Abstract
Endogenously formed reactive oxygen species continuously damage cellular constituents including DNA. These challenges, coupled with exogenous exposure to agents that generate reactive oxygen species, are both associated with normal aging processes and linked to cardiovascular disease, cancer, cataract formation, and fatty liver disease. Although not all of these diseases have been definitively shown to originate from mutations in nuclear DNA or mitochondrial DNA, repair of oxidized, saturated, and ring-fragmented bases via the base excision repair pathway is known to be critical for maintaining genomic stability. One enzyme that initiates base excision repair of ring-fragmented purines and some saturated pyrimidines is NEIL1, a mammalian homolog to Escherichia coli endonuclease VIII. To investigate the organismal consequences of a deficiency in NEIL1, a knockout mouse model was created. In the absence of exogenous oxidative stress, neil1 knockout (neil1−/−) and heterozygotic (neil1+/−) mice develop severe obesity, dyslipidemia, and fatty liver disease and also have a tendency to develop hyperinsulinemia. In humans, this combination of clinical manifestations, including hypertension, is known as the metabolic syndrome and is estimated to affect >40 million people in the United States. Additionally, mitochondrial DNA from neil1−/− mice show increased levels of steady-state DNA damage and deletions relative to wild-type controls. These data suggest an important role for NEIL1 in the prevention of the diseases associated with the metabolic syndrome.
Keywords: DNA repair, fatty liver disease, mitochondria, obesity, oxidative stress
After exposure to reactive oxygen species (ROS), the major purine lesions are 8-oxoguanine, 2,6-diamino-4-hydroxy-5-formamidopyrimidine, and 4,6-diamino-5-formamidopyrimidine (1–5). To reverse the potentially deleterious effects of oxidative DNA base lesions, cells primarily use the base excision repair (BER) pathway to restore the DNA to its undamaged state (6). The BER pathway is initiated by lesion-specific DNA glycosylases that catalyze bond scission and release the damaged base from the deoxyribose sugar.
Unlike nucleotide excision repair, which functions only on nuclear DNA, the BER pathway is operative on both nuclear DNA and mitochondrial DNA (mtDNA). Although BER in the mitochondria repairs normal endogenous oxidant-induced DNA damage, expression of mitochondrially targeted DNA glycosylases that are specific for the repair of oxidatively induced DNA lesions leads to enhanced repair and increased survival after ROS challenge (7–10). These data emphasize that maintenance of the mitochondrial genome is a delicate balance of mtDNA copy number and BER capacity.
To initiate repair of oxidative lesions, mammalian cells primarily use the products of the ogg1, nth1, neil1, and neil2 genes (11). These corresponding proteins have somewhat overlapping substrate specificities that may explain, at least partially, the absence of obvious phenotypes in ogg1- and nth1-null mice. NEIL1 has a strong substrate preference for 2,6-diamino-4-hydroxy-5-formamidopyrimidine and 4,6-diamino-5-formamidopyrimidine (12, 13), with reduced excision of other ROS-damaged bases (12, 14–16). In addition, it stimulates the activity of OGG1 on DNAs containing 8-oxoguanine substrates (17), and it is a major enzyme involved in excision of the 3′ end proximal lesions (18). Furthermore, in contrast to OGG1 and NTH1, which require duplex DNA for efficient repair, NEIL1 also operates on single-stranded and bubble-structured substrates (19). This distinct substrate specificity, as well as increased levels of NEIL1 during the S phase of the cell cycle, have given rise to the suggestion of its possible involvement in replication-associated and/or transcription-coupled repair (12, 19, 20). Recently, Das et al. (21) demonstrated that neil1 mRNA levels are elevated ≈3-fold after ROS exposure. Additionally, NEIL1 was shown to localize in both the nucleus and mitochondria (22), suggesting that this enzyme is involved in the overall maintenance of genomic stability.
The potential biological importance of NEIL1 was demonstrated in studies showing a correlation of inactivating mutations in neil1 with human gastric cancer (23). Furthermore, RNA interference experiments revealed that cells deficient in NEIL1 were significantly sensitized to the killing effects of ionizing radiation (24). These data also indicate a critical role for NEIL1 in long-term maintenance of genetic integrity. To determine the biological consequences resulting from deficiencies in NEIL1, we generated knockout (KO) mice with a frameshift deletion and insertion in the neil1 gene.
Results
Construction of neil1 KO Mice.
The neil1-targeting vector and chimeric neil1+/− heterozygotes were obtained as described in Materials and Methods. These mice were then bred to C57BL/6 mice to generate heterozygous founder neil1+/− mice (designated as sublines 4 and 6). Offspring of heterozygous mice were generated at the expected 1:2:1 Mendelian ratios. Subsequent generations of mice were confirmed to be neil1−/− by Southern blot analyses and/or PCR analyses that were specific for disruption of the neil1 gene. These mice did not show any obvious developmental or immediate postnatal defects.
