Abstract
The p53 tumor suppressor retains its wild-type conformation and transcriptional activity in half of all human tumors, and its activation may offer a therapeutic benefit. However, p53 function could be compromised by defective signaling in the p53 pathway. Using a small-molecule MDM2 antagonist, nutlin-3, to probe downstream p53 signaling we find that the cell-cycle arrest function of the p53 pathway is preserved in multiple tumor-derived cell lines expressing wild-type p53, but many have a reduced ability to undergo p53-dependent apoptosis. Gene array analysis revealed attenuated expression of multiple apoptosis-related genes. Cancer cells with mdm2 gene amplification were most sensitive to nutlin-3 in vitro and in vivo, suggesting that MDM2 overexpression may be the only abnormality in the p53 pathway of these cells. Nutlin-3 also showed good efficacy against tumors with normal MDM2 expression, suggesting that many of the patients with wild-type p53 tumors may benefit from antagonists of the p53–MDM2 interaction.
Keywords: apoptosis, inhibitor, cell cycle, tumor, xenograft
The p53 tumor suppressor plays a central role in protection from tumor development by inducing cell-cycle arrest or apoptosis in response to diverse stresses (1). p53 is a transcription factor that controls a complex signal transduction network referred to as the p53 pathway (2, 3). Because of its critical function, p53 is frequently targeted for inactivation and suffers disabling mutations or deletions in ≈50% of all malignant tumors (4, 5). The other half of human cancers express wild-type p53 protein, and its activation may offer a therapeutic benefit. One p53 activation strategy targets the interaction of p53 with its negative regulator MDM2 (6–9). MDM2 is an E3 ubiquitin ligase that binds the tumor suppressor and facilitates its ubiquitin-dependent degradation (10). The MDM2 binding domain overlaps with the transcriptional activation domain of p53, and therefore MDM2 binding also inhibits the transcriptional activity of p53, thus effectively impairing its function (11). Recently, we identified potent and selective small-molecule inhibitors of p53–MDM2 binding, the nutlins, which activate the p53 pathway in vitro and in vivo (12) and may provide a treatment option for tumors with wild-type p53.
To be effective as cancer therapeutics, MDM2 antagonists require not only wild-type p53 but also functional signaling in the p53 pathway. Many tumors retain wild-type p53 because they might have developed alternative mechanisms for disabling p53 function. One such mechanism is overproduction of MDM2 (13–16). Also, defects in p53 signaling can disable or attenuate specific p53 functions or the entire p53 pathway (2, 17). Upstream signaling aberrations can be compensated for by MDM2 antagonists that act directly on the p53–MDM2 interaction. However, defects in downstream p53 signaling may significantly diminish the activity of MDM2 antagonists. Although it has been postulated that such defects exist, systematic studies on the nature and the prevalence of signaling abnormalities downstream of p53 have not been reported to our knowledge.
Here, we use the MDM2 antagonist nutlin-3 as a specific p53 activator to probe downstream signaling in the p53 pathway of cancer cell lines representing diverse solid malignancies with wild-type p53. Our data suggest that the ability of p53 to induce cell-cycle arrest is preserved in solid tumors, but the apoptotic function of p53 is altered to varying extents in many tumor-derived cell lines. In vitro apoptotic response is strongest in cells with aberrant expression of MDM2 and correlates with the in vivo efficacy of nutlin-3, suggesting that MDM2 antagonists may offer promising agents for cancer therapy and that MDM2 status of tumors may provide a valuable response biomarker in the clinic.
Results
Nutlin-3 Is a Selective Inducer of the p53 Pathway.
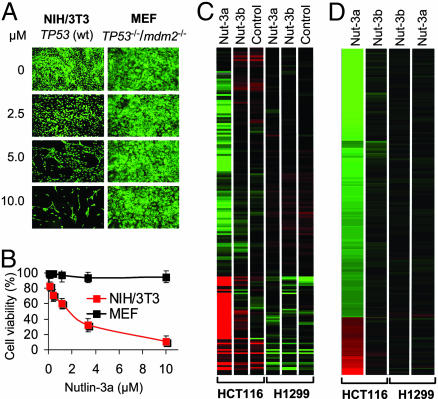
Nutlins are recently identified cis imidazoline inhibitors of the p53–MDM2 interaction (12). They bind at the p53 pocket on the surface of the MDM2 molecule and effectively disrupt p53–MDM2 binding, leading to stabilization of p53 and activation of the p53 pathway. Initial experiments demonstrated that nutlins are active in cells with wild-type p53, but not in cells in which p53 is disabled as a transcription factor (12). These data suggested that nutlins may provide novel molecular tools for studying p53 regulation and its deregulation in cancer. However, the value of nutlins as molecular probes depends on their ability to selectively modulate the p53–MDM2 interaction within living cells. Therefore, we investigated the target specificity of nutlin-3 in the cellular context. First, we tested the activity of nutlin-3a, the active enantiomer of nutlin-3 (12), on exponentially proliferating mouse NIH/3T3 fibroblasts that express wild-type p53 and mouse embryonic fibroblasts (MEFs) derived from TP53−/−/mdm2−/− mice (18). Seven days of incubation with 10 μM nutlin-3a led to >90% inhibition of NIH/3T3 cells’ growth but did not affect the proliferation of MEF in which both targets of the drug were eliminated (Fig. 1 A and B). The inactive enantiomer (nutlin-3b) that binds MDM2 with ≈200-fold lower affinity (12) did not show any effect on the proliferation of either cell line (data not shown). These experiments confirmed that nutlin-3a requires functional p53 and MDM2 for its biological activity.
