Abstract
An approach to understand quantitative traits was recently proposed based on the finding that nonsynonymous (NS) sequence variants in certain genes are preferentially enriched at one extreme of the population distribution. The NS variants, although individually rare, are cumulatively frequent and influence quantitative traits, such as plasma lipoprotein levels. Here, we use the NS variant technique to demonstrate that genetic variation in NPC1L1 contributes to variability in cholesterol absorption and plasma levels of low-density lipoproteins (LDLs). The ratio of plasma campesterol (a plant sterol) to lathosterol (a cholesterol precursor) was used to estimate relative cholesterol absorption in a population-based study. Nonsynonymous sequence variations in NPC1L1 were five times more common in low absorbers (n = 26 of 256) than in high absorbers (n = 5 of 256) (P < 0.001). The rare variants identified in low absorbers were found in 6% of 1,832 African-Americans and were associated with lower plasma levels of LDL cholesterol (LDL-C) (96 ± 36 mg/dl vs. 105 ± 36 mg/dl; P = 0.005). These data, together with prior findings, reveal a genetic architecture for LDL-C levels that does not conform to current models for quantitative traits and indicate that a significant fraction of genetic variance in LDL-C is due to multiple alleles with modest effects that are present at low frequencies in the population.
Keywords: cholesterol absorption, complex trait, genetic architecture, mutation, plant sterol
The plasma level of low-density lipoprotein cholesterol (LDL-C) is clinically important and genetically complex. This trait provides an excellent model system for genetic dissection: LDL-C levels are simple to measure, relatively stable within an individual, and strongly influenced by genetic variation. Moreover, many genes controlling LDL metabolism have been identified, providing candidates for genetic analysis. One factor that contributes to differences in plasma levels of LDL-C is the efficiency of intestinal cholesterol absorption, which ranges from 29% to 80% among healthy individuals (1). Pharmacological blockade of cholesterol absorption leads to reduced plasma LDL-C levels (2), raising the possibility that genetic variation may produce the same result.
Cholesterol absorption is heritable in humans (3), but the genetic basis of this variation is not known. Recently the Niemann–Pick Type C1 Like 1 (NPC1L1) protein was shown to facilitate cholesterol absorption in mice (4, 5), making it an excellent candidate gene for variation in cholesterol absorption. In the present study, we screened the gene encoding NPC1L1 for sequence variations in subsets of individuals estimated to have the highest and lowest rates of sterol absorption. To estimate sterol absorption we measured the ratio of campesterol to lathosterol (Ca:L ratio) in plasma (6). Campesterol, a plant sterol, is absorbed entirely from the diet. Lathosterol is an intermediate in cholesterol biosynthesis, and its concentration in plasma is correlated with rates of endogenous cholesterol synthesis (6, 7). A high Ca:L ratio was shown previously to indicate a high rate of intestinal absorption of cholesterol (6), whereas a low Ca:L ratio indicates a low absorption of cholesterol. Here, we show that rare, nonsynonymous (NS) sequence variants in NPC1L1 are cumulatively common among African-Americans and are associated with reduced Ca:L ratios and a significant reduction in mean plasma levels of LDL-C.
Results
As a first step in this study, we sequenced the coding regions of NPC1L1 in those individuals from the Dallas Heart Study who had the highest and lowest cholesterol absorption, as indicated by the Ca:L ratio. A total of 32 individuals from each of the following groups was sequenced: black men, black women, white men, and white women. Thus, the coding region of the NPC1L1 gene was sequenced in 128 high absorbers and 128 low absorbers. We found 13 NS variants, including a nonsense mutation, that were present only in the low absorbers and 3 NS sequence changes that were unique to the high absorbers (P < 0.01, Fisher’s exact test) (Table 1). The number of synonymous sequence changes unique to the low absorbers (n = 5) was similar to the number observed only in the high absorbers (n = 4). Twelve NS and eight synonymous sequence variants were found in both groups (Table 1).
Table 1.
