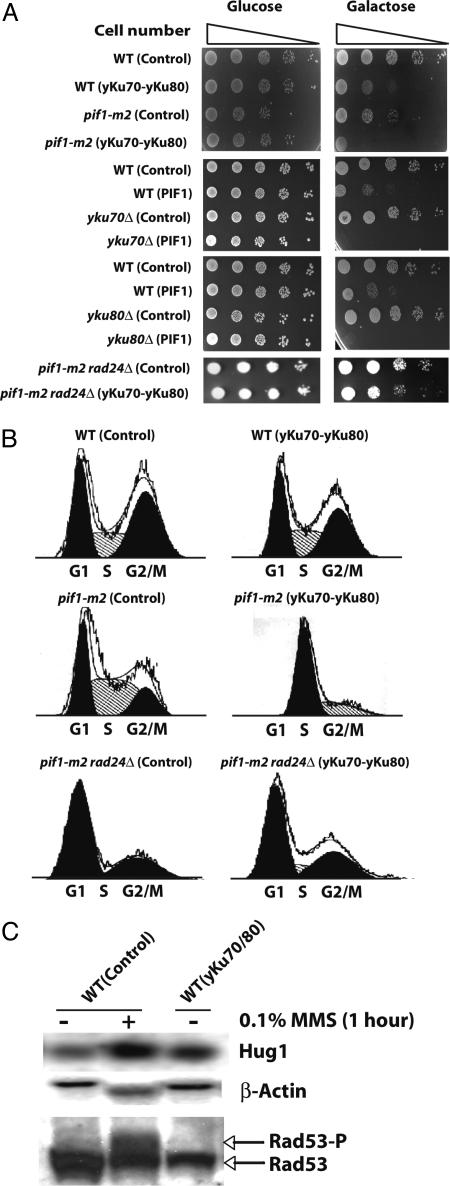
Interestingly, we could not measure the GCR rate from the pif1-m2 strain when Ku was overexpressed because Ku overexpression in the pif1-m2 strain completely arrested cell growth (Fig. 2A). FACS analysis revealed that the pif1-m2 strain arrested in S phase when the yKu70-yKu80 heterodimer was overexpressed (Fig. 2B). A complete growth defect was also observed when Pif1 was overexpressed either in the yku70 or yku80 strains (Fig. 2A). The S phase arrest was not mediated by telomere size change or the amount of single-stranded DNA at telomeres (Fig. 6, which is published as supporting information on the PNAS web site, and data not shown). Analysis of the telomere size of several clones that escaped this growth defect and which could be propagated for 10 days showed only a subtle decrease in telomere size (Fig. 6). Different pathways, which could also intuitively be perturbed by yKu70-yKu80 overexpression, were tested. A lig4 mutation did not suppress the growth defect caused by Ku overexpression in the pif1-m2 strain, suggesting that NHEJ is not the pathway causing the growth defect (Table 2, which is published as supporting information on the PNAS web site). Mutation of the YCA1 gene encoding a yeast caspase that might be involved in yeast programmed cell death (49) also did not attenuate the growth defect caused by Ku overexpression in the pif1-m2 strain (Table 2). Finally, we hypothesized that because Ku overexpression resulted in an S phase arrest in the pif1-m2 strain, aberrant regulation of the cell cycle checkpoint might be the cause of the growth defect. In support of this hypothesis, mutation in the RAD24 gene, which is a sensor for the intra-S checkpoint, abolished the growth defect caused by Ku overexpression in the pif1-m2 strain and resulted in a normal cell cycle (Fig. 2B). A slight retardation in cell growth remained that was similar to the delay observed in wild-type cells when the yKu70-yKu80 heterodimer was overexpressed. In summary, the growth defect caused by Ku overexpression in the pif1-m2 strain appeared to partially require the activation of the intra-S phase cell cycle checkpoint.
Fig. 2.