Fig. 1.
Nutlin-3 selectively activates p53. (A) Exponentially growing mouse NIH/3T3 fibroblasts (wt-TP53) and MEFs from TP53−/−/mdm2−/− knockout mice were incubated for 7 days in the presence of nutlin-3a and stained live by acridine orange. (B) Exponentially growing cells were treated with nutlin-3a for 5 days, and their viability was measured in three independent samples by the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide assay and expressed as percentage of control ± SD. (C) A heat map of probe sets differentially expressed between nutlin-3a and all other treatments (3-fold changed, t test P < 0.05 vs. untreated cells) represented as the median normalized expression of the condition. Red indicates higher than median expression; green indicates lower than median expression. Data were generated on Affymetrix HG-U133 Genechips. (D) A heat map of the same probe sets displayed as a ratio of the treated condition vs. the untreated control.
Second, we compared the global pattern of gene expression in cancer cells with wild-type p53 (HCT116) or p53-null status (H1299) after 24 h of treatment with nutlin-3a or nutlin-3b by using Affymetrix (Santa Clara, CA) HG-U133 GeneChips. This analysis revealed that nutlin-3a up-regulated or down-regulated multiple genes in HCT116 cells but not in p53-null H1299 cells (Fig. 1C). The inactive enantiomer had insignificant effect on gene expression in both lines. To reduce the effect of cell-to-cell variability in gene expression we normalized nutlin induction to a basal level of expression (Fig. 1D). Of ≈40,000 human transcripts represented on the chip, 143 genes (166 probe sets) were found differentially expressed in HCT116 cells treated with nutlin-3a (Fig. 1D and Table 1, which is published as supporting information on the PNAS web site). The list included multiple genes previously shown to be affected by p53 activation (19–21), including 32 transcripts reported as directly regulated by p53. Changes in the expression of many genes may reflect the altered cell-cycle distribution in cells that arrest predominantly in G1 and G2 phases as a result of p53 activation (12). With the exception of a few genes that were marginally down-regulated in HCT116 cells, the inactive enantiomer did not significantly change the gene expression profile of both cell lines. Taken together, the data in Fig. 1 indicate that nutlin-3a is a highly selective MDM2 antagonist and p53 inducer.
Cell-Cycle Arrest Function of Activated p53 Is Preserved in Cancer Cell Lines.
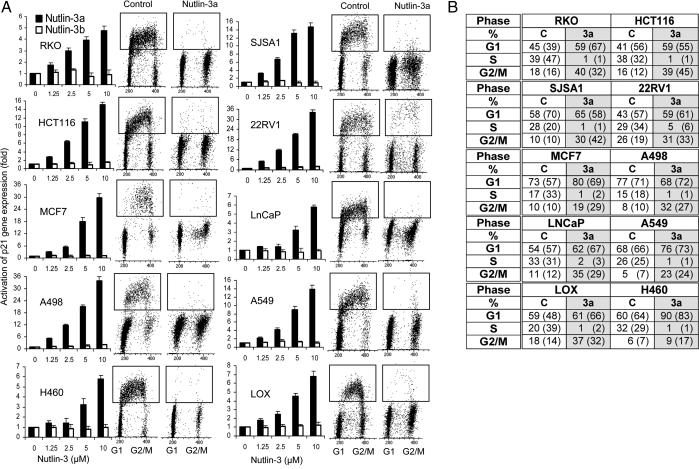
To investigate the functionality of the p53 pathway we randomly selected a panel of 10 cell lines derived from diverse human solid tumors including colorectal (HCT116 and RKO), lung (H460 and A549), breast (MCF7), prostate (LnCaP and 22Rv1), melanoma (LOX), osteosarcoma (SJSA-1), and renal cancer (A498). All cell lines have been reported to express wild-type p53. One of the main functions of the p53 pathway is blocking cell-cycle progression in response to DNA damage or other stresses (1, 2). The cyclin-dependent kinase inhibitor p21Waf1/Cip1, a direct transcriptional target of p53, is a major mediator of p53-dependent cell-cycle arrest (22, 23). To assess the ability of our cell panel to activate p53 in response to MDM2 antagonists we treated exponentially growing cells with nutlin-3a or nutlin-3b for 24 h and measured p21 expression (Fig. 2A). All cell lines showed a dose-dependent induction of p21 expression, confirming their wild-type p53 status and the ability of nutlin-3a to stabilize and activate p53. Nutlin-3b did not show significant induction of p21 expression at the highest dose used (10 μM). Therefore, we chose this concentration for cell-cycle analysis.
Fig. 2.
Nutlin-activated p53 induces G1 and G2 arrest in cancer cell lines with wild-type p53. (A) Human cancer lines in exponential growth were treated with nutlin-3a or nutlin-3b for 24 h, and the expression of the p53 target gene p21 was measured by quantitative real-time PCR and expressed as fold increase ± SD. Cell-cycle profile before and after treatment with 10 μM nutlin-3a or an equivalent amount of solvent for 24 h was analyzed by BrdUrd labeling and flow cytometry. (B) Cell-cycle distribution was calculated from the flow cytograms in A and expressed as the percentage of the total population rounded to the full percentage value. Cell-cycle distribution from an independent experiment is shown in parentheses.
