Abstract
To determine whether microbial chemosensors can be used to find new or better biocatalysts, we constructed Escherichia coli hosts that recognize the product of a biocatalytic conversion through the transcriptional activator NahR and respond by expression of a lacZ or tetA reporter gene. Equipped with a benzaldehyde dehydrogenase (XylC from Pseudomonas putida), the lacZ-based host responded to the oxidation of benzaldehyde and 2-hydroxybenzaldehyde to the corresponding benzoic acids by forming blue colonies, whereas XylC− cells did not. Similarly, the tetA-based host was able to grow under selective conditions only when equipped with XylC, enabling selection of biocatalytically active cells in inactive populations at frequencies as low as 10−6.
Keywords: enzyme selection, reporter genes
An increasing number of industrial products, ranging from pharma intermediates to bulk chemicals, are manufactured with processes that involve one or more biocatalytic steps (1–3), foreshadowing the emergence of a significant biotechnology-based chemical industry (4).
A key factor in this development is the availability of biocatalysts with the necessary regioselectivity and enantioselectivity, activity and stability under practical process conditions. The usual approach to developing such biocatalysts is to start with one or several known enzymes or corresponding DNA sequences or to screen for desired activities among enzyme or expressed DNA libraries in strain collections and in environmental samples. Promising enzymes can then be improved by design or with random mutagenesis techniques in attempts to meet desired functional criteria. The immense natural diversity from which enzymes are recruited and the impressive tool set for further improving native enzymes suggests that there are few conceptual limits to the enzyme activities that might be developed.
There are however practical limits to the testing of new enzyme activities. Typically, natural isolates or recombinants that contain enzyme variants are grown on solid media or in microcultures. Various methods are then used to screen for desired activities, including assays based on chromogenic or fluorogenic substrates (5, 6), which enable high throughput screening but often detect analogs rather than specific products. It is also possible to quantitate products with gas chromatography, high-performance liquid chromatography, mass spectrometry, or NMR (7, 8), but these methods are expensive and rather slow (8).
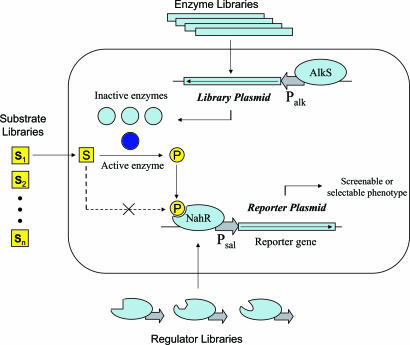
The limitations of screening techniques can in principle be circumvented with biocatalyst selection approaches, if host cell survival and growth are linked to the synthesis of specific biocatalysis products. This approach has been used successfully for products that are essential metabolites, such as prephenate (9, 10), pyruvate (11), and ammonia (12). To also use this approach with industrial products that are not essential metabolites, a linkage might be created by using native or modified regulatory systems as sensing–transducing systems that detect the formation of industrial products. Recently, several assays for the in vivo detection of small molecules with nuclear receptors (13) or riboswitches (14) and for the detection of bond-breaking or bond-forming reactions with a modified yeast-three hybrid system (15) have been reported. Our approach is to detect the reaction product of a desired intracellular biocatalyst with a bacterial regulatory protein that relays this signal to one or more reporter and selection systems (see Fig. 1). By coupling signal detection to survival under selective conditions, the system enables selection for biocatalytically active cells from a background of inactive cells.
Fig. 1.
Overview of the biocatalyst selection system.
Here we describe a system tailored to sense the production of benzoate and 2-hydroxybenzoate from their corresponding aldehydes with an appropriate dehydrogenase: XylC from Pseudomonas putida. As illustrated in Fig. 1, substrate is converted to product in those cells that contain active enzyme. For the detection of benzoate and 2-hydroxybenzoate, we used a previously described mutant of the transcriptional activator protein NahR from P. putida (16). This mutant recognizes both benzoic acids but not the corresponding aldehydes, and it activates transcription from its cognate salicylate promoter. The system reports the presence of an active biocatalyst on the basis of a screenable or selectable phenotype. The system easily selects biocatalyst containing cells present in test cultures at frequencies as low as 10−6.
Results
Construction of Reporter Vectors.
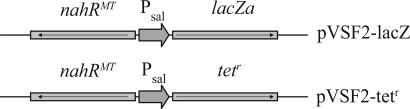
Two plasmids were constructed with different reporter genes fused to the salicylate promoter. Plasmid pVSF2-lacZ carries a gene coding for the α-fragment of β-galactosidase, whereas the tetracycline-resistant (tetr) plasmid pVSF2-tetr carries the tetr gene tetA. Both plasmids carry a constitutively expressed gene coding for a mutated form of the regulatory protein NahR (Fig. 2). In contrast to wild-type NahR, this mutant recognizes benzoate as an inducer and has an increased affinity for the natural inducer 2-hydroxybenzoate (16). Escherichia coli hosts that carry either plasmid should express the reporter genes, producing a screenable (lacZα) or selectable (tetA) phenotype when grown in the presence of benzoate and 2-hydroxybenzoate.
Fig. 2.