Single-nucleotide variants in the coding regions of NPC1L1
| Single-nucleotide variants found only in low or high Ca:L |
Single-nucleotide variants found in both low and high Ca:L ratio groups | |||||
|---|---|---|---|---|---|---|
| Low Ca:L ratio | High Ca:L ratio | |||||
| NS | S | NS | S | NS | S | |
| Group 1 (n = 256) | 13 | 5 | 3 | 4 | 12 | 8 |
| Group 2 (n = 256) | 10 | 1 | 2 | 1 | 13 | 8 |
Values represent the numbers of single-nucleotide variants identified in two groups of 256 individuals from the Dallas Heart Study; Group 1 consists of the 32 individuals in the Dallas Heart Study with the lowest and highest Ca:L ratio from each of the following groups (black men, black women, white men, and white women). Group 2 includes the individuals with the next-lowest Ca:L ratio from the same four groups. S, synonymous.
To determine whether this finding was reproducible or an artifact of chance variation in allele frequencies, we sequenced the coding region of NPC1L1 in the individuals in the Dallas Heart Study who had the next highest and the next lowest Ca:L ratios. Thus, the coding region of NPC1L1 was sequenced in an additional 128 high absorbers and 128 low absorbers. Again, we observed an excess of NS variants that were restricted to the low Ca:L group (Table 1, Group 2): A total of 10 NS sequence variations were restricted to the low absorbers in contrast to just two NS variations in the high absorber group (P < 0.025, Fisher’s exact test, Table 2). Three of the NS sequence variations (R306C, I647N, and R693C) were found in both groups of low absorbers (Table 2). Thus, 19 NS sequence variations and one nonsense mutation (26 individuals) were identified only in the low Ca:L ratio group compared with 5 in the high group.
Table 2.
NS sequence variations in NPC1L1 in Dallas Heart Study participants with low or high Ca:L ratios
| Low Ca:L Ratio |
High Ca:L Ratio |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Nucleotide | Amino acid | n | Ethnicity | Frequency | Nucleotide | Amino acid | n | Ethnicity | Frequency |
| Group 1 | |||||||||
| c.182C>T | T61M | 2 | AA | 0.007 | c.521G>A | R174H | 1 | W | 0.0003 |
| c.395A>G | N132S | 1 | AA | 0.004 | c.848G>A | G283D | 1 | AA | ND |
| c.916C>T | R306C | 2 | AA | 0.006 | c.3698T>A | I1233N | 1 | W | 0.001 |
| c.1193A>G | D398G | 1 | AA | 0.0003 | |||||
| c.1249C>T | R417W | 1 | W | 0.001 | |||||
| c.1300G>A | G434R | 1 | AA | 0.0005 | |||||
| c.1496C>T | T499M | 2 | both | 0.0008 | |||||
| c.1859C>G | S620C | 1 | AA | 0.0005 | |||||
| c.1940T>A | I647N | 1 | AA | 0.006 | |||||
| c.2077C>T | R693C | 1 | AA | 0.004 | |||||
| c.2642C>T | S881L | 1 | AA | 0.0005 | |||||
| c.3042G>A | W1014X | 1 | AA | 0.0008 | |||||
| c.3322C>T | R1108W | 1 | AA | 0.0005 | |||||
| Group 2 | |||||||||
| c.328C>T | L110F | 1 | AA | 0.001 | c.731A>G | N244S | 1 | AA | 0.002 |
| c.916C>T | R306C* | 1 | W | 0.006 | c.845C>T | P282L | 1 | W | 0.0003 |
| c.1184C>T | A395V | 1 | AA | 0.002 | |||||
| c.1204G>A | G402S | 1 | AA | 0.0005 | |||||
| c.1238C>T | T413M | 1 | AA | 0.0005 | |||||
| c.1940T>A | I647N* | 1 | AA | 0.006 | |||||
| c.2014G>A | G672R | 1 | AA | 0.0003 | |||||
| c.2077C>T | R693C* | 1 | AA | 0.004 | |||||
| c.3641G>A | R1214H | 1 | W | ND | |||||
| c.3803G>A | R1268H | 1 | AA | 0.0003 | |||||
n is the number of carriers identified in the 128 individuals sequenced in the low absorbers or in the high absorbers in Group 1 or Group 2. Frequency is the allele frequency in 1,831 African-American men and women in the Dallas Heart Study. ND, not determined because of assay failure; AA, African-American; W, white.