Onset of Obesity, Fatty Liver Disease, and Kidney Vacuolization Associated with Male neil1 KO Mice.
The progeny of a neil1−/− male (no. 218) with two neil1−/− females (nos. 257 and 260) generated 17 pups that were phenotypically normal through their first 4–6 months of life (Fig. 1). However, in a time interval covering ≈6–10 months of age, all male progeny developed severe obesity such that, on average, by 7 months of age male neil1−/− mice weighed ≈50 g, as compared with 28 g for the control C57BL/6 wild-type mice (Fig. 2A). In the same time frame, their female littermates were modestly overweight (≈32 g on average) as compared with wild-type female C57BL/6 (≈24 g).
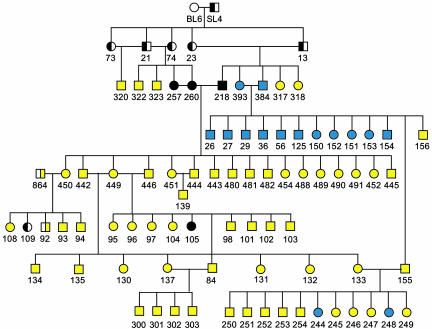
Fig. 1.
Pedigree of neil1−/− mice. Open symbols are wild-type mice, half-filled symbols are heterozygous mice, and filled symbols are homozygous KO mice. Yellow symbols represent obese mice, black symbols represent mice that were killed before disease manifestation, and blue symbols represent mice that did not or have not yet shown an obesity phenotype.
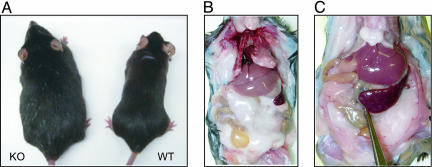
Fig. 2.
Images of neil1 KO mice. (A) Age-matched comparison of a morbidly obese KO male with a wild-type male at ≈7 months of age. Necropsies revealed extensive fat deposits in the abdominal cavity, completely encasing many organs for male (B) and female (C) KO mice.
Within the first generations of neil1−/− mice, the majority of these mice developed severe obesity, but isolated individuals remained visibly normal through at least their first year of life. However, all subsequent generations of neil1−/− crosses within founder subline 4 consistently manifest early–midlife obesity (Fig. 1), with males being severely more affected than females. Because all mice were housed in a common facility, fed a normal chow diet ad libitum (PicoLab Rodent Diet 20), and had continuous access to a common watering system, these factors were eliminated as variables in explaining the observed phenotype.
Autopsies of the obese male and female neil1−/− confirmed massive deposition of fat within the subcutis, retroperitoneum, mediastinum, and abdomen, with nearly complete encasement of most internal organs (Fig. 2 B and C, respectively). Typically, the livers from neil1−/− mice >9 months of age were significantly enlarged, granulated, and pale yellow in color (Fig. 3A). Histologic sections from wild-type (Fig. 3B) and neil1−/− (Fig. 3C) mice confirmed marked hepatic steatosis in the obese neil1−/− animals, with >50% of the cellular contents being a mixture of micro- and macrovesicular globules (Fig. 3C). These vacuoles were confirmed to contain lipids by carrying out oil red O staining of flash-frozen liver tissues (Fig. 3D). Interestingly, the steatosis was most prominent in the centrilobular regions (zone 3). There was no significant inflammation or fibrosis. The livers of female neil1−/− mice also showed steatosis, but in most cases it did not manifest with the same severity as was observed in male neil1−/− mice.
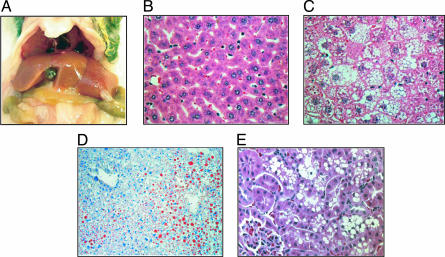
Fig. 3.
Alterations in liver and kidney tissues from neil1 KO mice. (A) Photograph of enlarged, pale yellow-colored liver from an obese KO male. (B and C) Comparison of hematoxylin and eosin-stained sections of liver from wild-type and male KO mice, respectively. Micro- and macrovesicular steatosis of centrilobular hepatocytes is prominent in the KO. (Original magnifications: ×400.) (D) Oil red O staining of sections of flash-frozen liver for an obese KO male. (Original magnification: ×400.) (E) Hematoxylin and eosin-stained section of kidney from an obese KO male showing vacuoles in proximal tubule epithelial cells.