Proliferating cells were treated with nutlin-3a for 24 h, and the cell-cycle distribution was determined by flow cytometry after 2 h of BrdUrd labeling. Nutlin-3a effectively arrested cell-cycle progression in all cell lines, depleting the S-phase compartment to 0.2–2% and increasing the G1- and G2/M-phase compartments, indicating G1 and G2 arrest (Fig. 2A). The slightly higher (5%) S-phase fraction in 22Rv1 cells was likely caused by the relatively longer cycling time of these cells. Overall, the panel of diverse cancer cell lines responded uniformly to p53 activation. These data indicate that the cell-cycle arrest function of the p53 pathway is preserved in all 10 randomly selected cell lines and therefore is likely preserved in the solid tumors from which they were derived.
Apoptotic Activity of p53 Could Be Attenuated by Defective Downstream Signaling.
Apoptosis is another major function of activated p53 that involves the induction of multiple and frequently overlapping pathways. It is believed that most of these pathways are induced at the transcriptional level but a transcription-independent role of p53 in the mitochondria has also been reported (24). Because of its critical function, p53-dependent apoptotic signaling is a likely target for inactivation in cancer. Cell-cycle analysis of the 10-cell panel demonstrated that nutlin-3a at 10 μM can penetrate diverse cell types and uniformly activates p53. Therefore, we chose this concentration to probe the functionality of p53-dependent apoptotic signaling. To this end, we determined the ability of the 10-cell panel to undergo apoptosis after treatment with nutlin-3a.
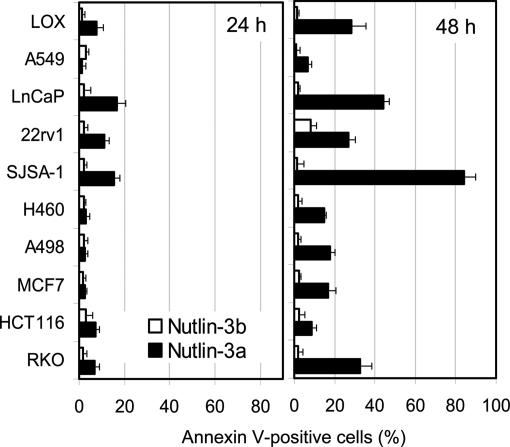
Proliferating cells were incubated with nutlin-3a or nutlin-3b for 24 and 48 h, and the Annexin V-positive fraction was measured by flow cytometry. Annexin V positivity is a relatively early indicator of apoptosis and one of the most accurate means for identifying cells destined to undergo programmed death (25). At 24 h, all cell lines showed a very small fraction of Annexin V-positive cells and only three (LnCaP, 22Rv1, and SJSA-1) >10% (Fig. 3). This finding is typical for solid tumor-derived cell lines in which p53-dependent apoptosis is usually delayed for 24 h (12). At 48 h, a wide range of Annexin V-positive cells was observed from as high as 80% (SJSA-1) to <10% (A549 and HCT116). Nutlin-3b did not induce measurable apoptosis at either 24 or 48 h. Only 22Rv1 cells had an increased fraction (10%), likely caused by the natural subfraction of cells with decreased viability. Although 48 h of drug treatment appeared the most adequate time point for comparison of cells’ ability to undergo apoptosis, these data could not exclude the possibility that the apoptotic response is delayed in some cells. Therefore, the apoptotic fraction was measured in selected cells with low (HCT116, A549, and H460) and high (SJSA-1) apoptotic index after 72 h of exposure to nutlin-3a. The results showed only marginal increase in the Annexin V-positive fraction (HCT116, 8.8 ± 2.1%; A549, 7.6 ± 1.1%; H460, 15.1 ± 1.8%; SJSA-1, 88.3 ± 3.2%) compared with those at 48 h but did not change the overall ranking of the cell lines.
Fig. 3.
Apoptotic response of cancer cells with wild-type p53 to nutlin-3. Cells in exponential phase were treated with 10 μM nutlin-3a or nutlin-3b for 24 or 48 h, and the percentage of Annexin V-positive cells (live plus dead) was determined by the Guava Nexin kit and expressed as a percentage of the total cell number as in ref. 28. Data bars represent the average of two independent experiments.
In principle, differences in the apoptotic response could reflect differences in the drug influx/efflux ratio and/or general impairment of the apoptotic machinery in certain cell lines. Comparable induction of p53 target genes (e.g., p21) and the uniformly effective cell-cycle arrest argue against the first possibility (Fig. 2). To address the general apoptotic competency of the cells, the Annexin V assay was conducted in three cell lines with low (HCT116), medium (RKO), and high (SJSA-1) apoptotic index after treatment with doxorubicin, a DNA-damaging agent that induces both p53-dependent and p53-independent apoptosis (26). Incubation with 250 nM doxorubicin for 48 h led to a dramatic increase of the Annexin V-positive cell fraction in all tested lines (HCT116, 77.7 ± 4.2%; RKO, 81.9 ± 3.7%; SJSA-1, 28.0 ± 2.2%), indicating that low p53-dependent apoptotic level is not caused by defects in the general components of the apoptotic machinery. Therefore, it is reasonable to conclude that nutlin-derived apoptotic index is a reflection of a cell’s ability to undergo p53-dependent apoptosis. These experiments revealed that cancer cell lines can differ significantly in their apoptotic response to similar levels of p53 activation, suggesting that some cell lines might have accumulated defects in p53-dependent apoptotic signaling.