*Also found in group 1.
To determine whether the excess of NS sequence variations found in low absorbers was caused by population stratification, we assayed 2,000 unlinked SNPs in the sample using the program structure (8). We found no evidence for differences in genetic ancestry between high and low absorbers (data not shown).
To assess the frequencies and phenotypic effects of the NS sequence variants identified, a 5′ nucleotidase procedure was used to assay the variants in the entire Dallas Heart Study (n = 3,553). NS sequence variations initially identified in only the 256 low absorbers were found in 114 blacks (6.2%), 19 whites (1.8%), and 10 Hispanics (1.7%). None of these sequence variants was sufficiently common (allele frequencies 0.03–0.6%) to allow for meaningful statistical analysis individually, but cumulatively, they were associated with a significant reduction in the Ca:L ratio and in the plasma LDL-C concentrations in African-American men and women (Fig. 1, Table 3). In contrast, the sequence variants were not associated with any significant differences in body mass index, plasma levels of triglyceride, or high-density lipoprotein cholesterol (Table 3). These data indicate that NPC1L1 alleles associated with impaired cholesterol absorption and lower LDL-C levels are individually rare but collectively common in African-Americans.
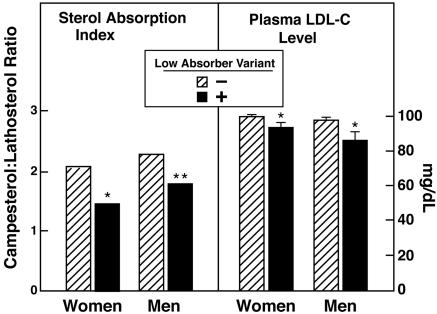
Fig. 1.
Plasma Ca:L ratios and mean plasma levels of LDL-C in African-American men and women in the Dallas Heart Study. Sequence variants found in only the low-absorber group were assayed by using 5′ nucleotidase assays (TaqMan, Applied Biosystems). All individuals in whom the NPC1L1 sequence variants were initially identified by sequencing and all study participants taking lipid-lowering medications were excluded from the analysis. Filled bars represent the median Ca:L ratios and mean (±SEM) levels of LDL-C in women (n = 66) and men (n = 39) who had at least one NS sequence variant in NPC1L1 identified in only the low absorber group. Hatched bars represent individuals who did not have one of these alleles (926 women and 674 men). ∗, P < 0.05; ∗∗, P < 0.01.
Table 3.
Plasma lipid profiles of African-American men and women with rare NS sequence variants in NPC1L1
| Men |
Women |
Both |
||||
|---|---|---|---|---|---|---|
| Normal | NPC1L1+/− | Normal | NPC1L1+/− | Normal | NPC1L1+/− | |
| n | 674 | 39 | 926 | 66 | 1,600 | 105 |
| Age, yr | 44 ± 10 | 45 ± 9 | 44 ± 10 | 44 ± 10 | 44 ± 10 | 45 ± 10 |
| BMI, kg/m2 | 28.1 ± 6.0 | 28.6 ± 7.3 | 31.8 ± 7.9 | 32.6 ± 9.9 | 30.2 ± 7.3 | 31.1 ± 9.2 |
| Total cholesterol, mg/dL | 177 ± 39 | 170 ± 40 | 178 ± 40 | 172 ± 42 | 178 ± 40 | 171 ± 41 |
| Triglyceride, mg/dL | 117 ± 111 | 132 ± 110 | 95 ± 71 | 106 ± 111 | 104 ± 91 | 116 ± 111 |
| HDL-C, mg/dL | 50 ± 16 | 53 ± 20 | 54 ± 14 | 54 ± 14 | 52 ± 15 | 54 ± 17 |
| LDL-C, mg/dL | 105 ± 37 | 92 ± 40∗ | 105 ± 36 | 98 ± 33∗ | 105 ± 37 | 96 ± 36∗∗ |
| Ca:L ratio | 2.3 (2.5) | 1.8∗ (1.2) | 2.1 (2.3) | 1.5∗∗ (1.2) | 2.2 (2.4) | 1.7∗∗ (1.2) |
Values are means ± SD, except for Ca:L ratios, which are shown as medians with interquartile ranges in parentheses. ∗, P < 0.05; ∗∗, P < 0.01 by one-sided Wilcoxon’s two-sample test. NPC1L1+/−, heterozygous for NS sequence variation in NPC1L1; BMI, body mass index.