Growth delays or cell cycle arrests are caused by Ku overexpression in different strains. (A) Ku overexpression delays cell growth or arrests the cell cycle in wild-type or pif1-m2 strains, respectively. Pif1 overexpression either in the yku70 or the yku80 strains also arrests the cell cycle. The S phase cell cycle arrest in the pif1-m2 strain by Ku overexpression was abolished by the rad24 mutation. (B) FACS analysis demonstrates that Ku overexpression in the wild-type strain did not change the cell cycle profile. However, Ku overexpression in the pif1-m2 strain arrests the cell cycle in S phase. The rescue of cell cycle arrest in the pif1-m2 strain with Ku overexpression by the rad24 mutation was also confirmed by FACS analysis. (C) Northern blot analysis of HUG1 showed slight mRNA level increase by Ku overexpression, in contrast to the steady expression of β-actin. In contrast, there was no noticeable phosphorylation of Rad53 was observed by Ku overexpression although the phosphorylation of Rad53 was enhanced by MMS treatment. WT, wild type; Control, control plasmids, pYES3CT and pRS425, transformed. A protein name indicated in parentheses represents the protein overexpressed in the strain written in front of it. “+” and “−” represent the presence and absence of MMS treatment. The two major populations in the FACS analysis represented as black indicate G1 and G2/M populations. The FACS profile of pik1-m2 rad24Δ (yku70-yku80) shows one major population presented as black, and its DNA contents represent the S phase.
GCR Suppression by Ku Overexpression Is Cell Cycle Checkpoint-Dependent and NHEJ-Independent.
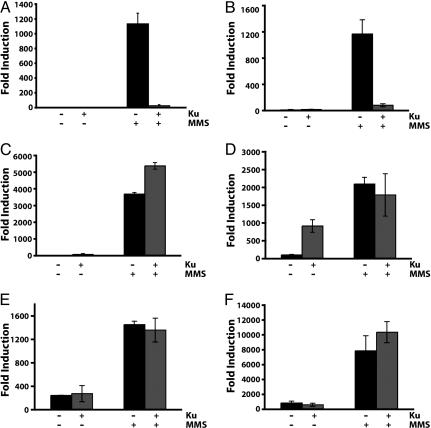
The link between Ku overexpression and cell cycle checkpoint observed at least in the pif1-m2 strain drove us to investigate the possibility of GCR suppression by Ku overexpression communicating with cell cycle checkpoints. First, the possibility of cell cycle checkpoint activation by Ku overexpression to suppress GCR formation was tested by investigating the expression level of HUG1, which encodes a protein responding to different types of DNA damage through cell cycle checkpoints activation (50). The HUG1 expression was slightly increased by Ku overexpression in wild type (Fig. 2C). These additional data suggest that cell cycle checkpoints could cause the GCR suppression by Ku overexpression. However, we could not detect noticeable enhancement of Rad53 phosphorylation when Ku was overexpressed in contrast to a significant enhancement of Rad53 phosphorylation upon MMS treatment (Fig. 2C). Intensive analysis revealed that the suppression of GCR formation by Ku overexpression was abolished by the inactivation of cell cycle checkpoint genes (Fig. 3 C–F). The rad24 mutation, which inactivates a branch of the intra-S checkpoint increased the GCR frequency up to 3, 700-fold upon MMS treatment. This high induction of GCR frequency in the rad24 strain was actually exacerbated by Ku overexpression (Fig. 3C). Similarly, the high induction of GCR frequencies by MMS treatment in different S phase checkpoint-defective sgs1, dpb11–1, and mec1 strains was not reduced by Ku overexpression (Fig. 3 D–F). There was an additional interesting observation that Ku overexpression alone in the sgs1 strain increased the GCR frequency up to 10-fold compared with the sgs1 strain without Ku overexpression (Fig. 3D). We envision that the spontaneous DNA damage by the sgs1 mutation might be channeled to GCR formation by the overexpression of Ku (see Discussion). The suppression of spontaneously generated GCRs in different mutator strains by Ku overexpression is also mediated through the cell cycle checkpoint (Fig. 1E). The inactivation of Rad24 in the rfa1-t33 strain completely impaired the suppression of GCRs by Ku overexpression. In summary, the impairment of GCR suppression only in a subset of cell cycle checkpoint-defective mutants strongly suggests that the GCR suppression by Ku overexpression is mediated through its signaling to cell cycle checkpoints.
Fig. 3.
GCR suppression by Ku overexpression after MMS treatment is mediated by its crosstalk with cell cycle checkpoints. (A) Ku overexpression suppressed GCR formation after MMS treatment. (B) GCRs were still suppressed by Ku overexpression in the lig4 strain. Mutations, which inactivate cell cycle checkpoints, abolished the suppression of GCR by Ku overexpression after MMS treatment. (C) rad24. (D) sgs1. (E) dpb11–1. (F) mec1. “+” and “−” represent the presence and absence of Ku overexpression or the treatment and no treatment of 0.05% MMS, respectively.