Histological analyses of kidney sections in obese mice revealed significant cytoplasmic vacuolization of tubule cells (Fig. 3E). In particular, clusters of clear, round vacuoles, variable in size, caused distention in many of the epithelial cells of the proximal convoluted tubules. The vacuoles were inferred to be aqueous in nature, based on negative periodic acid Schiff and oil red O staining. Similar to the steatosis being more pronounced in the neil1−/− males, vacuolization of the proximal tubules was also significantly more evident in male versus female neil1−/− mice.
Hyperleptinemia.
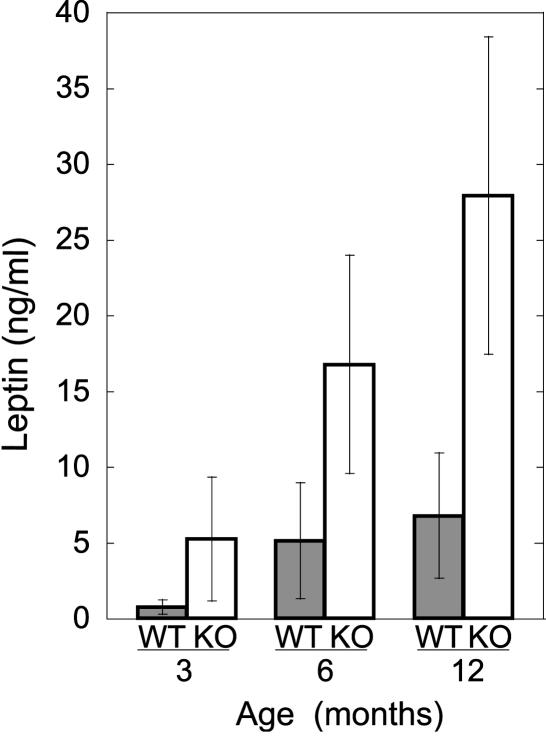
In addition to the observation of significant macro- and microscopic fat deposition, circulating leptin levels were assessed as an independent measure of adipose tissue burden, because plasma concentrations of leptin are generally proportional to total body fat. Male neil1−/− mice that were either obese or the progeny of obese matings (n = 22) ranging in age from 3 to 12 months showed significantly elevated levels of leptin, even at 3 months (Fig. 4). By 6 months, all male neil1−/− mice displayed characteristics of hyperleptinemia, presumably through the acquisition of hypothalamic leptin resistance. Consistent with the observations that female neil1−/− mice also display fatty liver disease, albeit to a lesser degree, leptin values in 1-year-old neil1−/− females showed increased levels (≈2.5-fold) over wild-type controls (data not shown). Additionally, circulating triglycerides were measured in the male (n = 21) and female (n = 10) neil1−/− mice and compared with age-matched control mice (n = 10 for each sex). With P < 0.05, they were 1.4- and 1.3-fold elevated at 3 months and 1.7- and 1.4-fold elevated at 7 months in males and females, respectively (data not shown).
Fig. 4.
Leptin levels are elevated in neil1 KO mice. Circulating levels of leptin were measured in male wild-type and neil1−/− mice that ranged in age from 3 to 12 months. Error bars indicate SDs.
Hyperinsulinemia.
Another disease manifestation that frequently cosegregates with obesity and dyslipidemia is insulin resistance, which can ultimately manifest as type 2 diabetes. Data analyzed from 1-year-old male obese neil1−/− mice showed that insulin levels were distributed over a wide range, with the highest values being ≈13-fold higher than those for age-matched control mice. On average, insulin was 4-fold elevated relative to the control with a P value of 0.078 as calculated by the Wilcoxon–Mann–Whitney test. The large range of insulin values for male neil1−/− mice may reflect interpopulation differences along the development of full insulin resistance. Insulin levels in offspring of obese mice at 3 and 6 months of age are indistinguishable from age-matched controls. neil1−/− females did not show a statistically significant increase in insulin (data not shown). Additionally, blood glucose levels were monitored in control and neil1−/− mice ranging in age from 3 to 12 months under both fasting and fed conditions. There were no statistically significant differences in glucose levels among any of the mice (data not shown).
Increased mtDNA Damage and Deletions.