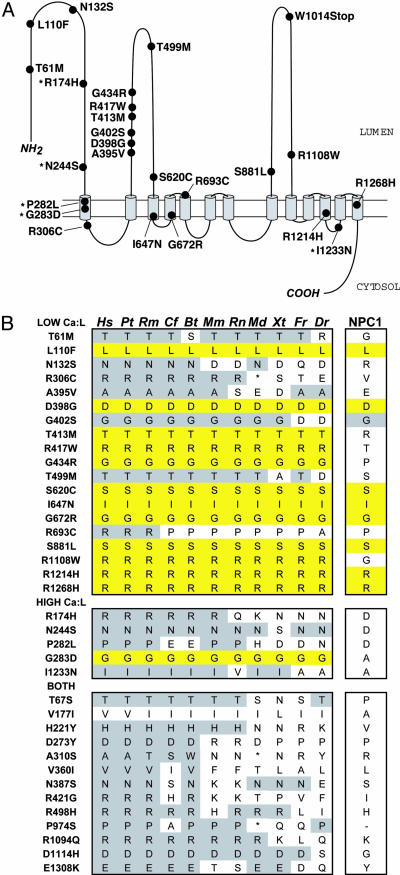
The NS sequence variations identified in only the low absorbers or in only the high absorbers were distributed throughout the protein, but a cluster of sequence variants associated with a low Ca:L ratio (6 of the 19 missense variants) was located within a 39-aa stretch in the first extracellular loop (Fig. 2A), suggesting that this region may be of particular functional importance. We also examined the evolutionary conservation of the NS sequence variations identified in this study by comparing the human sequence to that of NPC1L1 from 10 other species and the closely related human protein NPC1 (Fig. 2B). Sequence variants found in only the low absorbers tended to be more highly conserved than those found in only high absorbers or those identified in both groups. Twelve of the 19 NS variants identified among only low absorbers were completely conserved from human to zebrafish, whereas only 1 of the 5 variants found in only the high absorbers was similarly conserved (Fig. 2B). Of the 12 amino acids that were conserved in all 11 species, 8 were also conserved in NPC1.
Fig. 2.
NS sequence variants in NPC1L1. (A) The membrane topology of NPC1L1 was predicted by using the program phd (31) and the model published for NPC1 (32). The positions of sequence variants found exclusively in low absorbers or in high absorbers (indicated with an asterisk) are provided. (B) Sequence conservation of NS sequence variants in NPC1L1 found in only low absorbers (Top), high absorbers (Middle), or both high and low absorbers (Bottom). The alignment includes human (hs), chimpanzee (pt), rhesus monkey (rm), dog (cf), cow (bt), mouse (mm), rat (rt), opossum (md), frog (xt), pufferfish (fr), and zebrafish (dr). The GenBank accession no. for the human sequence in this alignment is AAS56939.