The yKu70-yKu80 heterodimer is known to function in the maintenance of genomic integrity through its ability to bind DNA DSBs and facilitate repair by NHEJ. To experimentally test whether the effect on the reduction of GCRs initiated by DNA damage was due to the activation of NHEJ, a strain carrying a mutation in LIG4 was created, and the effect on GCR formation upon MMS treatment by the overexpression of yKu70-yKu80 was tested (Fig. 3B). The lig4 mutation did not attenuate the suppression of GCRs caused by the overexpression of the yKu70-yKu80 heterodimer. Therefore, it is clear that the suppression of GCRs by Ku overexpression is not mediated through Ku’s role in NHEJ.
Slow Growth by Chronic Overexpression of Ku Depends on Rad27.
Lastly, we observed a retarded cell growth when Ku protein was chronically overexpressed (Fig. 2A). To identify pathway(s) responsible for the growth delay caused by Ku overexpression in the wild-type strain, selected strains defective in different aspects of DNA metabolism were investigated. Using this survey approach, we found that a rad27 mutation completely abolished the growth defect caused by Ku overexpression (Table 3, which is published as supporting information on the PNAS web site). RAD27 encodes a flap endonuclease, which functions in the processing and maturation of Okazaki fragments during DNA replication, telomere replication, and long patch base excision repair (48, 51). To determine whether there is a link between the growth defect caused by Ku overexpression and specific role of Rad27, two other mutations, elg1 or pol3–01, which has partially overlapping defect with rad27, were tested. Both strains partially alleviate the growth defect caused by Ku overexpression (Table 3).
Discussion
The overexpression of the yKu70-yKu80 heterodimer decreased GCR formation either generated spontaneously or induced by treatments with different DNA damaging agents. The suppression of GCRs was not due to the cells’ capability to increase NHEJ activity or random DNA binding. Instead, we found that the suppression of GCRs induced by the overexpression of the yKu70-yKu80 heterodimer is mediated through cell cycle checkpoints. In higher eukaryotes, the Ku70-Ku80 heterodimer is thought to participate in DNA repair, primarily through its association with the DNA-dependent protein kinase (DNA-PK) complex, which is an important signal transduction kinase regulating downstream targets (29). Although the direct role of DNA-PK in cell cycle checkpoints was not observed, DNA-PK might function in cell cycle checkpoints through its downstream target that is redundantly regulated by other kinases. In contrast, in S. cerevisiae, a homolog of the DNA-PK catalytic subunit has not been identified. Therefore, the role of yKu70-yKu80 in cell cycle checkpoints observed in this study is unlikely to be mediated as a subunit of the DNA-PK complex. Intriguingly, a study in mouse Ku80−/− cells demonstrated that DNA damage delayed the cell cycle in S phase independently from the DNA-PK catalytic subunit (52). Therefore, even in mammalian cells, Ku may function in cell cycle checkpoints (or at least cell cycle regulation) in a DNA-PK complex-independent fashion. The genetic interaction of yKu70-yKu80 with cell cycle checkpoint factors for the GCR suppression observed in this study could presumably occur in a similar manner. Because S. cerevisiae has other kinases, such as Mec1 and Tel1 (53, 54), that function in the cell cycle checkpoints, it is still possible that the overexpressed Ku functions to produce a signal along with these kinases to stall the cell cycle and activate proper DNA repair to suppress GCRs.
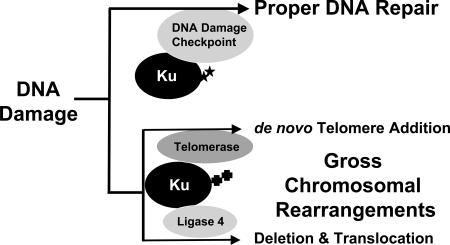
Proteins suppressing GCR formation function in different types of DNA metabolisms such as cell cycle checkpoints, DNA replication, DNA repair, and telomere maintenance (1). Interestingly, proteins required to produce GCRs also function in similar types of DNA metabolisms. For example, the nucleotide excision and recombination repair proteins, Rad1 and Rad10, and the mitotic checkpoints, which normally ensure correct chromosome segregation during cell division, are required for the generation of GCRs (25, 26). Ku, which functions in end protection at telomeres and in NHEJ, has likewise been shown to participate in the production of GCRs (11, 41). In particular, translocations require the NHEJ function of Ku whereas de novo telomere addition requires Ku’s role as a telomerase accessory factor. However, our present study demonstrates that Ku also functions to protect the genome from the production of GCRs through communication with cell cycle checkpoints. It is not clear how Ku functions in at least two opposite mechanisms. We propose that Ku might function at two different stages to deal with DNA damage. In cells, when DNA damage occurs, Ku plays a role to repair the damage properly. At this stage, Ku communicates with proteins functioning at cell cycle checkpoints (Fig. 4). Some portion of the DNA damage, however can escape from proper DNA repair, which in turn is deleterious to cells. Ku might function to change DNA damage to GCRs at this stage either by participating in de novo telomere addition or catalyzing translocation-type GCR through its NHEJ function. Different cell cycle phases and/or different modifications of Ku could influence in which direction Ku will participate. A recent observation that the Ku protein can be SUMO-ylated (SUMO is small ubiquitin-like modifier) suggests different modifications of the Ku protein for changing its roles (55).