Reporter vector pVSF2-lacZ and selector vector pVSF2-tetr carry lacZα and tetA fused to the salicylate promoter. The divergently and constitutively expressed NahR mutant regulates expression of the reporter and selector genes.
Screening for Functional Biocatalysts Based on β-Galactosidase.
To show that expression of the lacZα reporter gene was induced by benzoate and 2-hydroxybenzoate, plasmid pVSF2-lacZ was introduced into E. coli DH10B. We determined the response of the resulting reporter strain by streaking recombinants on plates containing chloramphenicol, X-Gal, and various concentrations of benzoate or 2-hydroxybenzoate (see Tables 1 and 2, which are published as supporting information on the PNAS web site). Blue colonies of DH10B-pVSF2-lacZ were detected at benzoate concentrations of >100 μM and 2-hydroxybenzoate concentrations at >10 μM, with a saturating concentration of ≈500 μM benzoate or ≈100 μM 2-hydroxybenzoate, respectively. Blue colonies were not formed in the absence of inducers.
To demonstrate that benzoate and 2-hydroxybenzoate, which are formed enzymatically from the corresponding aldehydes, could also induce expression of the reporter gene, we introduced benzaldehyde dehydrogenase (XylC) into the strain. Strain DH10B-pVSF2-lacZ was transformed with pCom10-XylC, streaked on plates containing chloramphenicol, kanamycin, X-Gal, and various concentrations of benzaldehyde or 2-hydroxybenzaldehyde and grown exposed to decane vapor to induce XylC. Strain DH10B-pVSF2-lacZ carrying an empty pCom10 vector served as control. On plates containing the XylC substrates, the biocat reporter strain, DH10B-pVSF2-lacZ/pCom10-XylC, formed blue colonies at benzaldehyde concentrations of >50 μM and 2-hydroxybenzaldehyde concentrations of >25 μM. In the absence of substrate, the colonies remained white. The control strain (DH10B-pVSF2-lacZ/pCom10) showed no blue colonies on addition of aldehydes (see Tables 3 and 4, which are published as supporting information on the PNAS web site).
Development of a Biocatalyst Selector Host.
Having shown that catalysis can be coupled to reporter gene expression, we replaced lacZα with tetA to go from a β-galactosidase-based screening system to a tetr-based selection system. In preliminary experiments with E. coli DH10B harboring pVSF2-tetr, we observed benzoate-dependent growth on selective plates containing various concentrations of benzoate. However, only ≈0.1% of the plated cells were able to grow under these conditions. Of the colonies that appeared on the plates, some grew faster than others, resulting in an inhomogeneous population of small and large colonies. When DH10B-pVSF2-tetr was transformed with pCom10-XylC, we observed benzaldehyde-dependent growth, again of only ≈0.1% of plated cells forming small and large colonies. We repeated the plate assay with several of these small and large colonies. Cells that previously formed small colonies performed very poorly in the plate assay, with only ≈0.01–0.05% of plated cells able to grow. However, cells picked from large colonies showed the desired behavior: All cells plated were able to grow and formed similar sized colonies. When tetracycline was omitted from the assay, no difference could be seen between the small and large colony-forming cells.
Some cells displayed a higher tetracycline tolerance. This tolerance is independent of XylC because DH10B-pVSF2-tetr with and without pCom10-XylC showed this effect. To investigate whether a higher tetracycline tolerance was caused by mutation(s) of the host or of the reporter plasmid, we extracted pVSF2-tetr from one slow- and one fast-growing DH10B-pVSF2-tetr/pCom10-XylC strain. Fresh DH10B transformants transformed with these different pVSF2-tetr plasmids grew equally well, indicating that the higher tetracycline tolerance of the fast-growing DH10B-pVSF2-tetr/pCom10-XylC strain was related to a mutation in the host rather than the reporter plasmid.
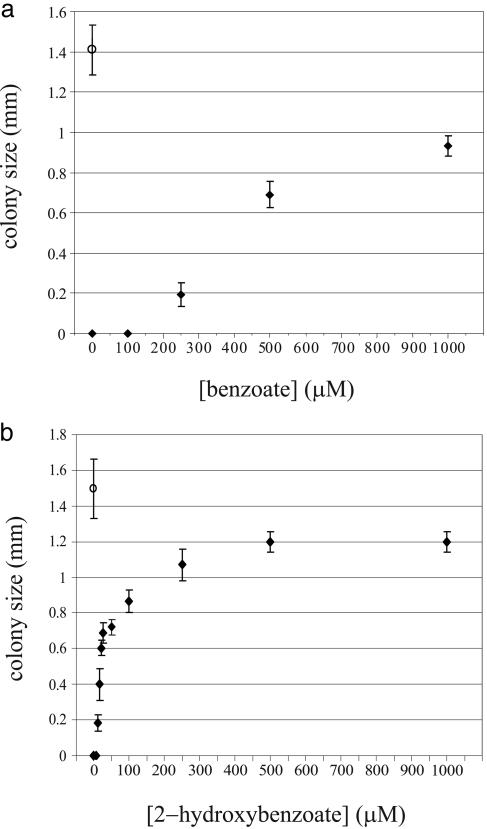
The fast-growing DH10B-pVSF2-tetr/pCom10-XylC strain was then cured of plasmid pCom10-XylC. The resulting DH10B-pVSF2-tetr strain was able to grow on nonselective plates without inducers, whereas growth on selective plates required addition of benzoate or 2-hydroxybenzoate. The effect of benzoate and 2-hydroxybenzoate on growth is dose-dependent (Fig. 3): All cells plated were able to grow on plates with benzoate concentrations of >500 μM or 2-hydroxybenzoate concentrations of >15 μM. On plates containing 250 μM benzoate or 10 μM 2-hydroxybenzoate, however, we observed not only smaller but also fewer colonies, providing us with a suitable biocatalyst selection host.