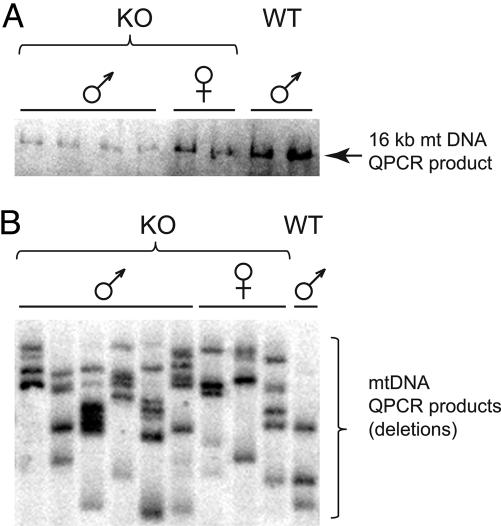
It is well established that the onset and progression of fatty liver disease correlates well with exogenous and endogenous cellular stressors that lead to increased ROS: alcohol consumption (25), exposure to bacterial lipopolysaccharides (26), or excessive caloric intake (27). Although these ROS-generating environments may directly modify proteins and lipids and interfere with mitochondrial signaling pathways, we hypothesize that, in the absence of NEIL1, conditions may be generated in which there is increased ROS damage to the mitochondrial genome leading to impaired metabolism versus that found in wild-type mice. To test this hypothesis, steady-state mtDNA damage and deletions from liver tissues of control and neil1−/− mice were compared by using quantitative PCR (QPCR), which measures the relative level of DNA strand breaks and/or polymerase-blocking lesions in a gene-specific manner. Fig. 5A shows that neil1−/− male mice had significantly greater (P < 0.05) levels of mtDNA damage compared with control males or neil1−/− female mice, a result that is consistent with the observation that the neil1−/− male mice were the most severely affected animals in terms of both phenotype and pathology. To examine whether differences existed in the amount of mtDNA deletions between neil1−/− animals and controls, QPCR conditions were modified to enhance the amplification of shorter mtDNA molecules and thus favor mtDNAs that contained deletions. Fig. 5B illustrates that mtDNA deletions were significantly (P < 0.05) more prevalent among the neil1−/− animals when compared with wild-type controls. These data are consistent with either a deficiency of mtDNA repair of oxidatively induced DNA lesions in the neil1−/− animals or an increased level of ROS-induced DNA damage that the steady-state BER enzymes are unable to repair as efficiently as in the wild-type mice.
Fig. 5.
mtDNA damage and deletions are increased in neil1−/− mice. (A) QPCR products from neil1−/− and control mice; less product indicates increased mtDNA damage. (B) Southern blot of amplification products from a modified QPCR designed to enrich for mtDNA molecules containing deletions, probed with radiolabeled mtDNA.
Disease Manifestation in C57BL6 neil1−/− and neil1+/− Mice That Were Extensively Backcrossed.
All of the above characterizations were made on mice that are a first-generation backcross with C57BL/6. Even though we considered it unlikely that the disease phenotype that we describe for the neil1−/− mice is the result of the number of generations that these KOs were backcrossed to C57BL/6 mice, we carried out multiple backcrosses of neil1+/− to wild-type C57BL/6 to test this assumption (Fig. 6, which is published as supporting information on the PNAS web site).
Fourth-generation heterozygotes were mated to produce additional KO mice, and at 8 months of age all neil1−/− mice compared with wild-type neil1+/+ littermates were >50% heavier, suggesting that the original obesity phenotype was not due to the mice being in a mixed background of C57BL/6 and 129/SV. Furthermore, sixth-generation backcrossed heterozygotic males also manifest a significant obesity phenotype relative to their wild-type littermates, thus demonstrating that there are disease implications for even the heterozygotic mice. Although we had previously observed obese heterozygotic males from selected matings (see Fig. 1, nos. 864 and 92), we had not maintained a sufficient number of these mice to confirm disease manifestation in the neil1+/− males. However, these data are strongly suggestive that both first- and sixth-generation neil1+/− become obese.
Sporadic Phenotypes.
In addition to the phenotypes described above, we also observed many additional phenotypes only in our KO colony, including reduced subcutaneous fat, skin ulcerations, joint inflammation, infertility, and cancers. Although we would classify these phenotypes as sporadic, we find it of interest that these sporadic symptoms often occur in clusters within litters. For example, all males (nos. 26, 27, 29, 36, and 56) from a mating of neil1−/− nos. 393 and 384 had low body fat, low leptin, normal liver histology, and normal insulin values (Fig. 1). However, in contrast, tumors were only sporadically observed, and these tumors were neither clustered in litters nor affected the same organ. Thus, it does not appear that a deficiency in NEIL1 alone is sufficient to induce high-frequency cancers.
Analyses of neil1−/− male mice from another founder mouse, subline 6 (Fig. 7, which is published as supporting information on the PNAS web site), which were continuously maintained at the University of Texas Medical Branch in collaboration with Jonathan Ward, also revealed manifestations of severe obesity as observed for the majority of subline 4 neil1−/− mice. However, in collaboration with George Teebor (New York University, New York), a breeding pair of neil1−/− mice from subline 6 gave progeny that appear to be phenotypically normal, even at 1 year of age. At the present time we are not able to explain the differences in phenotype of animals housed at New York University as compared with Oregon Health & Science University and University of Texas Medical Branch because of dietary or stress factors.