Of the 10 cell lines, the most effective apoptotic response was observed in SJSA-1 osteosarcoma cells. We have shown previously that nutlin-induced apoptosis in this cell line is p53-dependent and involves caspase activation (12). This cell line differs from the rest of the panel by the level of MDM2 expression. Whereas the other nine cell lines have a single copy of the mdm2 gene, the gene is amplified ≈25-fold in SJSA-1, leading to correspondingly high expression of the MDM2 protein. Overproduction of MDM2 has been postulated as one of the principal mechanisms for inactivation of p53. Restoration of p53 activity by MDM2 antagonists should therefore restore its function if there are no other defects in the pathway downstream of p53. The fact that SJSA-1 cells have a highly effective apoptotic response suggests that MDM2 amplification could be the only major abnormality in the p53 pathway of this cell line.
Amplification of the mdm2 Gene May Indicate Intact p53 Signaling.
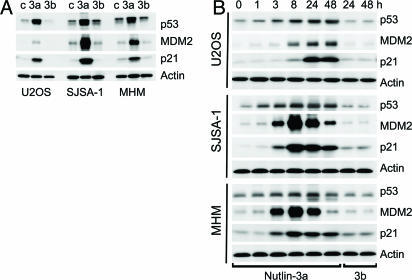
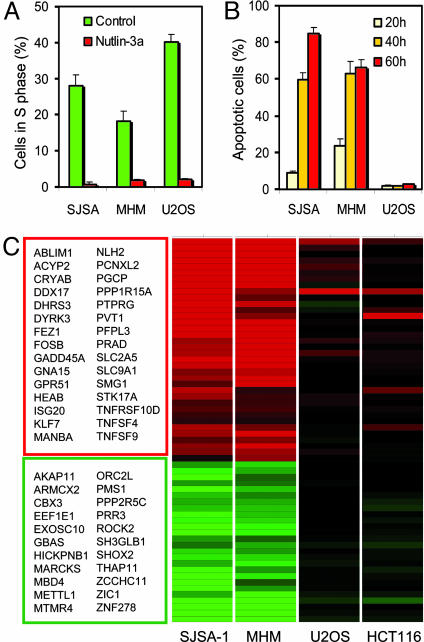
The strong apoptotic response of SJSA-1 cells to nutlin-3a suggested that these cells might be free from defects downstream of p53. To investigate the significance of this observation we tested a second osteosarcoma cell line (MHM) shown to have an ≈10-fold amplification of the mdm2 gene (27). Because this phenomenon could be tumor-specific we also added a third osteosarcoma cell line (U2OS) that has a single copy of the mdm2 gene and expresses normal levels of MDM2 protein. Western blot analysis showed an increased level of p53 that was elevated to a similarly high level in all three cell lines after 24-h exposure to nutlin-3a but not to nutlin-3b (Fig. 4A). The p53 targets MDM2 and p21 were also elevated in an enantiomer-specific manner. The high level of MDM2 expression in the control SJSA-1 and MHM cells correlated with the level of gene amplification. Nutlin-3b did not show activity in any of the cell lines. The uninduced level of p53 differed in the three osteosarcoma lines in correlation with MDM2 expression. SJSA-1 and MHM cells had elevated basal p53 levels compared with U2OS, which is why treatment with nutlin-3a led to a faster reactivation of the p53 pathway in SJSA-1 and MHM2 cells (Fig. 4B). The p53 targets p21 and MDM2 were elevated significantly 3 h after nutlin-3a addition and reached maximal levels at 8 h. Their activation in U2OS cells was delayed until p53 accumulated to comparable levels. After 24 h of treatment, all three cell lines showed comparable elevation of p21, indicating that p53 was maximally activated. Cell-cycle analysis at the 24-h time point revealed equally effective cell-cycle arrest with <2% S-phase cells (Fig. 5A), indicating that the cell-cycle arrest function of p53 is preserved in all three osteosarcoma cell lines.
Fig. 4.
Nutlin-3 stabilizes p53 and induces the p53 pathway in osteosarcoma cells. (A) Exponentially proliferating SJSA-1, MHM, and U2OS cells were treated with 10 μM nutlin-3a, nutlin-3b, or DMSO (c) for 24 h, and p53, p21, and MDM2 proteins in the cell lysates were analyzed by Western blotting. (B) Cells were treated with 10 μM nutlin-3a or nutlin-3b for the indicated times and analyzed by Western blotting. Representative results from two independent experiments are shown.
Fig. 5.
Nutlin-3-induced gene profiles differ between cells with high and low apoptotic index. (A) Nutlin-3a effectively arrests cell-cycle progression in the osteosarcoma cell lines SJSA-1, MHM, and U2OS. Cell-cycle analysis was performed as in Fig. 2. (B) Apoptosis induced by nutlin-3a in osteosarcoma cells. Treatment with nutlin-3a and determination of Annexin V-positive cell fraction was as in Fig. 3. (C) Differentially expressed genes in cells with high and low apoptotic index after 24 h incubation with 10 μM nutlin-3a. Expression profile was determined by Affymetrix gene array analysis (HG-U95 GeneChips) in cells treated with nutlin-3a or nutlin-3b. A heat map of the 62 probe sets found to be differentially expressed in SJSA-1 and MHM cells, but not in U2OS or HCT116, is shown. Differential expression was determined by a 3-fold change compared with untreated controls and an ANOVA P < 0.001 under a simple model including treatment, cell line, and their interaction. Red indicates activated genes; green indicates inhibited genes.