Fig. 4.
Ku protein functions in two different ways in dealing with DNA damage. When DNA damage (presumably DSB) occurs, Ku binds at the end of DNA and protects it, and alarms DNA damage checkpoints to protect genome. However, if DNA damage persists, Ku changes its role to recruit other DNA metabolism machinery to generate GCRs. In this stage, Ku might be modified differently compared with when it protects genome through DNA repair, telomere maintenance, and DNA damage checkpoints. Ku might interact with telomerase directly to generate de novo telomere additions or Ku could participate in the production of large deletions or translocations with ligase 4. Stars and crosses represent putative different modifications that might direct divergent roles of Ku.
The overexpression of Ku proteins in an sgs1 strain induced GCR formation (Fig. 3D). Sgs1 is a yeast homolog of the BLM and WRN helicases, mutations of which have been found in the cancer-predisposed genetic diseases Bloom’s and Werner’s syndromes, respectively (56). It has been shown that these helicases function to promote proper DNA repair through their interaction with Ku and are presumably recruited to DNA by Ku (57). Certain DNA damage generated during DNA replication or recombination is repaired by an Sgs1-dependent pathway (58). The increase of repair centers, which are characterized by the localizations of recombination repair proteins (59), and the increase of homologous recombination (60) in the sgs1 strain support the hypothesis that in the absence of Sgs1, DNA damage will be repaired by other proper DNA repair mechanisms. To negate such compensation, we propose that the overexpressed Ku might bind to DNA damage in the sgs1 strain and block the access of the compensating repair machinery resulting in unrepaired DNA damage subject for GCR formation.
Ku overexpression retarded cell growth (Fig. 2A and Table 3). Interestingly, the growth delay was rescued by the rad27 mutation (Table 3). The rad27 mutation increases the amount of single stranded DNA at telomeres, although the length of the telomere is not changed (51). Defects at telomeres caused by the rad27 mutation might recruit excess Ku, which causes cell growth delay, to telomeres. As a result, the growth delay phenotype observed by Ku overexpression might be compensated. In accordance with this, the overexpression of Ku in the rad27 strain decreased the single-stranded DNA amount at telomeres (data not shown). The pol3–01 or elg1 mutation also relieved the growth delay caused by Ku overexpression although the effect was much weaker compared with that by rad27 (Table 3). Because the elg1 and pol3–01 mutations change telomere size (3, 5, 61, 62), they might affect telomere homeostasis by redirecting excess Ku to telomere and relieving the growth delay. However, we cannot exclude a possibility that defects in the lagging strand synthesis during DNA replication might redirect excess Ku from its effect in cell growth.
The overexpression or at least the activation of Ku in mammalian cells confers a resistance to γ-irradiation treatment (63). However, irradiation itself generates a strong mutagenic effect in cells, which sometimes causes secondary cancer formation after various therapies (64). Present studies detailing the reduction of genome rearrangements induced by DNA damaging agents treatment by Ku overexpression strongly argue that in chemo- or radiotherapies of cancer cells, the induction of Ku could be used to reduce the possibility of cells obtaining other mutations through genome rearrangements. Although there still needs to be more research performed before the application of such a protocol is used in the treatment of cancers, we believe that further understanding of the detailed mechanisms underlying Ku overexpression and its suppression of GCRs in different model organisms may ultimately provide avenues for better cancer treatment.
Materials and Methods
General Genetic and Molecular Biology Methods.