Discussion
Although there is growing public awareness of the health dangers associated with long-term human obesity, its prevalence continues to increase, with >60 million Americans and 300 million worldwide being affected (28, 29). The deleterious health effects frequently associated with obesity include hypertension, dyslipidemia, fatty liver disease, and insulin resistance, which in combination comprise the metabolic syndrome. Although the pathogenesis of this syndrome remains controversial, evidence suggests that repeated exposures to ROS may play a central role (30). Based on the data presented herein, a mouse with symptomology resembling the metabolic syndrome was created as a consequence of knocking out the gene encoding the DNA repair enzyme NEIL1.
The neil1−/− mice are not the first mice reported to manifest the combination of diseases associated with the metabolic syndrome. Recently, obesity and the metabolic syndrome have also been linked to a defect in the circadian CLOCK transcription factor (31). The CLOCK-deficient mice displayed attenuated diurnal feeding patterns, and this misalignment of food intake and feeding rhythmicity is postulated to result in the metabolic syndrome. To determine whether neil1−/− mice display defects in circadian rhythms, mouse wheel-running activity was recorded in the laboratory of Charles Allen (Center for Research on Occupational and Environmental Toxicology, Oregon Health & Science University). The neil1−/− mice showed normal entrainment to a 12:12 light:dark cycle and demonstrated circadian wheel running activity when switched into total darkness, thus ruling out defects in the circadian clocks of these mice.
Another mouse model that shows a very similar overall phenotype to the neil1−/− is the peroxisome proliferator-activated receptor (PPAR)-α KO mouse (32). PPAR-α, in association with retinoid X receptors, controls gene expression of proteins that regulate free fatty acid transport and oxidation. Between 6 and 12 months of age, PPAR-α KO females and, to a much lesser extent, males become obese with profound fatty liver disease and elevated triglycerides. Histopathology of female liver sections shows marked centrilobular steatosis that appear to be identical to that observed in the male neil1−/− mice. Based on similar disease progression and pathology, we considered that, in addition to its role in DNA repair, NEIL1 could be functioning as a transcription factor regulating PPAR-α. To investigate this possibility we carried out Affymetrix analyses of mRNA levels in control and neil1−/− livers. In contrast to our hypothesis, values of PPAR-α mRNA were indistinguishable in the three wild-type and three neil1−/− samples. Additionally, no other known PPAR family member or members of the sterol regulatory element-binding proteins that regulate the enzymes for synthesis of cholesterol, fatty acids, and triglycerides were significantly changed.
Overall, the central question that remains to be answered in explaining the observed phenotype in the neil1−/− and neil1+/− mice is: How does a defect in a DNA glycosylase lead to the manifestation of the metabolic syndrome? In the following, we consider two mechanisms to rationalize these data. One mechanism focuses on the implications for an insufficient repair of ROS-induced DNA lesions in the mitochondria. Recently, mitochondrial localization of NEIL1 was demonstrated, thus linking NEIL1 to repair of oxidative damage in mtDNA (22). Our data demonstrate significantly elevated levels of mtDNA damage and deletions, suggesting that at least a subset of lesions may not be repaired in the mtDNA genome. Furthermore, because NEIL1 can initiate repair in single-stranded and bubble structures, the lack of this repair may lead to severe decreases in mitochondrial replication and transcription rates and, thus, an overall disruption of energy homeostasis. The hypothesis that impaired DNA repair capacity of oxidative damage could increase susceptibility to fatty liver disease has previously been proposed by the Diehl group (33). They demonstrated that the greatest accumulation of 8-oxoguanine lesions and the greatest amount of hepatocyte cell death occurred in their models in which the levels of MYH, the glycosylase responsible for initiating the removal of adenine in the A·8-oxoguanine mispair, was the least. These data were interpreted to conclude that the levels of oxidative DNA damage and MYH expression are inversely related in mouse models of nonalcohol-induced fatty liver disease.
The link between excessive ROS in mitochondria and aging and disease has received strong additional support from a recent report by Schriner et al. (34), who demonstrated an increase in murine lifespan by the overexpression of mitochondrially targeted catalase. Median lifespans were increased, on average, 5 months, with corresponding delays in cardiac pathology and cataract development and reduced oxidative damage and mtDNA deletions. Collectively, these data demonstrate a central role for mitochondrial functions in overall organismal fitness.