Next, we compared the ability of activated p53 to induce apoptosis in osteosarcoma cells with different MDM2 status. Because of the kinetic differences in p53 induction we extended the treatment period to 60 h. Cells were treated with 10 μM nutlin-3a, and Annexin V-positive cells were counted after 20, 40, or 60 h (Fig. 5B). Nutlin-3a induced apoptosis in ≈60% of SJSA-1 and MHM cells after 40 h, which increased further after 60 h (85% and 65%, respectively). Under the same treatment conditions, U2OS cells had <5% Annexin V-positive fraction compared with the controls, indicating severe impairment of the apoptotic function of p53.
To gain further insight into the resistance to p53-dependent apoptosis we studied p53-induced genes by microarray analysis. The expression profiles of two cell lines with amplified mdm2 gene and strong apoptotic response (SJSA-1 and MHM) were compared with the profiles of two cell lines with normal MDM2 levels and weak apoptotic response (U2OS and HCT116). Cells were treated with 10 μM nutlin-3a or nutlin-3b for 24 h, and the level of gene transcription was assessed by Affymetrix HG-U95a expression array, covering ≈12,000 probe sets. To reduce cell line-specific noise, expression profiles of nutlin-3a-treated cells were normalized to both untreated cells and cells treated with nutlin-3b. This comparison revealed a striking clustering of genes between apoptosis-prone (SJSA1 and MHM) and apoptosis-resistant cells (U2OS and HCT116). A total of 62 probe sets representing 53 individual genes were found differentially expressed (>3-fold) between these two groups, ranging from 3- to 30-fold (Fig. 5C). Of these, 31 genes were induced and 22 suppressed in SJSA-1 and MHM in comparison to U2OS and HCT116. The list included genes reported as direct transcriptional targets of p53 (e.g., FEZ1, GADD45a, KPNB1, NHLH2, PEG3, and SMG1) as well as genes the expression of which is likely indirectly affected by p53 activation. Fourteen genes (CYRAB, DDX17, FEZ1, FOSB, GADD45a, HIC, KPNB1, PEG3, PTPRG, ROCK2, RRAD, SLC9A1, STK17a, and TNFRSF10D) were annotated as apoptosis-related. In addition, other well known apoptotic p53 target genes (e.g., PUMA, NOXA, and BAX) were found to be induced to a higher level in SJSA-1 and MHM cells but not U2OS cells. However, some induction was found in HCT116 cells. These data are consistent with the hypothesis that the proapoptotic function of wild-type p53 in cancer cells can be disabled by defective apoptotic signaling and that mdm2 gene amplification and overexpression indicates an intact downstream p53 signaling.
Nutlin-3 Is a Potent Antitumor Agent in Vivo.
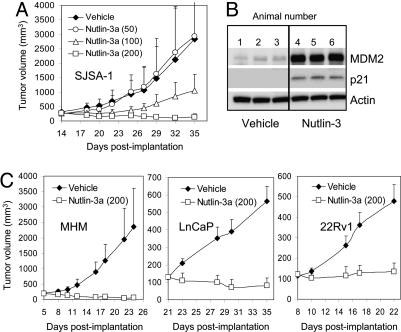
We investigated how MDM2 status could influence the response of established human tumor xenografts in nude mice. We have shown previously that nutlin-3 (racemic) inhibits the growth of SJSA-1 tumors (12). To increase its potency we purified the active enantiomer nutlin-3a and performed a 3-week antitumor efficacy study in SJSA-1 tumor-bearing nude mice (Fig. 6A). Nutlin-3a suppressed xenograft growth in a dose-dependent fashion with the highest dose (200 mg/kg) showing a substantial tumor shrinkage (eight partial and one full regressions). No weight loss or significant pathological changes were recorded during the course of the study (data not shown). To show that the antitumor effect of nutlin-3a is caused by activation of the p53 pathway we analyzed the level of p53 targets p21 and MDM2 in nutlin-treated SJSA-1 xenografts. Western blot analysis revealed accumulation of both proteins in tumors from nutlin-treated animals but not vehicle controls (Fig. 6B). This experiment is consistent with the hypothesis that the in vivo antitumor effect of nutlin-3 is derived from activation of the p53 pathway. To exclude any possibility for an in vivo-related artifact we also tested nutlin-3a on cells with mutant p53. Colon cancer cell line HT29 expresses mutant p53 and does not respond to nutlin-3a treatment in vitro (data not shown). Nude mice bearing established HT29 xenografts were treated orally with 200 mg/kg nutlin-3 for 3 weeks. In agreement with the in vitro results, HT29 xenografts showed undistinguishable growth characteristics in the presence or absence of nutlin-3 (data not shown). These experiments confirmed that nutlin-3 is a selective activator of the p53 pathway in vivo and highly efficacious against SJSA-1 osteosarcoma tumors.
Fig. 6.
In vivo activity of nutlin-3a. (A) Nude mice bearing established SJSA-1 tumor xenografts received nutlin-3a for 3 weeks, and tumor growth was followed periodically. Error bars show SD from the mean volume (P < 0.01 for 200 mg/kg). (B) Nude mice with established SJSA-1 xenografts (three animals per group) received three oral doses of nutlin-3a (150 mg/kg) or vehicle and were killed 3 h after the last dose. Tumors were harvested and analyzed for the presence of the p53 targets MDM2 and p21 by Western blotting. (C) Nude mice with established tumor xenografts (100–200 mm3) were treated orally twice daily with 200 mg/kg nutlin-3a for 2 weeks (LnCaP and 22Rv1) or 3 weeks (MHM), and tumor volumes were recorded periodically. Error bars show SD from the mean volume. P was < 0.01 for all treatments.