Fig. 3.
Effect of various concentrations of benzoate (a) and 2-hydroxybenzoate (b) on the growth of DH10B-pVSF2-tetr. Under nonselective conditions, DH10B-pVSF2-tetr is able to grow in the absence of an inducer (○), whereas, under selective conditions, an inducer is required for growth (♦). Error bars indicate SDs. Note that the tetracycline concentrations for the benzoate and 2-hydroxybenzoate assays were 22.5 and 25 μg/ml, respectively.
Response of the Selector Host to XylC Substrates.
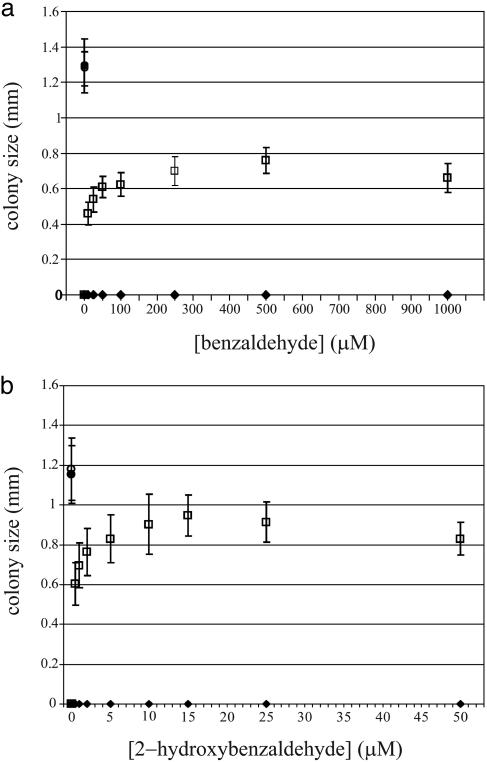
Plasmid pCom10-XylC was reintroduced into DH10B-pVSF2-tetr to produce the desired biocat selector strain DH10B-pVSF2-tetr/pCom10-XylC. This strain grew on tetracycline containing selective plates only after addition of benzaldehyde or 2-hydroxybenzaldehyde. As previously observed with the benzoic acids, the effect of aldehydes on growth also showed a dose dependence (Fig. 4). On plates containing >100 μM benzaldehyde or 2 μM 2-hydroxybenzaldehyde, all plated cells grew. As the aldehyde concentration was lowered, colony diameter and number also decreased. The control strain DH10B-pVSF2-tetr/pCom10, which contains no biocatalyst, failed to grow on addition of aldehydes. As expected, both strains were able to grow on nonselective plates without aldehydes.
Fig. 4.
Effect of various concentrations of benzaldehyde (a) and 2-hydroxybenzaldehyde (b) on the growth of cells containing an active enzyme. Under nonselective conditions, cells with (○) and without (•) active enzyme are able to grow. Growth under selective conditions requires an active enzyme and an aldehyde substrate. Cells containing an active enzyme are able to grow upon addition of benzaldehyde or 2-hydroxybenzaldehyde (□), whereas cells lacking an active enzyme fail to grow in the presence of benzaldehyde or 2-hydroxybenzaldehyde (♦). Error bars indicate SDs. Note that the tetracycline concentrations for the benzaldehyde and 2-hydroxybenzaldehyde assays were 22.5 and 25 μg/ml, respectively
We compared the sensitivity of the selector host for substrates and products by plotting the data of Figs. 3 and 4 as inverse plots of 1/[colony diameter] vs. 1/[compound concentration] and estimating the concentration required for half maximal colony size. Approximately 400 μM benzoate and 20 μM 2-hydroxybenzoate were needed to produce colonies with diameters equal to 50% of the maximum diameters seen in the experiments of Fig. 3. When cells containing XylC were plated with the corresponding substrates, ≈6 μM benzaldehyde and 0.3 μM 2-hydroxybenzaldehyde were needed to produce colonies with half-maximal diameters (Fig. 4). Thus, the aldehyde substrates were ≈65 times more effective than the acid products in triggering the antibiotic resistance response and the 2-hydroxy-substituted compounds were ≈15- to 20-fold more effective than the parent compounds in triggering a response.
Selection of Functional Biocatalysts.
Our goal was to enable the selection of cells that contain active enzymes from a large background population containing no enzyme activity. To demonstrate that this selection is possible with our system, DH10B cells containing the active enzyme XylC were mixed with cells lacking the enzyme in ratios of 1:104, 1:105, and 1:106 and plated on selective and nonselective plates. The number of colonies that appeared on selective plates containing benzaldehyde or 2-hydroxybenzaldehyde corresponded to the number of enzyme containing cells plated. For each ratio and substrate, 10 colonies were tested for pCom10-XylC.