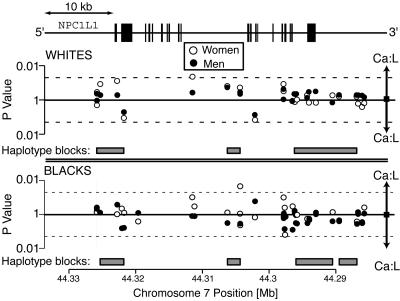
To determine whether common variants in NPC1L1 are associated with Ca:L ratios and plasma levels of LDL-C, we assayed 30 common SNPs at the NPC1L1 locus (minor allele frequency ≥1%) in the Dallas Heart Study. The four major race–sex groups in the study (black men, black women, white men, and white women) were analyzed separately. Two SNPs were associated with the plasma Ca:L ratio at a nominal significance threshold of 0.05 in one of the four groups, but neither was replicated in any of the other groups (Fig. 3). We also analyzed men and women together in the two ethnic groups. None of the SNPs was associated with a significant difference in Ca:L ratio in both blacks and whites. Next, we examined whether haplotype blocks were associated with the Ca:L ratio. Haplotypes were constructed by using an accelerated expectation maximization algorithm (9), and global P values for association with log transformed Ca:L ratios (10) were calculated (Fig. 3). None of the blocks was consistently associated with the Ca:L ratios (data not shown). These analyses were repeated by using SNPs with minor allele frequencies >0.05 and >0.1, and essentially identical results were obtained. These data did not reveal a significant role for common NPC1L1 SNPs in determining cholesterol absorption or plasma LDL-C levels, consistent with the findings of Hegele et al. (11) and Simon et al. (12), who found no relationship between common sequence variants in NPC1L1 and baseline plasma levels of LDL-C. We cannot exclude the possibility that common NPC1L1 SNPs not examined in this study affect cholesterol absorption.
Fig. 3.
Associations between common sequence variants in NPC1L1 and plasma Ca:L ratios. A total of 30 common sequence variants in NPC1L1 (minor allele frequency ≥0.01) were genotyped in the Dallas Heart Study population by using chip-based oligonucleotide hybridization and 5′ nucleotidase assays and tested for association with plasma Ca:L ratios by using ANOVA. Ca:L ratios were log transformed before analysis, and separate analyses were performed in white men (Upper , filled circles), white women (Upper, filled circles), black men (Lower, filled circles), and black women (Lower, open circles). SNPs associated with increased Ca:L ratios are shown above the solid lines, and SNPs associated with lower Ca:L rations are shown below the solid lines. Dotted lines represent P values of 0.05. Haplotype blocks (gray blocks) were constructed in whites (Upper) and blacks (Lower) by using the expectation-maximization algorithm as implemented in haploview (12) and tested for association with Ca:L ratios by using haplo.stats (17). Schematic of NPC1L1 is shown to scale, with boxes indicating exons and lines introns and flanking noncoding sequences. The identification numbers, minor allele frequencies, and the Hardy–Weinberg equilibrium P values are provided in Table 4.
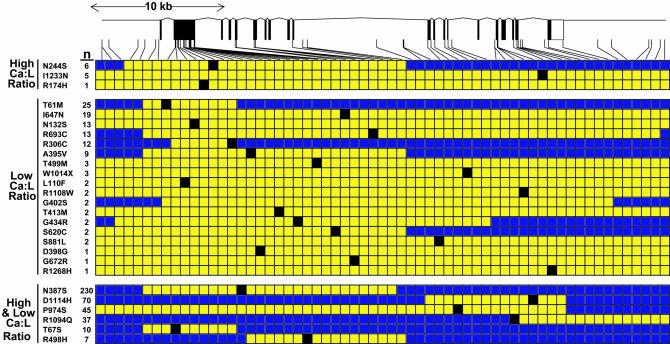
To determine whether each of the NS sequence variants in NPC1L1 arose from a single founder mutation or from recurrent mutations, we constructed haplotypes using the common SNPs at the NPC1L1 locus. For 15 of the NS variants, haplotypes were successfully constructed in at least two unrelated individuals. Each of these NS sequence variants was flanked by regions of haplotype identity that were shared by all carriers, consistent with a single founder for each mutation (Fig. 4). The majority of the sequence variants found in only the low absorber group had haplotypes that were larger than the genomic region analyzed (40 kb), but for six of these variants, the region of haplotype identity was <40 kb. This finding indicates that some of these alleles are ancient.
Fig. 4.
Haplotype-sharing among NPC1L1 alleles. Haplotypes for the alleles bearing the NS variants were determined in the African-Americans in the Dallas Heart Study by using the partition-ligation method described in ref. 30 and then manually curated to maximize the extent of haplotype-sharing among carriers. Yellow blocks represent the largest haplotype that could be shared among carriers. Numbers next to the amino acid substitutions indicate the number of individuals in each group. High and low Ca:L ratios denote sequence variants found in both the high and the low Ca:L groups. The identification numbers, minor allele frequencies, and the Hardy–Weinberg equilibrium P values are provided as Table 4.