Next, we tested the in vivo activity of nutlin-3a in three additional mouse models: MHM osteosarcoma with amplified mdm2 gene and two prostate cancer cell lines (LnCaP and 22Rv1) with normal MDM2 expression and medium p53-dependent apoptosis index (Fig. 3). All tumors were established (100–200 cm3) before dosing was initiated, and mice were treated with 200 mg/kg oral doses twice daily for 2 weeks (LnCaP and 22Rv1) or 3 weeks (MHM). Nutlin-3a was efficacious in all models with average tumor growth inhibition ≥98% (Fig. 6C). Tumor regressions were observed in all tumor models (MHM, 15/15; LnCaP, 9/10; 22Rv1, 4/10). The relative tumor shrinkage calculated for each group as a percent reduction in the median tumor volume was highest in MHM (71.4 ± 10.6%, P < 0.0001), followed by SJSA-1 (48.6 ± 35.3%, P = 0.002), LnCaP (37 ± 33.7%, P = 0.008), and 22Rv1 (−10 ± 35.7%, P = 0.39). Although likely influenced by the differences in the average tumor volume at the start of treatment (MHM, 193 mm3; SJSA-1, 271 mm3; LnCaP, 128 mm3; 22Rv1, 121 mm3) and treatment duration, these data show a reasonable correlation between in vitro and in vivo tumor response. Based on these results one can speculate that tumors with wild-type p53 and mdm2 gene amplification would respond best to therapy with MDM2 antagonists. However, tumors with no MDM2 abnormalities and partially preserved apoptotic signaling may also represent good targets for p53 activation in the clinic.
Discussion
Recently discovered small-molecule inhibitors of the p53–MDM2 interaction, the nutlins, effectively activate the p53 pathway in cancer cells with wild-type p53 and may offer a therapeutic benefit (12). However, their clinical utility would depend on the functionality of the p53 pathway. Nutlin-3 disrupts the physical interaction between p53 and MDM2 in a highly selective manner, leading to stabilization and accumulation of p53. Despite the lack of serine phosphorylation on its N-terminal domain, nutlin-induced p53 appears to be fully capable as a transcription factor and apoptotic inducer (28) and provides a unique molecular tool for studying p53 signaling in the natural cellular context of tumor cell lines without genetic manipulation. In addition, nutlin-3 allows for specifically probing downstream signaling events in the p53 pathway. It has been speculated that cancer cells that retain wild-type p53 must inactivate p53 function by other means, including defective signaling (2, 17). Although several possible ways for p53 inactivation have been described (e.g., overexpression of MDM2, viral oncogenes, and defects in Apaf-1 and p14ARF), very little is known about the functionality of downstream p53 signaling in the majority of tumors with wild-type p53 (2, 17, 29).
We randomly selected 10 tumor cell lines with wild-type p53 representing several solid tumor types and investigated two major p53 functions, cell-cycle arrest and apoptosis, in response to activation by nutlin-3. Cell-cycle distribution in the presence of activated p53 revealed an efficient and relatively uniform arrest of all cell lines in G1 and G2 phases. Although the mechanisms of p53-induced cell-cycle block are not fully understood, it is well established that the pan-cyclin-dependent kinase inhibitor p21 plays a major role (23, 30). p21 is a transcriptional target of p53 that is rarely mutated in cancer, and this circuit appears to be well preserved in all 12 cancer cell lines tested. However, it is possible that defects in the upstream p53 signaling cascade may attenuate the cell-cycle arrest function of p53 in response to different stress factors. Defects in radiation-induced p53 activation have been reported in melanoma (31). Such defects could be compensated for by the nutlins that increase p53 stability.
In contrast to cell-cycle arrest, apoptosis induction by p53 is more complex, involves multiple pathways, and is thus more likely to be a target for inactivation (24, 29, 32). Indeed, the Annexin V assay detected a wide variety of apoptotic responses in the 12 cell lines tested. Cell lines that overexpress MDM2 because of amplification of the mdm2 gene locus (e.g., SJSA-1 and MHM) had the highest apoptotic index most likely caused by intact p53 signaling. Overproduction of MDM2 can effectively disable p53 function. MDM2 amplification is frequently found in sarcomas and correlates with wild-type p53 status of the tumors (33). Because more than one genetic defect rarely occurs on the same pathway, it is likely that p53 signaling in these cells is intact and MDM2 antagonists may fully restore p53 function. In support of this notion, treatment of established SJSA-1 and MHM osteosarcoma xenografts with nutlin-3a caused extensive tumor regression. Therefore, MDM2 amplification status may provide a valuable response indicator in future clinical exploration of MDM2 antagonists.
Several tumor cell lines in our panel (A549, HCT116, H460, MCF7, A498, and U2OS) showed a marginal p53-dependent apoptotic response, indicating that defective apoptotic signaling is not an infrequent event in solid tumors. Gene array analysis of nutlin-induced expression patterns revealed that the expression profile of cells with low apoptotic index differs significantly from that of highly apoptotic cell lines (Fig. 5C), suggesting that some of the p53-dependent signaling components may be defective in these cells. Nearly one-third of 53 genes unaffected in low apoptotic cells (14 genes) have been implicated directly or indirectly in apoptosis, suggesting a causal link between low apoptotic index and defective p53 signaling. Although one could not exclude the possibility that some of these defects have been acquired during in vitro propagation of the cell lines, it is more likely that many of these signaling aberrations have been selected for in the process of tumor development. Nutlins could facilitate the identification of the most prevalent abnormalities in the downstream components of the p53 pathway and might help in the development of diagnostic tools for patient selection.