In this study, we established that mtDNA derived from liver tissue from neil1−/− mice contain increased damage, but we did not determine whether these lesions ultimately result in a mtDNA mutator phenotype. It is of interest to note that the phenotype of the neil1−/− mice does not match that of transgenic mice in which a proofreading-deficient polymerase γ A subunit was knocked-in (35). These mice had a 3- to 5-fold increase in point mutations and increased mtDNA deletions and developed symptoms of premature aging. These phenotypes included reduced lifespan, weight loss, hair loss, spinal curvature, anemia, and reduced fertility. Thus, there is no direct correlation of phenotypes between these exonucleolytic–defective polymerase γ mice and those of the neil1−/−, even though both may have an underlying disruption of the respiratory chain pathway.
An alternative mechanism could place elevated mtDNA damage in liver of neil1−/− mice as a consequence of disease progression, whereas impaired capacity to repair oxidatively induced DNA base lesions in nuclear DNA is a primary cause. Cumulative data clearly suggest that NEIL1 is not simply a backup glycosylase for OGG1 and NTH1, but that it has very distinct substrate specificity and other biochemical and biological properties. The laboratory of Sankar Mitra has put forth compelling arguments for a primary role of NEIL1 in transcription and replication-coupled repair (19). It is reasonable to speculate that the organs in which NEIL1 is maximally expressed would be those organs that might be most drastically affected by a deficiency in NEIL1. Because Northern blot analyses reveal maximal expression in liver, pancreas, and thymus (12), we consider it significant that liver and pancreatic functions are altered in the neil1−/− mice. Thus, in the absence of NEIL1, critical cellular processes may be compromised in these highly differentiated cells, leading to the diseases of the metabolic syndrome.
Materials and Methods
For details, see Supporting Materials and Methods, which is published as supporting information on the PNAS web site.
Construction of the neil1 Targeting Vector and Generation of KO Mice.
The neil1 gene was cloned from a mouse genomic library (ES129SvJ), and the gene organization was determined by restriction endonuclease digestion and DNA sequence analyses (Fig. 8A, which is published as supporting information on the PNAS web site). To construct the null-gene KO plasmid, the neomycin (neo) resistance gene under the transcriptional control of the phosphoglycerate kinase promoter was inserted into the neil1-coding region (Fig. 8B and Supporting Materials and Methods).
The neil1-targeting construct was introduced into 129/SV-derived CJ7 ES that were then selected and screened for homologous recombination (Supporting Materials and Methods). Chimeric mice were generated by microinjection of KO ES cells into C57BL/6 blastocysts, and embryos developed in foster mothers. Successful chimeras were mated with C57BL/6 to generate heterozygous mice. Genotyping was performed by Southern blot analyses and PCR amplification by using standard procedures (Supporting Materials and Methods).
Histopathology.
Mice were euthanized with CO2, and organs were immediately harvested and fixed in 10% neutral-buffered formalin for at least 24 h before processing, paraffin embedding, sectioning, and staining with hematoxylin and eosin, periodic acid Schiff, or periodic acid Schiff with diastase after standard histology protocols. Alternatively, portions of fresh liver and kidney were flash-frozen, and cryostat sections were prepared for staining with oil red O. Photomicrographs were taken on an Olympus BH2 microscope equipped with a Nikon CoolPix 4500 digital camera.
Circulating Disease Marker Analyses.
Whole blood was removed, and glucose and triglyceride levels were analyzed by using the CardioCheck PA Analyzer (Test Medical Systems at Home, Maria Stein, OH). For leptin and insulin analyses, blood was collected from euthanized mice into 5 mM EDTA, and whole cells were removed by centrifugation at 6,000 × g for 10 min. Plasma samples were frozen at −80°C until the time of analyses. Leptin and insulin levels were measured in duplicate by using diagnostic kits from Linco Research (St. Charles, MO) following the manufacturer’s protocol. Sample values that exceeded the linear range of the assays (0.2–30 ng/ml leptin and 0.2–10 ng/ml insulin) were diluted, and the assay was repeated.
QPCR for Evaluating DNA Damage and mtDNA Deletions.
A 16,059-bp QPCR product that encompasses all but 236 bp of the NADH5/6 genes in the mouse mtDNA genome was amplified by using primer set M13597 forward (13,597–13,620 bp) and 13361 reverse (13,361–13,337 bp). To assess levels of mtDNA deletions, the extension time of the QPCR was reduced to 3 min favoring amplification of shorter mtDNA molecules. Deletion amplification products were subjected to electrophoresis and Southern transfer and were probed with radiolabeled mtDNA for verification. Copy number differences were normalized by using a short QPCR that yields products directly related to gene copy numbers. QPCR was performed as described in ref. 36.