We found 100% positives for ratios of 1:104 and 1:105 for both substrates (Fig. 5), and 90% (benzaldehyde) or 70% (2-hydroxybenzaldehyde) positives for a ratio of 1:106. In addition, the 60 picked colonies were subjected to a second assay to verify that they did indeed show aldehyde-dependent growth. All candidates confirmed as positives were able to grow in the presence of aldehydes and failed to grow in the absence of aldehydes. The four false-positives found on plates with 10−6 positive/inactive cells grew in the presence and in the absence of aldehydes and could thus be singled out by this secondary selection round.
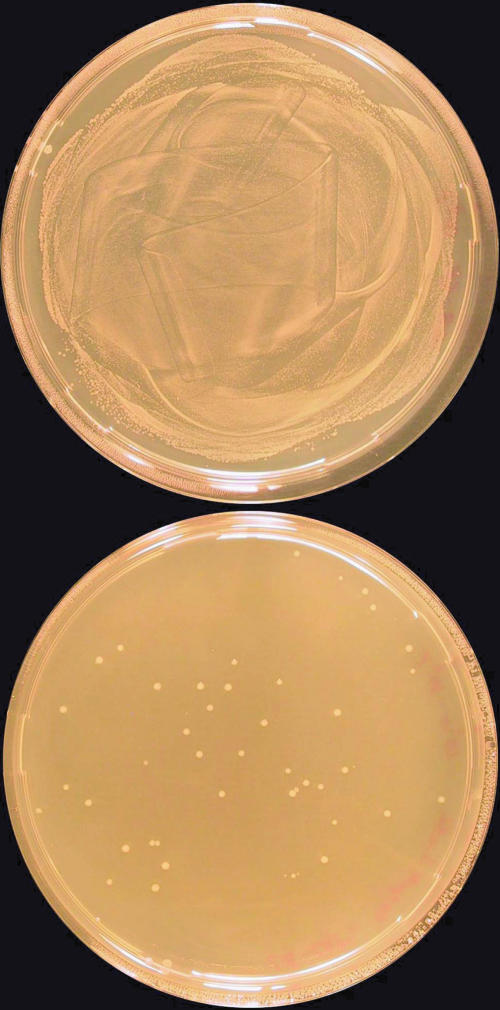
Fig. 5.
Selection of biocatalytically active cells from a background of inactive cells. Biocatalytically active cells that contain the active enzyme XylC were mixed with inactive cells in a ratio of 1:105. Approximately 107 cells were plated and incubated on nonselective (Upper) and selective (Lower) plates. Ten randomly chosen colonies from the selective plate were tested for XylC; all contained the plasmid.
Discussion
The Biocatalyst Selector System.
Colonies developed when test cells were plated on selective media containing benzoic or 2-hydroxybenzoic acid. These XylC products bound the NahR regulator and induced tetA expression with consequent tetracycline antiporter activity, enabling cell growth in the presence of tetracycline. Colonies were formed with minimum concentrations of 10 μM 2-hydroxybenzoate or 100 μM benzoate. Cells plated on selective media containing the substrates benzaldehyde or 2-hydroxybenzaldehyde formed colonies only when they expressed xylC and produced the corresponding products.
Differential Effects of Extracellularly Added Substrates and Products.
The substrate concentrations required to produce half-maximal colony diameters were 65-fold lower than the corresponding product concentrations needed for the same effect. This result can be explained by considering the transport of benzaldehyde and benzoate through the E. coli cell envelope, which for this organism is believed to be based on unlimited passive diffusion (16, 17). When benzaldehyde is oxidized to benzoate by XylC, the intracellular benzoate concentration will increase until the benzoate efflux rate, which increases with increasing intracellular benzoate concentrations, equals the benzoate synthesis rate. Thus, when benzaldehyde is added externally to the concentration needed for half-maximal colony development (6 μM) (Fig. 4), the resulting intracellular benzoate concentration is likely to be close to 400 μM. This value is the concentration needed for half-maximal colony development (Fig. 4) and the one most likely needed for the intracellular benzoate concentration because, based on earlier work, we have established that intracellular and extracellular benzoate equilibrate within 1 or 2 min (18). Interestingly, the 400 μM benzoate concentration is similar to the apparent affinity of benzoate for the NahR variant used here and is estimated at Kd = 500 μM by de Lorenzo and coworkers (16).
At extracellular benzaldehyde concentrations below 6 μM, the rate of benzoate synthesis decreases. The corresponding lower efflux rates result in lower steady state benzoate concentrations, lower tetracycline antiporter activities, higher intracellular tetracycline concentrations, and smaller colony diameters, as seen in Figs. 3 and 4. For benzaldehyde concentrations of >6 μM, benzoate will be synthesized faster, resulting in a higher steady-state benzoate concentration and the formation of larger colonies.
Tuning the Biocatalyst Selection System for Optimal Dynamic Range and Resolution.