Prostate cancer cell lines with normal MDM2 expression, LnCaP and 22Rv1, showed decreased in vitro apoptotic index in comparison with MDM2 overexpressing MHM and SJSA-1 osteosarcoma lines. However, despite possible attenuation of the apoptotic signaling in the prostate cells, nutlin-3 treatment caused partial regression of LnCaP and tumor stasis of 22Rv1 xenografts. These results suggest that MDM2 antagonists may be efficacious not only against tumors with MDM2 abnormalities but also against tumors with wild-type p53 that have retained critical downstream circuits of p53-dependent apoptotic signaling. The observations that nutlin-induced p53 is fully activated without the need for genotoxic damage to the cells (28) and that growth suppressive function of the p53 pathway is well preserved in solid tumors (this study) further strengthens the concept that small-molecule MDM2 antagonists may provide an efficacious single-agent therapy for cancers with wild-type p53.
Methods
Cells and Drug Treatment.
RKO cells were a gift from B. Vogelstein (Johns Hopkins Oncology Center, Baltimore). LOX IMVI melanoma cells were provided by the Biological Testing Branch, National Cancer Institute, Frederick, MD. H460a cells used in this study, a clone of H460 cancer cell line, were a gift from J. Roth (M. D. Anderson Cancer Center, Houston). MHM cells, also known as NCS2x, were provided by the Norwegian Radium Hospital. All other cell lines were purchased from the American Type Culture Collection and maintained as described (12). Tested drugs were dissolved in DMSO and kept as 10 mM stock solutions at −20°C. For Western blotting, the cells were grown in 75-cm2 flasks (106 cells per flask) and processed as described (12). For cell-cycle analysis, 1 × 106 cells were treated with nutlins for 24 h, labeled with BrdUrd (20 μM) during the last 2 h, and processed as described (12). Apoptotic index was determined by the Annexin V method using the GuavaNexin kit and the Guava Personal Cell Analyzer (Guava Technologies, Hayward, CA) as described (28). For PCR analysis, cells were seeded in 96-well plates (104 cells per well) 24 h before drug treatment. Cells were lysed, total RNA was extracted by using the ABI 6700 robotic workstation (Applied Biosystems), and the level of p21 transcript was determined by TaqMan (Applied Biosystems) as described (34).
Affymetrix Analysis.
Cells were seeded in 6-well plates (3 × 105 cells per well) 24 h before treatment. There were three replicates per condition, nine samples per cell line. RNA extracted from each replicate (one well) was used for each Affymetrix array. Data analysis was performed by using a simple t test between treated and untreated samples. Differential expression was determined as 3-fold altered in HCT116 treated with nutlin-3a vs. vehicle and P < 0.05, and no change (<1.5-fold change) in the H1299 cells or HCT116 treated with nutlin-3b.
In Vivo Studies.
Nude mice bearing s.c. tumor xenografts (10 mice per group in the SJSA-1, LnCaP, and 22Rv1 study and 15 mice per group in the MHM study) were dosed orally twice daily with nutlin-3a (50–200 mg/kg) or vehicle (1% Klucel, 0.1% Tween 80) for 2 weeks (22Rv1 and LnCap) or 3 weeks (SJSA-1 and MHM). Tumor volume was measured with a caliper and calculated as described (12). For Western blot analysis, nude mice with established SJSA-1 tumors (200–400 mm3, three animals per group) were treated with three doses of nutlin-3a at 150 mg/kg (at 0, 8, and 24 h), and tumors were harvested 3 h after the last dose. Tumor samples were flash-frozen and processed as described (34).
Additional Methods.
For additional details on methods, see Supporting Methods, which is published as supporting information on the PNAS web site.
Supplementary Material
Acknowledgments
We thank J. Chen and L. Loistro for help with the in vivo studies, A. Ryan for help with the ABI 6700 workstation, R. Margolis for the formulation work, G. Lozano (M. D. Anderson Cancer Center) for the MEFs, and M. Simcox for critically reading the manuscript.
Abbreviation
- MEF
mouse embryonic fibroblast.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
See Commentary on page 1659 .