Discussion
The major finding of this study is that multiple rare sequence variants in NPC1L1 are collectively associated with variations in sterol absorption and plasma levels of LDL-C. NS variants were significantly more common among low absorbers than among high absorbers, and the excess could not be explained by population structure or by chance fluctuations in allele frequency. The excess of NS sequence variants was largely confined to African-Americans: 6% of the men and women in this ethnic group were heterozygous for NPC1L1 sequence variants associated with reduced cholesterol absorption and LDL-C levels. These data indicate that sequence variations in NPC1L1 contribute to variation in sterol absorption and LDL-C levels in the population.
DNA sequencing of low absorbers revealed 20 NS variants with allele frequencies <0.01. Of these, 18 were detected in the African-Americans. The probability of detecting an allele in a sample of 64 individuals (128 alleles) is 72% for alleles with frequencies of 0.01 (1%) and 47% for alleles with a frequencies of 0.005 (0.5%). Therefore, it is likely that other rare alleles associated with low cholesterol absorption are present in African-Americans. The significant excess of rare NPC1L1 alleles with NS sequence variants among individuals with low cholesterol absorption is consistent with the notion that these mutations decrease NPC1L1 function. However, the absence of a specific assay for NPC1L1 function precludes direct functional analysis of the sequence variants identified. Thus, we cannot directly determine which of these alleles impair NPC1L1 function and which were present in low absorbers simply by chance. Alignment of the NPC1L1 amino acid sequence from human, mouse, frog, and pufferfish indicated that the NS sequence variants found only in the low absorbers tended to involve more highly conserved residues than those found in only high absorbers or in both groups (Fig. 2). Because replacement of conserved residues is more likely to alter protein function (13), this finding supports the notion that the mutations found in low absorbers are more likely to reduce NPC1L1 function.
Cholesterol absorption and plasma LDL-C levels are complex traits that are strongly influenced by both genetic and environmental factors. Two general models of complex diseases have been proposed. The common disease–common variant model holds that genetic susceptibility to common diseases is conferred primarily by alleles that are common in the population and have modest phenotypic effects (14, 15). This model is supported by a small number of well validated examples (16). An alternative model is that susceptibility to common diseases is the result of multiple rare alleles with large phenotypic effects (common disease–rare variant model). Although individually rare, these alleles may be collectively common in the population. Few direct tests of this model have been reported, but we (17, 18) and others (19) have shown that rare alleles with large effects on plasma levels of lipoproteins are collectively common in the population. The results of the present study provide further evidence that multiple rare variants contribute to variations in plasma lipoprotein levels in the population, but, in this case, the effect on phenotype is more modest. The average reduction in plasma LDL-C levels associated with the variant NPC1L1 alleles was ≈10%, which resembles the magnitude of the effect of common genetic variants in APOE (20) but is less than the 40% reductions in LDL-C we observed in heterozygotes with null mutations in PCSK9 (18). Taken together, these data suggest that the genetic architecture of plasma LDL-C levels is not adequately described by the common disease–common variant model or by the common disease–rare variant model. Rather, heritable variation in the population is conferred by sequence variation in at least three loci, with a small number of common APOE alleles and multiple rare alleles in PCSK9 and NPC1L1 contributing to the phenotype. This architecture is congruent with the evolutionary model of complex disease proposed by Pritchard (21), in which the major fraction of genetic variance is attributable to loci where high overall mutation rates generate a large number of mildly deleterious alleles that only occasionally achieve high frequencies in the population.
The predictions of the Pritchard model have important implications for the selection of strategies to identify sequence variants underlying complex traits. Both association and linkage-disequilibrium mapping studies are predicated on the assumption that each locus has few functional alleles. Alleles with modest functional effects are difficult to detect by linkage, and the presence of multiple low-frequency alleles at a locus will greatly reduce the power of conventional statistical tests for association. Under these conditions, direct sequencing of targeted populations may be the optimal way to identify alleles associated with complex phenotypes. The strategy of comparing NS sequence variations in individuals at the extremes of the population distribution to identify genes (and sequence variations) contributing to variation in plasma lipoprotein levels can be readily applied to other phenotypes (17).