The system was tuned by optimizing the specific substrate concentration and the extracellular tetracycline concentration used for selection. The effect of the substrate concentration is illustrated by the differences in the substrate concentrations needed in experiments with the 2-hydroxy-substituted and unsubstituted compounds. Compared with a concentration of 400 μM benzoate, only 20 μM 2-hydroxybenzoate was needed to trigger a half-maximal colony diameter. This result is consistent with the 10-fold higher affinity of the NahR sensor for 2-hydroxybenzoate compared to benzoate (16). To maintain a similar 15- to 20-fold lower intracellular product concentration in selection experiments requires a significantly lower 2-hydroxybenzoate synthesis rate than seen for benzoate synthesis, especially if 2-hydroxybenzoate diffusion rates through E. coli cell membranes are similar to or lower than the benzoate diffusion rates. This supposition probably explains why the 2-hydroxybenzaldehyde concentration needed for half-maximal colony development was only 0.3 μM compared with 6 μM needed in experiments with benzaldehyde as the substrate (Fig. 4). Although there will be differences in kinetic constants between different substrates, a lower product synthesis rate can always be obtained empirically through a substantial lowering of the XylC substrate concentration.
The system can also be modulated via the tetracycline concentration used for selection. In initial experiments with 22.5 μg/ml tetracycline, selection of active biocatalyst-containing hosts against large backgrounds of inactive hosts led to many false positives. When we increased the tetracycline concentration to 25 μg/ml and decreased the substrate concentration, we observed 100% positives at frequencies of 1 in 104 and 1 in 105 biocatalyst containing cells and 70–90% positives for frequencies of 1 in 106.
Additional Biocatalyst Selectors.
A single selection system can be used to monitor several enzymatic reactions because it depends on the synthesis of product and not on how product is made. Thus, the NahR mutant used here can detect nitrilase, amidase, aldehyde oxidase, and aldehyde dehydrogenase activities yielding differently substituted benzoates. In addition, sensing systems such as the NahR and Psal combination used here can be modified to recognize a wider range of biocatalysis products (16, 19). Mutagenesis directed at the effector-binding region produced mutants with higher sensitivity, lower basal reporter gene expression, stronger induction, and the ability to recognize nonnatural effectors while retaining the ability to bind to DNA and activate transcription (16, 19).
Most importantly, however, NahR is far from being an exception. Examples of successful modifications of effector specificity by directed mutagenesis, shuffling, domain swapping, and structure-based design can be found for members of almost all known bacterial transcriptional regulator families (17, 20–32). All of these approaches yielded regulators with altered effector profiles, which is consistent with the notion that structural information can be helpful, but is not required for directed evolution of proteins. In addition, new regulators that respond to chosen effectors have been sought and isolated from bacterial genomes (33, 34). Finally, the conserved domain architecture, the typical small-molecule-binding domains and strongly conserved DNA-binding domains serve as markers to retrieve potential regulators from genomic sequence databases (35–37).
Perspectives of Biocatalyst Selectors.
Given sensors for specific substrate-product pairs, both screening and selection (with tetA or other antibiotic resistance genes) can be used for the identification of relevant biocatalysts. The charm of the selection approach is that it can be carried out with classical and very simple microbiological methods, such as culturing entire libraries (106 to 109 cells) in 1–100 ml of liquid or soft agar cultures and enriching for only those cells that survive antibiotics. A few parallel cultures or plates with different antibiotic levels enable simultaneous biocatalyst searches with different stringencies. Selected cells contain the most active biocatalysts, thus enabling rapid selection of improved biocatalysts among shuffled or otherwise modified variants. In cases for which screening is preferred for the isolation of biocatalysts, additional flexibility can be achieved with fluorescent reporter proteins (38) or cell-surface-displayed binding proteins (39), producing sensor systems compatible with powerful and established analytical methods like FACS (40) and affinity chromatography (41, 42). We believe that biocatalyst screening and selection strains will help accelerate the development of high-quality biocatalysts for many industrial reactions of interest.
Methods
Details on enzymes and materials, all of which were purchased from major suppliers, and information on strains, plasmids (Table 5, which is published as supporting information on the PNAS web site), media, and general procedures are described in the Supporting Methods, which is published as supporting information on the PNAS web site.
Construction of Plasmids pVSF2-tetr and pVSF2-lacZ.
Plasmid pVSF2 was constructed from pMG209 and pCK01. Plasmid pMG209 (10) is a dual-promoter plasmid with a salicylate promoter (Psal) and a T7 RNA polymerase promoter (PT7). In addition, it contains the divergently transcribed nahR gene coding for the wild-type transcriptional regulator NahR. Plasmid pCK01 (43) is a low-copy cloning vector that confers chloramphenicol resistance. The mutation N169D (16) and a PstI restriction site were introduced into the nahR gene of pMG209 with a QuikChange mutagenesis kit (Stratagene). To construct pVSF2-lacZ, lacZα was generated on template pCK01. To construct pVSF2-tetr, tetA was amplified from pUCP26 (44) and ligated into pVSF2. See Supporting Methods for further details.
Plasmids pCom10 and pCom10-XylC.
XylC was placed under the control of the Palk promoter in pCom10 (45), a high-copy expression vector that is based on the alkane-responsive promoter PalkB and its positive regulator, AlkS (45), and is compatible with the low-copy reporter plasmids. Expression of XylC from the Alk promoter is regulated by the transcriptional activator AlkS and can be induced by decane. To construct pCom10-XylC, the xylC gene coding for benzaldehyde dehydrogenase (XylC) from P. putida was amplified from pCK04AXylR (46) and ligated into pCom10. See Supporting Methods for further details.