References
- 1.Levine A. J. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 2.Vogelstein B., Lane D., Levine A. J. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 3.Harris S. L., Levine A. J. Oncogene. 2005;24:2899–2908. doi: 10.1038/sj.onc.1208615. [DOI] [PubMed] [Google Scholar]
- 4.Hollstein M., Sidransky D., Vogelstein B., Harris C. C. Science. 1991;253:49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- 5.Hainaut P., Hollstein M. Adv. Cancer Res. 2000;77:81–137. doi: 10.1016/s0065-230x(08)60785-x. [DOI] [PubMed] [Google Scholar]
- 6.Picksley S. M., Lane D. P. BioEssays. 1993;15:689–690. doi: 10.1002/bies.950151008. [DOI] [PubMed] [Google Scholar]
- 7.Lane D. P. Br. J. Cancer. 1999;80(Suppl. 1):1–5. [PubMed] [Google Scholar]
- 8.Freedman D. A., Wu L., Levine A. J. Cell Mol. Life Sci. 1999;55:96–107. doi: 10.1007/s000180050273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chene P. Nat. Rev. Cancer. 2003;3:102–109. doi: 10.1038/nrc991. [DOI] [PubMed] [Google Scholar]
- 10.Honda R., Tanaka H., Yasuda H. FEBS Lett. 1997;420:25–27. doi: 10.1016/s0014-5793(97)01480-4. [DOI] [PubMed] [Google Scholar]
- 11.Momand J., Zambetti G. P., Olson D. C., George D., Levine A. J. Cell. 1992;69:1237–1245. doi: 10.1016/0092-8674(92)90644-r. [DOI] [PubMed] [Google Scholar]
- 12.Vassilev L. T., Vu B. T., Graves B., Carvajal D., Podlaski F., Filipovic Z., Kong N., Kammlott U., Lukacs C., Klein C., et al. Science. 2004;303:844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 13.Momand J., Wu H., Dasgupta G. Gene. 2000;242:15–29. doi: 10.1016/s0378-1119(99)00487-4. [DOI] [PubMed] [Google Scholar]
- 14.Ashkroft M., Vousden K. Oncogene. 1999;18:7637–7643. doi: 10.1038/sj.onc.1203012. [DOI] [PubMed] [Google Scholar]
- 15.Michael D., Oren M. Semin. Cancer Biol. 2003;13:49–58. doi: 10.1016/s1044-579x(02)00099-8. [DOI] [PubMed] [Google Scholar]
- 16.Bond G. L., Hu W., Levine A. J. Curr. Cancer Drug Targets. 2005;5:3–8. doi: 10.2174/1568009053332627. [DOI] [PubMed] [Google Scholar]
- 17.Vousden K. H., Prives C. Cell. 2005;120:7–10. doi: 10.1016/j.cell.2004.12.027. [DOI] [PubMed] [Google Scholar]
- 18.Montes de Oca Luna R., Wagner D. S., Lozano G. Nature. 1995;378:203–206. doi: 10.1038/378203a0. [DOI] [PubMed] [Google Scholar]
- 19.Zhao R., Gish K., Murphy M., Yin Y., Notterman D., Hoffman W. H., Tom E., Mack D. H., Levine A. J. Genes Dev. 2000;14:981–993. [PMC free article] [PubMed] [Google Scholar]
- 20.Kannan K., Amariglio N., Rechavi G., Jakob-Hirsch J., Kela I., Kaminski N., Getz G., Domany E., Givol D. Oncogene. 2001;20:2225–2234. doi: 10.1038/sj.onc.1204319. [DOI] [PubMed] [Google Scholar]
- 21.Kannan K., Kaminski N., Rechavi G., Jakob-Hirsch J., Amariglio N., Givol D. Oncogene. 2001;20:3449–3455. doi: 10.1038/sj.onc.1204446. [DOI] [PubMed] [Google Scholar]
- 22.el-Deiry W. S., Tokino T., Velculescu V. E., Levy D. B., Parsons R., Trent J. M., Lin D., Mercer W. E., Kinzler K. W., Vogelstein B. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 23.Bunz F., Dutriaux A., Lengauer C., Waldman T., Zhou S., Brown J. P., Sedivy J. M., Kinzler K. W., Vogelstein B. Science. 1998;282:1497–1501. doi: 10.1126/science.282.5393.1497. [DOI] [PubMed] [Google Scholar]
- 24.Schuler M., Green D. R. Trends Genet. 2005;21:182–187. doi: 10.1016/j.tig.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 25.Steensma D. P., Timm M., Witzig T. E. Methods Mol. Med. 2003;85:323–332. doi: 10.1385/1-59259-380-1:323. [DOI] [PubMed] [Google Scholar]
- 26.Perego P., Corna E., De Cesare M., Gatti L., Polizzi D., Pratesi G., Supino R., Zunino F. Curr. Med. Chem. 2001;8:31–37. doi: 10.2174/0929867013373994. [DOI] [PubMed] [Google Scholar]
- 27.Berner J.-M., Forus A., El Kahloun A., Meltzer P. S., Fodstad ∅, Myklebost O. Genes Chromosomes Cancer. 1996;17:254–259. doi: 10.1002/(SICI)1098-2264(199612)17:4<254::AID-GCC7>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 28.Thompson T., Tovar C., Yang H., Carvajal D., Vu B. T., Xu Q., Wahl G. M., Heimbrook D. C., Vassilev L. T. J. Biol. Chem. 2004;279:53015–53022. doi: 10.1074/jbc.M410233200. [DOI] [PubMed] [Google Scholar]
- 29.Vousden K. H., Lu X. Nat. Rev. Cancer. 2002;2:594–604. doi: 10.1038/nrc864. [DOI] [PubMed] [Google Scholar]
- 30.Xiong Y., Hannon G. J., Zhang H., Casso D., Kobayashi R., Beach D. Nature. 1993;366:701–704. doi: 10.1038/366701a0. [DOI] [PubMed] [Google Scholar]
- 31.Satyamoorthy K., Chehab N. H., Waterman M. J., Lien M. C., El-Deiry W. S., Herlyn M., Halazonetis T. D. Cell Growth Differ. 2000;11:467–474. [PubMed] [Google Scholar]
- 32.Oren M. Cell Death Differ. 2003;10:431–442. doi: 10.1038/sj.cdd.4401183. [DOI] [PubMed] [Google Scholar]
- 33.Momand J., Jung D., Wilczynski S., Niland J. Nucleic Acids Res. 1998;26:3453–3459. doi: 10.1093/nar/26.15.3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang H., Filipovic Z., Brown D., Breit S. N., Vassilev L. T. Mol. Cancer Ther. 2003;2:1023–1029. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.