The excess of rare NS alleles among low absorbers in this study was most striking in African-Americans. The reason for this population-specific difference is not known. One possibility is that sequence variants that decrease NPC1L1 function confer a selective advantage under some circumstances, although it seems unlikely that selection would result in the accumulation of multiple rare alleles rather than a small number of more common alleles. Alternatively, the spectrum of NPC1L1 alleles may reflect the allelic spectrum of the population (22). Africans are far more genetically diverse than are other populations, particularly for rare variants, but most of these variants are presumed to be functionally silent, because deleterious alleles tend to be removed from the population by purifying selection (23). Functional alleles may persist if they are only mildly deleterious; therefore, African populations may be enriched for alleles that affect gene function but do not confer strong selective disadvantage.
Materials and Methods
Study Subjects.
Study subjects were obtained from the Dallas Heart Study, a population-based probability sample of Dallas County residents that includes 1,043 whites, 1,832 African-Americans, and 601 Hispanics (24). Individuals who identified themselves as black (African-American) or white (European-Caucasian) were stratified by sex and by Ca:L ratio. DNA samples from 256 individuals with the highest Ca:L ratios (high absorbers), and 256 individuals with the lowest ratios (low absorbers) were selected for sequencing. High absorbers and low absorbers included 64 men and 64 women from each ethnic group.
DNA Sequencing.
The exons and flanking intron sequences of NPC1L1 were amplified by PCR. PCR products were treated with recombinant exonuclease I and shrimp alkaline phosphatase (Exo-Sap; United States Biochemicals), and both strands of each product were sequenced on an ABI3730 automated sequencer using BigDye terminator cycle sequencing reagents (Applied Biosystems). The sequences of the oligonucleotides used for sequencing are available upon request from the corresponding author.
Measurement of Plasma Lipids and Lipoproteins.
Plasma and lipoprotein cholesterol and triglyceride concentrations were determined colorimetrically by using commercial enzymatic reagents (25). Plasma lathosterol and campesterol levels were determined by gas chromatography as described in ref. 26, except that sterol levels were detected by using flame ionization rather than mass spectrometry.
Assay of Sequence Variations.
Specific 5′ nucleotidase assays for the NS sequence variants in NPC1L1 were developed by using the TaqMan system (Applied Biosystems). The assays were performed on a HT7900 Real-Time PCR system by using probes and reagents purchased from Applied Biosystems.
Testing for Population Substructure.
A panel of 2,000 SNPs that are in linkage equilibrium was used to test for differences in population ancestry in the high absorbers and the low absorbers by using the program structure (27). African-Americans and whites were analyzed separately. The Dirichlet parameter λ was set at 1, and structure was run with the number of populations (K) set at 2, 3, 4, and 5.
Statistical Analysis.
The prevalence of NS variants in the high absorbers and low absorbers was compared by using Fisher’s exact test. For rare variants, values for the Ca:L ratio and plasma levels of LDL-C were compared in carriers and noncarriers by using the Wilcoxon rank-sum test with age and gender as covariates. For common variants in which all three genotype groups were represented, the values for the Ca:L ratio and the plasma levels of LDL-C were compared by using the Kruskal–Wallis test. LDL-C levels and log-transformed Ca:L ratios were also compared across genotypes by using one-way ANOVA with age and gender as covariates. Only the 1,722 subjects who were not on lipid-lowering therapy were included in the analysis.
Haplotype Analysis of NPC1L1.
Haplotype blocks were constructed by using 30 SNPs (see Table 4, which is published as supporting information on the PNAS web site) genotyped by oligonucleotide array hybridization (Perlegen Sciences, Mountain View, CA) and 5′ nucleotidase assays. All SNPs available from dbSNP and the Perlegen Sciences database as of October, 2004 were assayed, and those that had a minor allele frequency ≥1% and were in Hardy–Weinberg equilibrium were included in the analysis. We estimated haplotype blocks in each ethnic group by using the expectation-maximization algorithm as implemented in haploview (28) and the Gabriel et al. (29) definition for haplotype blocks and calculated the association of each haplotype block with the log-transformed Ca:L ratios by using the haplo.stats package. The results are presented as P values based on the global statistic for the association of haplotype blocks with Ca:L ratios.