Supplementary Material
Acknowledgments
We acknowledge Drs. H. Q. Wang and John Papaconstantinou (University of Texas Medical Branch, Galveston) for preliminary histopathologic analyses and consultations, respectively; Dr. Donald Houghton (Oregon Health & Science University) for assistance with the manuscript and interpretations of kidney tissue sections; Dr. Charles Allen for collaboration on the circadian rhythm studies; and Todd Downes and Toni Brennan for help with manuscript preparation. This work was partially funded through the Houston Endowment of the University of Texas Medical Branch (to R.S.L.) and Oregon Opportunity Funds (to Oregon Health & Science University).
Abbreviations
- ROS
reactive oxygen species
- BER
base excision repair
- KO
knockout
- QPCR
quantitative PCR
- PPAR
peroxisome proliferator-activated receptor.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Freely available online through the PNAS open access option.
References
- 1.Evans M. D., Dizdaroglu M., Cooke M. S. Mutat. Res. 2004;567:1–61. doi: 10.1016/j.mrrev.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 2.Cooke M. S., Evans M. D., Dizdaroglu M., Lunec J. FASEB J. 2003;17:1195–1214. doi: 10.1096/fj.02-0752rev. [DOI] [PubMed] [Google Scholar]
- 3.Greenberg M. M. Biochem. Soc. Trans. 2004;32:46–50. doi: 10.1042/bst0320046. [DOI] [PubMed] [Google Scholar]
- 4.Cadet J., Douki T., Frelon S., Sauvaigo S., Pouget J. P., Ravanat J. L. Free Radical Biol. Med. 2002;33:441–449. doi: 10.1016/s0891-5849(02)00820-1. [DOI] [PubMed] [Google Scholar]
- 5.Dizdaroglu M., Jaruga P., Birincioglu M., Rodriguez H. Free Radical Biol. Med. 2002;32:1102–1115. doi: 10.1016/s0891-5849(02)00826-2. [DOI] [PubMed] [Google Scholar]
- 6.Sancar A., Lindsey-Boltz L. A., Unsal-Kacmaz K., Linn S. Annu. Rev. Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- 7.Dobson A. W., Xu Y., Kelley M. R., LeDoux S. P., Wilson G. L. J. Biol. Chem. 2000;275:37518–37523. doi: 10.1074/jbc.M000831200. [DOI] [PubMed] [Google Scholar]
- 8.Druzhyna N. M., Hollensworth S. B., Kelley M. R., Wilson G. L., Ledoux S. P. Glia. 2003;42:370–378. doi: 10.1002/glia.10230. [DOI] [PubMed] [Google Scholar]
- 9.Rachek L. I., Grishko V. I., Musiyenko S. I., Kelley M. R., LeDoux S. P., Wilson G. L. J. Biol. Chem. 2002;277:44932–44937. doi: 10.1074/jbc.M208770200. [DOI] [PubMed] [Google Scholar]
- 10.Rachek L. I., Grishko V. I., Alexeyev M. F., Pastukh V. V., LeDoux S. P., Wilson G. L. Nucleic Acids Res. 2004;32:3240–3247. doi: 10.1093/nar/gkh648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dizdaroglu M. Mutat. Res. 2003;531:109–126. doi: 10.1016/j.mrfmmm.2003.07.003. [DOI] [PubMed] [Google Scholar]
- 12.Hazra T. K., Izumi T., Boldogh I., Imhoff B., Kow Y. W., Jaruga P., Dizdaroglu M., Mitra S. Proc. Natl. Acad. Sci. USA. 2002;99:3523–3528. doi: 10.1073/pnas.062053799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jaruga P., Birincioglu M., Rosenquist T. A., Dizdaroglu M. Biochemistry. 2004;43:15909–15914. doi: 10.1021/bi048162l. [DOI] [PubMed] [Google Scholar]
- 14.Takao M., Kanno S., Kobayashi K., Zhang Q. M., Yonei S., van der Horst G. T., Yasui A. J. Biol. Chem. 2002;277:42205–42213. doi: 10.1074/jbc.M206884200. [DOI] [PubMed] [Google Scholar]
- 15.Morland I., Rolseth V., Luna L., Rognes T., Bjoras M., Seeberg E. Nucleic Acids Res. 2002;30:4926–4936. doi: 10.1093/nar/gkf618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bandaru V., Sunkara S., Wallace S. S., Bond J. P. DNA Repair (Amst.) 2002;1:517–529. doi: 10.1016/s1568-7864(02)00036-8. [DOI] [PubMed] [Google Scholar]