Media for propagating yeast strains used in this study are as described in refs. 6 and 17. All S. cerevisiae strains were propagated at 30°C except for strains containing the dpb11–1 mutation, which were propagated at 25°C. All S. cerevisiae strains used in this study were derived from the S288c parental strain RDKY3615 [MATa, ura3–52, leu2Δ1, trp1Δ63, his3Δ200, lys2ΔBgl, hom3–10, ade2Δ1, ade8, hxt13::URA3] for the general GCR assay and YKJM941 [MATa, ura3::KAN, leu2Δ1, trp1Δ63, his3Δ200, lys2ΔBgl, hom3–10, ade2Δ1, ade8, sit1::URA3-HO] for an HO-inducible GCR assay. The yKU70 or yKU80 genes regulated under the galactose-inducible promoter in the pYES2 (URA3, 2μ) plasmid (named as pKJM6) or in the pPM231 (LEU2, 2μ) plasmid (named as pKJM5) were used to overexpress the yKu70-yKu80 heterodimer (65). The yKU70 gene was subcloned into the pYES3CT (TRP1, 2μ) plasmid, which also contained a galactose-inducible promoter and was named as pKJM16. The plasmids were transformed into different strains used for this study. Detailed genotypes and plasmids in each strain are listed in Table 4, which is published as supporting information on the PNAS web site. The Northern blot analysis of HUG1 expression was performed by using the ExpressHyb kit (BD Bioscience) according to the manufacturer’s protocol. The full-length HUG1 gene was used for probe. Western blot analysis of Rad53 or Ku proteins was performed with antibodies against Rad53 protein (Santa Cruz Biotechnology) and yKu70 and yKu80 proteins (gift from Sang Eun Lee, University of Texas, San Antonio) by the ECL detection method (GE Healthcare).
Characterization of GCR Rates and Breakpoints.
All GCR rates were determined independently by fluctuation analysis with the method of the median with at least two independent clones analyzed two or more times using either 5 or 11 cultures for each clone, and the average value is reported as described in refs. 6 and 17. The breakpoint spectra from mutants carrying independent rearrangements were determined and classified as described in refs. 6 and 17.
Cell Growth Defect by the Overexpression of yKu70 and yKu80.
The cell growth defect caused by the overexpression of yKu70 and yKu80 was determined by spotting 5 μl of serially diluted yeast cultures onto synthetic drop-out plates containing either 2% (wt/vol) glucose (control) or 2% (wt/vol) galactose (yKu70 and yKu80 overexpression). Yeast cells on plates were then incubated for 2 days for glucose plates or 5 days for galactose plates at 30°C or 25°C as indicated, and photographs were taken.
Induction of GCR by Treatment with MMS or a Single DSB by HO Endonuclease.
GCR assays after the induction of a single DSB by HO endonuclease or after MMS treatment were performed as described in ref. 41 with minor modifications. Before MMS treatment, Ku proteins were induced for 2 h at 30°C by galactose, followed by the procedure as described. The induction of HO endonuclease expression that introduces a single DSB was achieved simultaneously with Ku overexpression by galactose. Then, the GCR frequency was determined as described in ref. 41.
FACS Analysis to Determine Cell Cycle.
To monitor the cell cycle progression of indicated strains carrying either control plasmids or yKu70-yKu80 overexpression plasmids, FACS analysis was carried out. Indicated yeast strains were grown in 2 ml of synthetic drop-out media with 2% glucose and all amino acids except leucine and tryptophan to reach log phase. One milliliter of the culture was collected and washed three times with sterile water. Then, the cells were resuspended with 1 ml of synthetic drop-out media with 2% galactose and all amino acids except leucine and tryptophan and allowed to grow for an additional 2 h for induction of the yKu70-yKu80 heterodimer. Cells (0.5 ml; 1–2 × 106) were washed and resuspended in cold 70% (wt/vol) ethanol followed by a 2-h incubation on ice. Then, the cells were collected by centrifugation and resuspended in 1 ml of 50 mM Tris·HCl (pH 7.5), followed by sonication. Cells were then incubated with 1 mg/ml RNase overnight and resuspended in 0.5 ml of propidium iodide solution (0.05 mg/ml propidium iodide/0.2 M Tris·HCl/0.2 M NaCl/0.1 M MgCl2, pH 7.5). After a 15-min incubation, the cells were resuspended with 1 ml of 50 mM Tris·HCl (pH 7.5), and the cellular DNA content was determined by FACS analysis.
Supplementary Material
Acknowledgments
We thank C. Chen (Dana–Faber Cancer Institute, Boston), D. Gottschling (Fred Hutchinson Cancer Research Center, Seattle), S. E. Lee, A. Nussenzweig (National Cancer Institute, Bethesda), and R. Rostein (Columbia University, New York) for helpful discussions; R. Kolodner (Ludwig Institute for Cancer Research, New York), S. E. Lee, and V. Zakian (Princeton University, Princeton) for antibody, strains, and plasmids; and D. Bodine [National Human Genome Research Institute (NHGRI)], E. Hendrickson (University of Minnesota), A. Nussenzweig, J. Puck (NHGRI), and members of the Myung laboratory for comments on the manuscript. K.M. especially thanks E. Cho. We also thank S. Anderson (NHGRI) for FACS analysis. This work was supported by the Intramural Research Program of the NHGRI/National Institutes of Health (K.M.).