Plate-Based Assay for β-Galactosidase Activity.
A 2% X-Gal solution (40 μl) in dimethylformamide was spread on LB/agar plates containing chloramphenicol (30 μg/ml) and various concentrations of benzoate or 2-hydroxybenzoate. E. coli DH10B cells harboring reporter plasmid pVSF2-lacZ were streaked on the plates from a liquid culture that had been grown at 30°C to an OD600 of 0.1. Cells were grown for 36 h at 30°C followed by incubation at 4°C for 24 h after which blue colony color formation was evaluated visually.
E. coli DH10B cells harboring the reporter plasmid pVSF2-lacZ together with either the catalyst containing plasmid pCom10-XylC or, as a control, the empty pCom10 vector, grown in a liquid culture at 30°C to an OD600 of 0.1, were similarly streaked on identical X-Gal/chloramphenicol plates supplemented with 25 μg/ml kanamycin and various concentrations of benzaldehyde or 2-hydroxybenzaldehyde. Cells were plated and grown as above under decane vapor, followed by incubation at 4°C and testing for blue colony color as above.
Plate-Based Assay for tetr.
Approximately 103 E. coli DH10B cells harboring the reporter plasmid pVSF2-tetr grown in liquid culture at 30°C to an OD600 of 0.1 were spread onto nonselective LB/agar plates containing only 30 μg/ml chloramphenicol and onto selective plates containing 30 μg/ml chloramphenicol, 22.5 μg/ml tetracycline, and various concentrations of benzoate. Identical nonselective and selective plates were supplemented with various concentrations of 2-hydroxybenzoate, except that the tetracycline concentration was 25 μg/ml. Plates were incubated for 36 h at 30°C. Each plate assay was conducted in triplicate, and growth was determined by measuring the colony diameter of 10 solitary colonies for each plate. Colony diameter differences provide significant amplification of small differences in colony growth rates. These reflect differences in tetA expression, which in turn depends on the intracellular concentrations of a biocatalysis product.
Approximately 103 E. coli DH10B cells harboring the reporter plasmid pVSF2-tetr and either the catalyst-containing plasmid pCom10-XylC or, as a control, the empty pCom10 vector, were similarly grown and spread onto nonselective LB/agar plates containing only 30 μg/ml chloramphenicol and 25 μg/ml kanamycin and onto selective plates containing 30 μg/ml chloramphenicol, 25 μg/ml kanamycin, 22.5 μg/ml tetracycline, and various concentrations of benzaldehyde. Plates were incubated for 36 h at 30°C under decane vapor, and colony size was assessed in triplicate as above. The assay was similarly performed with various concentrations of 2-hydroxybenzaldehyde, except that the tetracycline concentration was 25 μg/ml.
Plate-Based Selection for a Functional Enzyme.
Strain DH10B harboring pVSF2-tetr and either pCom10 or pCom10-XylC was grown in liquid culture to an OD600 of 0.1. The two cultures were diluted and mixed in ratios of 104:1, 105:1, and 106:1, keeping the amount of cells containing pCom10-XylC constant at 102 while increasing the amount of cells containing pCom10 from 106 to 107 to 108. The cells were spread onto selective LB/agar plates containing 30 μg/ml chloramphenicol, 25 μg/ml kanamycin, 25 μg/ml tetracycline, and either 200 μM benzaldehyde or 5 μM 2-hydroxybenzaldehyde and incubated for 36 h at 30°C, during which time the cells were exposed to decane vapor. For each ratio and substrate, the plate assay was conducted in triplicate, and per ratio and substrate the plasmids of 10 colonies were isolated and analyzed by restriction digest with NdeI and HindIII for the presence of the xylC gene. In addition, the 60 colonies were subjected to a second assay. The cells were restreaked onto selective LB/agar plates containing 30 μg/ml chloramphenicol, 25 μg/ml kanamycin, 25 μg/ml tetracycline, and no substrate, 200 μM benzaldehyde, or 5 μM 2-hydroxybenzaldehyde. Plates were incubated for 36 h at 30°C under decane vapor.
Curing DH10B-pVSF2-tetr/pCom10-XylC of pCom10-XylC.
DH10B-pVSF-tetr/pCom10-XylC was cured of pCom10-XylC by three successive rounds of growth in liquid LB containing only 30 μg/ml chloramphenicol and 0.05% dicyclopropylketone. Because plasmid pCom10-XylC confers kanamycin resistance, kanamycin was omitted from the medium. Addition of dicyclopropylketone-induced expression of xylC and resulted in selection pressure against cells containing pCom10-XylC. After three rounds, cells were analyzed on LB/agar plates containing only 30 μg/ml chloramphenicol or containing 30 μg/ml chloramphenicol and 25 μg/ml kanamycin. Plasmids were isolated from cells that were able to grow on plates containing chloramphenicol but had lost the ability to grow in the presence of kanamycin. Restriction analysis of plasmids confirmed that cells were cured of pCom10-XylC and retained pVSF2-tetr.