- 17.Mokkapati S. K., Wiederhold L., Hazra T. K., Mitra S. Biochemistry. 2004;43:11596–11604. doi: 10.1021/bi049097i. [DOI] [PubMed] [Google Scholar]
- 18.Parsons J. L., Zharkov D. O., Dianov G. L. Nucleic Acids Res. 2005;33:4849–4856. doi: 10.1093/nar/gki816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dou H., Mitra S., Hazra T. K. J. Biol. Chem. 2003;278:49679–49684. doi: 10.1074/jbc.M308658200. [DOI] [PubMed] [Google Scholar]
- 20.Hazra T. K., Kow Y. W., Hatahet Z., Imhoff B., Boldogh I., Mokkapati S. K., Mitra S., Izumi T. J. Biol. Chem. 2002;277:30417–30420. doi: 10.1074/jbc.C200355200. [DOI] [PubMed] [Google Scholar]
- 21.Das A., Hazra T. K., Boldogh I., Mitra S., Bhakat K. K. J. Biol. Chem. 2005;280:35272–35280. doi: 10.1074/jbc.M505526200. [DOI] [PubMed] [Google Scholar]
- 22.Hu J., de Souza-Pinto N. C., Haraguchi K., Hogue B. A., Jaruga P., Greenberg M. M., Dizdaroglu M., Bohr V. A. J. Biol. Chem. 2005;280:40544–40551. doi: 10.1074/jbc.M508772200. [DOI] [PubMed] [Google Scholar]
- 23.Shinmura K., Tao H., Goto M., Igarashi H., Taniguchi T., Maekawa M., Takezaki T., Sugimura H. Carcinogenesis. 2004;25:2311–2317. doi: 10.1093/carcin/bgh267. [DOI] [PubMed] [Google Scholar]
- 24.Rosenquist T. A., Zaika E., Fernandes A. S., Zharkov D. O., Miller H., Grollman A. P. DNA Repair (Amst.) 2003;2:581–591. doi: 10.1016/s1568-7864(03)00025-9. [DOI] [PubMed] [Google Scholar]
- 25.Cahill A., Cunningham C. C., Adachi M., Ishii H., Bailey S. M., Fromenty B., Davies A. Alcohol. Clin. Exp. Res. 2002;26:907–915. [PMC free article] [PubMed] [Google Scholar]
- 26.Suliman H. B., Carraway M. S., Piantadosi C. A. Am. J. Respir. Crit. Care Med. 2003;167:570–579. doi: 10.1164/rccm.200206-518OC. [DOI] [PubMed] [Google Scholar]
- 27.Lopez-Torres M., Gredilla R., Sanz A., Barja G. Free Radical Biol. Med. 2002;32:882–889. doi: 10.1016/s0891-5849(02)00773-6. [DOI] [PubMed] [Google Scholar]
- 28.Reaven G., Abbasi F., McLaughlin T. Recent Prog. Horm. Res. 2004;59:207–223. doi: 10.1210/rp.59.1.207. [DOI] [PubMed] [Google Scholar]
- 29.Bays H. E. Obes. Res. 2004;12:1197–1211. doi: 10.1038/oby.2004.151. [DOI] [PubMed] [Google Scholar]
- 30.Droge W. Physiol. Rev. 2002;82:47–95. doi: 10.1152/physrev.00018.2001. [DOI] [PubMed] [Google Scholar]
- 31.Turek F. W., Joshu C., Kohsaka A., Lin E., Ivanova G., McDearmon E., Laposky A., Losee-Olson S., Easton A., Jensen D. R., et al. Science. 2005;308:1043–1045. doi: 10.1126/science.1108750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Costet P., Legendre C., More J., Edgar A., Galtier P., Pineau T. J. Biol. Chem. 1998;273:29577–29585. doi: 10.1074/jbc.273.45.29577. [DOI] [PubMed] [Google Scholar]
- 33.Gao D., Wei C., Chen L., Huang J., Yang S., Diehl A. M. Am. J. Physiol. 2004;287:G1070–G1077. doi: 10.1152/ajpgi.00228.2004. [DOI] [PubMed] [Google Scholar]
- 34.Schriner S. E., Linford N. J., Martin G. M., Treuting P., Ogburn C. E., Emond M., Coskun P. E., Ladiges W., Wolf N., Van Remmen H., et al. Science. 2005;308:1909–1911. doi: 10.1126/science.1106653. [DOI] [PubMed] [Google Scholar]
- 35.Trifunovic A., Wredenberg A., Falkenberg M., Spelbrink J. N., Rovio A. T., Bruder C. E., Bohlooly Y. M., Gidlof S., Oldfors A., Wibom R., et al. Nature. 2004;429:417–423. doi: 10.1038/nature02517. [DOI] [PubMed] [Google Scholar]
- 36.Knight-Lozano C. A., Young C. G., Burow D. L., Hu Z. Y., Uyeminami D., Pinkerton K. E., Ischiropoulos H., Ballinger S. W. Circulation. 2002;105:849–854. doi: 10.1161/hc0702.103977. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.