Abbreviations
- DSB
double-strand break
- GCR
gross chromosomal rearrangement
- MMS
methyl methane sulfonate
- NHEJ
nonhomologous end-joining
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Freely available online through the PNAS open access option.
References
- 1.Kolodner R. D., Putnam C. D., Myung K. Science. 2002;297:552–557. doi: 10.1126/science.1075277. [DOI] [PubMed] [Google Scholar]
- 2.Lengauer C., Kinzler K. W., Vogelstein B. Nature. 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- 3.Banerjee S., Myung K. Eukaryot. Cell. 2004;3:1557–1566. doi: 10.1128/EC.3.6.1557-1566.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang D., Koshland D. Genes Dev. 2003;17:1741–1754. doi: 10.1101/gad.1089203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kanellis P., Agyei R., Durocher D. Curr. Biol. 2003;13:1583–1595. doi: 10.1016/s0960-9822(03)00578-5. [DOI] [PubMed] [Google Scholar]
- 6.Myung K., Datta A., Kolodner R. D. Cell. 2001;104:397–408. doi: 10.1016/s0092-8674(01)00227-6. [DOI] [PubMed] [Google Scholar]
- 7.Myung K., Kolodner R. D. Proc. Natl. Acad. Sci. USA. 2002;99:4500–4507. doi: 10.1073/pnas.062702199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lengronne A., Schwob E. Mol. Cell. 2002;9:1067–1078. doi: 10.1016/s1097-2765(02)00513-0. [DOI] [PubMed] [Google Scholar]
- 9.Tanaka S., Diffley J. F. Genes Dev. 2002;16:2639–2649. doi: 10.1101/gad.1011002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou J., Monson E. K., Teng S., Schulz V. P., Zakian V. A. Science. 2000;289:771–774. doi: 10.1126/science.289.5480.771. [DOI] [PubMed] [Google Scholar]
- 11.Myung K., Chen C., Kolodner R. D. Nature. 2001;411:1073–1076. doi: 10.1038/35082608. [DOI] [PubMed] [Google Scholar]
- 12.Myung K., Pennaneach V., Kats E. S., Kolodner R. D. Proc. Natl. Acad. Sci. USA. 2003;100:6640–6645. doi: 10.1073/pnas.1232239100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chan S. W., Blackburn E. H. Mol. Cell. 2003;11:1379–1387. doi: 10.1016/s1097-2765(03)00174-6. [DOI] [PubMed] [Google Scholar]
- 14.Pennaneach V., Kolodner R. D. Nat. Genet. 2004;36:612–617. doi: 10.1038/ng1359. [DOI] [PubMed] [Google Scholar]
- 15.Myung K., Datta A., Chen C., Kolodner R. D. Nat. Genet. 2001;27:113–116. doi: 10.1038/83673. [DOI] [PubMed] [Google Scholar]
- 16.Huang M. E., Rio A. G., Nicolas A., Kolodner R. D. Proc. Natl. Acad. Sci. USA. 2003;100:11529–11534. doi: 10.1073/pnas.2035018100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith S., Hwang J.-Y., Banerjee S., Majeed A., Gupta A., Myung K. Proc. Natl. Acad. Sci. USA. 2004;101:9039–9044. doi: 10.1073/pnas.0403093101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou B. B., Elledge S. J. Nature. 2000;408:433–439. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]
- 19.Weinert T. Cell. 1998;94:555–558. doi: 10.1016/s0092-8674(00)81597-4. [DOI] [PubMed] [Google Scholar]
- 20.Paulovich A. G., Toczyski D. P., Hartwell L. H. Cell. 1997;88:315–321. doi: 10.1016/s0092-8674(00)81870-x. [DOI] [PubMed] [Google Scholar]
- 21.Lowndes N., Murguia J. Curr. Opin. Genet. Dev. 2000;10:17–25. doi: 10.1016/s0959-437x(99)00050-7. [DOI] [PubMed] [Google Scholar]
- 22.Michelson R. J., Weinert T. BioEssays. 2000;22:966–969. doi: 10.1002/1521-1878(200011)22:11<966::AID-BIES2>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 23.Santocanale C., Diffley J. F. Nature. 1998;395:615–618. doi: 10.1038/27001. [DOI] [PubMed] [Google Scholar]