Supplementary Material
Acknowledgments
We thank Peter Kast and Donald Hilvert (both from the Swiss Federal Institute of Technology, Zürich) for the kind gift of plasmid pMG209, Sven Panke (Swiss Federal Institute of Technology) for plasmid pCK04AxylR, and Andreas Meyer (Swiss Federal Institute of Technology) for plasmid pUCP26.
Abbreviation
- tetr
tetracycline-resistant.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office. J.D.S. is a guest editor invited by the Editorial Board.
References
- 1.Faber K., editor. Biotransformations in Organic Chemistry. 3rd Ed. New York: Springer; 1997. [Google Scholar]
- 2.Schmid A., Hollmann F., Park J. B., Bühler B. Curr. Opin. Biotechnol. 2002;13:359–366. doi: 10.1016/s0958-1669(02)00336-1. [DOI] [PubMed] [Google Scholar]
- 3.Straathof A. J. J., Panke S., Schmid A. Curr. Opin. Biotechnol. 2002;13:548–556. doi: 10.1016/s0958-1669(02)00360-9. [DOI] [PubMed] [Google Scholar]
- 4.Schmid A., Dordick J. S., Hauer B., Kiener A., Wubbolts M., Witholt B. Nature. 2001;409:258–268. doi: 10.1038/35051736. [DOI] [PubMed] [Google Scholar]
- 5.Goddard J.-P., Reymond J.-L. Trends Biotechnol. 2004;22:363–370. doi: 10.1016/j.tibtech.2004.04.005. [DOI] [PubMed] [Google Scholar]
- 6.Goddard J.-P., Reymond J.-L. Curr. Opin. Biotechnol. 2004;15:314–322. doi: 10.1016/j.copbio.2004.06.008. [DOI] [PubMed] [Google Scholar]
- 7.Reetz M. T. Angew. Chem. Int. Ed. 2001;40:284–310. [PubMed] [Google Scholar]
- 8.Eggert T., Jaeger K.-E., Reetz M. T. In: Enzyme Functionality: Design, Engineering, and Screening. Svendsen A, editor. New York: Marcel Dekker; 2004. pp. 559–598. [Google Scholar]
- 9.MacBeath G., Kast P., Hilvert D. Science. 1998;279:1958–1961. doi: 10.1126/science.279.5358.1958. [DOI] [PubMed] [Google Scholar]
- 10.Gamper M., Hilvert D., Kast P. Biochemistry. 2000;39:14087–14094. doi: 10.1021/bi0016570. [DOI] [PubMed] [Google Scholar]
- 11.Griffiths J. S., Cheriyan M., Corbell J. B., Pocivavsek L., Fierke C. A., Toone E. J. Bioorg. Med. Chem. 2004;12:4067–4074. doi: 10.1016/j.bmc.2004.05.034. [DOI] [PubMed] [Google Scholar]
- 12.Robertson D. E., Chaplin J. A., DeSantis G., Podar M., Madden M., Chi E., Richardson T., Milan A., Miller M., Weiner D. P. Appl. Environ. Microbiol. 2004;70:2429–2436. doi: 10.1128/AEM.70.4.2429-2436.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schwimmer L. J., Rohatgi P., Azizi B., Seley K. L., Doyle D. F. Proc. Natl. Acad. Sci. USA. 2004;101:14707–14712. doi: 10.1073/pnas.0400884101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Buskirk A. R., Landrigan A., Liu D. R. Chem. Biol. 2004;11:1157–1163. doi: 10.1016/j.chembiol.2004.05.017. [DOI] [PubMed] [Google Scholar]
- 15.Baker K., Bleczinski C., Lin H., Salazar-Jimenez G., Sengupta D., Krane S., Cornish V. W. Proc. Natl. Acad. Sci. USA. 2002;99:16537–16542. doi: 10.1073/pnas.262420099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cebolla A., Sousa C., de Lorenzo V. J. Biol. Chem. 1997;272:3986–3992. doi: 10.1074/jbc.272.7.3986. [DOI] [PubMed] [Google Scholar]
- 17.Ramos J. L., Michan C., Rojo F., Dwyer D., Timmis K. J. Mol. Biol. 1990;211:373–382. doi: 10.1016/0022-2836(90)90358-S. [DOI] [PubMed] [Google Scholar]
- 18.Buhler B., Schmid A., Hauer B., Witholt B. J. Biol. Chem. 2000;275:10085–10092. doi: 10.1074/jbc.275.14.10085. [DOI] [PubMed] [Google Scholar]
- 19.Park H. H., Lee H. Y., Lim W. K., Shin H. J. Arch. Biochem. Biophys. 2005;434:67–74. doi: 10.1016/j.abb.2004.10.020. [DOI] [PubMed] [Google Scholar]
- 20.Skarfstad E., O’Neill E., Garmendia J., Shingler V. J. Bacteriol. 2000;182:3008–3016. doi: 10.1128/jb.182.11.3008-3016.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Salto R., Delgado A., Michan C., Marques S., Ramos J. L. J. Bacteriol. 1998;180:600–604. doi: 10.1128/jb.180.3.600-604.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garmendia J., Devos D., Valencia A., De Lorenzo V. Mol. Microbiol. 2001;42:47–59. doi: 10.1046/j.1365-2958.2001.02633.x. [DOI] [PubMed] [Google Scholar]