- 24.Shirahige K., Hori Y., Shiraishi K., Yamashita M., Takahashi K., Obuse C., Tsurimoto T., Yoshikawa H. Nature. 1998;395:618–621. doi: 10.1038/27007. [DOI] [PubMed] [Google Scholar]
- 25.Myung K., Smith S., Kolodner R. D. Proc. Natl. Acad. Sci. USA. 2004;101:15980–15985. doi: 10.1073/pnas.0407010101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hwang J.-Y., Smith S., Myung K. Genetics. 2005;169:1927–1937. doi: 10.1534/genetics.104.039768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bailey S. M., Meyne J., Chen D. J., Kurimasa A., Li G. C., Lehnert B. E., Goodwin E. H. Proc. Natl. Acad. Sci. USA. 1999;96:14899–14904. doi: 10.1073/pnas.96.26.14899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Myung K., Ghosh G., Fattah F. J., Li G., Kim H., Dutia A., Pak E., Smith S., Hendrickson E. A. Mol. Cell. Biol. 2004;24:5050–5059. doi: 10.1128/MCB.24.11.5050-5059.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Downs J. A., Jackson S. P. Nat. Rev. Mol. Cell Biol. 2004;5:367–378. doi: 10.1038/nrm1367. [DOI] [PubMed] [Google Scholar]
- 30.Hsu H. L., Gilley D., Blackburn E. H., Chen D. J. Proc. Natl. Acad. Sci. USA. 1999;96:12454–12458. doi: 10.1073/pnas.96.22.12454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Samper E., Goytisolo F. A., Slijepcevic P., van Buul P. P., Blasco M. A. EMBO Rep. 2000;1:244–252. doi: 10.1093/embo-reports/kvd051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mimori T., Hardin J. A. J. Biol. Chem. 1986;261:10375–10379. [PubMed] [Google Scholar]
- 33.Falzon M., Fewell J. W., Kuff E. L. J. Biol. Chem. 1993;268:10546–10552. [PubMed] [Google Scholar]
- 34.Riha K., Watson J. M., Parkey J., Shippen D. E. EMBO J. 2002;21:2819–2826. doi: 10.1093/emboj/21.11.2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nussenzweig A., Chen C., da Costa Soares V., Sanchez M., Sokol K., Nussenzweig M. C., Li G. C. Nature. 1996;382:551–555. doi: 10.1038/382551a0. [DOI] [PubMed] [Google Scholar]
- 36.Ouyang H., Nussenzweig A., Kurimasa A., Soares V. C., Li X., Cordon-Cardo C., Li W., Cheong N., Nussenzweig M., Iliakis G., Chen D. J., Li G. C. J. Exp. Med. 1997;186:921–929. doi: 10.1084/jem.186.6.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gu Y., Jin S., Gao Y., Weaver D. T., Alt F. W. Proc. Natl. Acad. Sci. USA. 1997;94:8076–8081. doi: 10.1073/pnas.94.15.8076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boulton S. J., Jackson S. P. EMBO J. 1998;17:1819–1828. doi: 10.1093/emboj/17.6.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nugent C. I., Bosco G., Ross L. O., Evans S. K., Salinger A. P., Moore J. K., Haber J. E., Lundblad V. Curr. Biol. 1998;8:657–660. doi: 10.1016/s0960-9822(98)70253-2. [DOI] [PubMed] [Google Scholar]
- 40.Goytisolo F. A., Samper E., Edmonson S., Taccioli G. E., Blasco M. A. Mol. Cell. Biol. 2001;21:3642–3651. doi: 10.1128/MCB.21.11.3642-3651.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Myung K., Kolodner R. D. DNA Repair (Amsterdam) 2003;2:243–258. doi: 10.1016/s1568-7864(02)00216-1. [DOI] [PubMed] [Google Scholar]
- 42.Tye B. K., Chang V. K. Front. Biosci. 2004;9:2548–2555. doi: 10.2741/1416. [DOI] [PubMed] [Google Scholar]
- 43.Chen C., Kolodner R. D. Nat. Genet. 1999;23:81–85. doi: 10.1038/12687. [DOI] [PubMed] [Google Scholar]
- 44.Umezu K., Sugawara N., Chen C., Haber J. E., Kolodner R. D. Genetics. 1998;148:989–1005. doi: 10.1093/genetics/148.3.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Petrini J. H., Stracker T. H. Trends Cell Biol. 2003;13:458–462. doi: 10.1016/s0962-8924(03)00170-3. [DOI] [PubMed] [Google Scholar]
- 46.Smith S., Gupta A., Kolodner R. D., Myung K. DNA Repair (Amsterdam) 2005;4:606–617. doi: 10.1016/j.dnarep.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 47.Symington L. S. Microbiol. Mol. Biol. Rev. 2002;66:630–670. doi: 10.1128/MMBR.66.4.630-670.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tishkoff D. X., Filosi N., Gaida G. M., Kolodner R. D. Cell. 1997;88:253–263. doi: 10.1016/s0092-8674(00)81846-2. [DOI] [PubMed] [Google Scholar]
- 49.Madeo F., Herker E., Maldener C., Wissing S., Lachelt S., Herlan M., Fehr M., Lauber K., Sigrist S. J., Wesselborg S., Frohlich K. U. Mol. Cell. 2002;9:911–917. doi: 10.1016/s1097-2765(02)00501-4. [DOI] [PubMed] [Google Scholar]
- 50.Basrai M. A., Velculescu V. E., Kinzler K. W., Hieter P. Mol. Cell. Biol. 1999;19:7041–7049. doi: 10.1128/mcb.19.10.7041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Parenteau J., Wellinger R. J. Mol. Cell. Biol. 1999;19:4143–4152. doi: 10.1128/mcb.19.6.4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Park S. J., Ciccone S. L., Freie B., Kurlmasa A., Chen D. J., Li G. C., Clapp D. W., Lee S. H. J. Biol. Chem. 2004;279:6046–6055. doi: 10.1074/jbc.M311054200. [DOI] [PubMed] [Google Scholar]
- 53.Greenwell P. W., Kronmal S. L., Porter S. E., Gassenhuber J., Obermaier B., Petes T. D. Cell. 1995;82:823–829. doi: 10.1016/0092-8674(95)90479-4. [DOI] [PubMed] [Google Scholar]
- 54.Sanchez Y., Desany B. A., Jones W. J., Liu Q., Wang B., Elledge S. J. Science. 1996;271:357–360. doi: 10.1126/science.271.5247.357. [DOI] [PubMed] [Google Scholar]
- 55.Zhao X., Blobel G. Proc. Natl. Acad. Sci. USA. 2005;102:4777–4782. doi: 10.1073/pnas.0500537102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.van Brabant A. J., Stan R., Ellis N. A. Annu. Rev. Genomics Hum. Genet. 2000;1:409–459. doi: 10.1146/annurev.genom.1.1.409. [DOI] [PubMed] [Google Scholar]
- 57.Karmakar P., Snowden C. M., Ramsden D. A., Bohr V. A. Nucleic Acids Res. 2002;30:3583–3591. doi: 10.1093/nar/gkf482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Barbour L., Xiao W. Mutat. Res. 2003;532:137–155. doi: 10.1016/j.mrfmmm.2003.08.014. [DOI] [PubMed] [Google Scholar]
- 59.Shor E., Gangloff S., Wagner M., Weinstein J., Price G., Rothstein R. Genetics. 2002;162:647–662. doi: 10.1093/genetics/162.2.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Onoda F., Seki M., Miyajima A., Enomoto T. Mol. Gen. Genet. 2001;264:702–708. doi: 10.1007/s004380000358. [DOI] [PubMed] [Google Scholar]
- 61.Carson M. J., Hartwell L. Cell. 1985;42:249–257. doi: 10.1016/s0092-8674(85)80120-3. [DOI] [PubMed] [Google Scholar]
- 62.Smolikov S., Mazor Y., Krauskopf A. Proc. Natl. Acad. Sci. USA. 2004;101:1656–1661. doi: 10.1073/pnas.0307796100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Komuro Y., Watanabe T., Hosoi Y., Matsumoto Y., Nakagawa K., Tsuno N., Kazama S., Kitayama J., Suzuki N., Nagawa H. Cancer. 2002;95:1199–1205. doi: 10.1002/cncr.10807. [DOI] [PubMed] [Google Scholar]
- 64.Friesland S., Kanter-Lewensohn L., Tell R., Munck-Wikland E., Lewensohn R., Nilsson A. Head Neck. 2003;25:313–321. doi: 10.1002/hed.10199. [DOI] [PubMed] [Google Scholar]
- 65.Chen L., Trujillo K., Ramos W., Sung P., Tomkinson A. E. Mol. Cell. 2001;8:1105–1115. doi: 10.1016/s1097-2765(01)00388-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.