- 23.Smirnova I. A., Dian C., Leonard G. A., McSweeney S., Birse D., Brzezinski P. J. Mol. Biol. 2004;340:405–418. doi: 10.1016/j.jmb.2004.04.071. [DOI] [PubMed] [Google Scholar]
- 24.Scholz O., Kostner M., Reich M., Gastiger S., Hillen W. J. Mol. Biol. 2003;329:217–227. doi: 10.1016/s0022-2836(03)00427-3. [DOI] [PubMed] [Google Scholar]
- 25.Henssler E.-M., Scholz O., Lochner S., Gmeiner P., Hillen W. Biochemistry. 2004;43:9512–9518. doi: 10.1021/bi049682j. [DOI] [PubMed] [Google Scholar]
- 26.Hellinga H. W., Marvin J. S. Trends Biotechnol. 1998;16:183–189. doi: 10.1016/s0167-7799(98)01174-3. [DOI] [PubMed] [Google Scholar]
- 27.Ramos J. L., Stolz A., Reineke W., Timmis K. N. Proc. Natl. Acad. Sci. USA. 1986;83:8467–8471. doi: 10.1073/pnas.83.22.8467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Devos D., Garmendia J., de Lorenzo V., Valencia A. Environ. Microbiol. 2002;4:29–41. doi: 10.1046/j.1462-2920.2002.00265.x. [DOI] [PubMed] [Google Scholar]
- 29.Wise A. A., Kuske C. R. Appl. Environ. Microbiol. 2000;66:163–169. doi: 10.1128/aem.66.1.163-169.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schumacher Maria A., Brennan Richard G. Mol. Microbiol. 2002;45:885–893. doi: 10.1046/j.1365-2958.2002.03039.x. [DOI] [PubMed] [Google Scholar]
- 31.Grkovic S., Hardie K. M., Brown M. H., Skurray R. A. Biochemistry. 2003;42:15226–15236. doi: 10.1021/bi035447+. [DOI] [PubMed] [Google Scholar]
- 32.Bertram R., Kraft C., Wisshak S., Mueller J., Scholz O., Hillen W. J. Mol. Microbiol. Biotechnol. 2005;8:104–110. doi: 10.1159/000084565. [DOI] [PubMed] [Google Scholar]
- 33.Uchiyama T., Abe T., Ikemura T., Watanabe K. Nat. Biotechnol. 2005;23:88–93. doi: 10.1038/nbt1048. [DOI] [PubMed] [Google Scholar]
- 34.Noda K., Watanabe K., Maruhashi K. Biotechnol. Lett. 2003;25:1875–1882. doi: 10.1023/a:1020926107636. [DOI] [PubMed] [Google Scholar]
- 35.Perez-Rueda E., Collado-Vides J. Nucleic Acids Res. 2000;28:1838–1847. doi: 10.1093/nar/28.8.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Anantharaman V., Koonin E. V., Aravind L. J. Mol. Biol. 2001;307:1271–1292. doi: 10.1006/jmbi.2001.4508. [DOI] [PubMed] [Google Scholar]
- 37.Babu M. M., Teichmann S. A. Nucleic Acids Res. 2003;31:1234–1244. doi: 10.1093/nar/gkg210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cormack B. P., Valdivia R. H., Falkow S. Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- 39.Samuelson P., Gunneriusson E., Nygren P.-A., Stahl S. J. Biotechnol. 2002;96:129–154. doi: 10.1016/s0168-1656(02)00043-3. [DOI] [PubMed] [Google Scholar]
- 40.Daugherty P. S., Iverson B. L., Georgiou G. J. Immunol. Methods. 2000;243:211–227. doi: 10.1016/s0022-1759(00)00236-2. [DOI] [PubMed] [Google Scholar]
- 41.Patel D., Vitovski S., Senior H. J., Edge M. D., Hockney R. C., Dempsey M. J., Sayers J. R. Biochem. J. 2001;357:779–785. doi: 10.1042/0264-6021:3570779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ferenci T., Lee K. S. J. Mol. Biol. 1982;160:431–444. doi: 10.1016/0022-2836(82)90306-0. [DOI] [PubMed] [Google Scholar]
- 43.Fernandez S., de Lorenzo V., Perez-Martin J. Mol. Microbiol. 1995;16:205–213. doi: 10.1111/j.1365-2958.1995.tb02293.x. [DOI] [PubMed] [Google Scholar]
- 44.West S. E. H., Schweizer H. P., Dall C., Sample A. K., Runyen-Janecky L. J. Gene. 1994;148:81–86. doi: 10.1016/0378-1119(94)90237-2. [DOI] [PubMed] [Google Scholar]
- 45.Smits T. H. M., Seeger M. A., Witholt B., van Beilen J. B. Plasmid. 2001;46:16–24. doi: 10.1006/plas.2001.1522. [DOI] [PubMed] [Google Scholar]
- 46.Panke S., Sanchez-Romero J. M., de Lorenzo V. Appl. Environ. Microbiol. 1998;64:748–751. doi: 10.1128/aem.64.2.748-751.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.