The haplotypes of the rare alleles were estimated by using the partition-ligation–expectation-maximization (PL–EM) algorithm (30). The single most probable haplotype pair for each individual was calculated by using atomistic segments of 5–8 loci, keeping 500 of the most frequent partial haplotypes to the next ligation step and performing 10 independent runs in each of the EM steps. For each NS substitution, we determined the extent of the haplotype shared among all carriers.
Supplementary Material
Acknowledgments
We thank the Dallas Heart Study Investigators Barbara Gilbert, Holly Brookman, and Teresa Eversole for collecting blood and processing samples; Tommy Hyatt, Sijing Niu, and Biman Pramanik for excellent technical assistance; Cheng Hui Lee for assistance with structure; Michael Brown and Joseph Goldstein for helpful discussions; and Andrew Sparks and David Cox (Perlegen Sciences, Mountain View, CA) for helpful discussions. This work was supported by grants from the Donald W. Reynolds Foundation, The LeDucq Foundation, The W. M. Keck Foundation, The Perot Family Fund, a Veterans Affairs Merit Research Grant, and the National Institutes of Health.
Abbreviations
- Ca:L
campesterol-to-lathosterol ratio
- LDL
low-density lipoprotein
- LDL-C
LDL cholesterol
- NS
nonsynonymous
- NPC1L1
Niemann–Pick Type C1 Like 1
- PCSK9
proprotein convertase subtilisin/kexin type 9 precursor.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Freely available online through the PNAS open access option.
References
- 1.Bosner M. S., Lange L. G., Stenson W. F., Ostlund R. E., Jr J. Lipid Res. 1999;40:302–308. [PubMed] [Google Scholar]
- 2.Ezzet F., Wexler D., Statkevich P., Kosoglou T., Patrick J., Lipka L., Mellars L., Veltri E., Batra V. J. Clin. Pharmacol. 2001;41:943–949. doi: 10.1177/00912700122010915. [DOI] [PubMed] [Google Scholar]
- 3.Gylling H., Miettinen T. A. J. Lipid Res. 2002;43:1472–1476. doi: 10.1194/jlr.m200155-jlr200. [DOI] [PubMed] [Google Scholar]
- 4.Altmann S. W., Davis H. R., Jr, Zhu L. J., Yao X., Hoos L. M., Tetzloff G., Iyer S. P., Maguire M., Golovko A., Zeng M., et al. Science. 2004;303:1201–1204. doi: 10.1126/science.1093131. [DOI] [PubMed] [Google Scholar]
- 5.Davis H. R., Jr, Zhu L. J., Hoos L. M., Tetzloff G., Maguire M., Liu J., Yao X., Iyer S. P., Lam M. H., Lund E. G., et al. J. Biol. Chem. 2004;279:33586–33592. doi: 10.1074/jbc.M405817200. [DOI] [PubMed] [Google Scholar]
- 6.Miettinen T. A., Tilvis R. S., Kesaniemi Y. A. Am. J. Epidemiol. 1990;131:20–31. doi: 10.1093/oxfordjournals.aje.a115479. [DOI] [PubMed] [Google Scholar]
- 7.Bjorkhem I., Miettinen T., Reihner E., Ewerth S., Angelin B., Einarsson K. J. Lipid Res. 1987;28:1137–1143. [PubMed] [Google Scholar]
- 8.Pritchard J. K., Stephens M., Donnelly P. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Niu T., Qin Z. S., Xu X., Liu J. S. Am. J. Hum. Genet. 2002;70:157–169. doi: 10.1086/338446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schaid D. J., Rowland C. M., Tines D. E., Jacobson R. M., Poland G. A. Am. J. Hum. Genet. 2002;70:425–434. doi: 10.1086/338688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hegele R. A., Guy J., Ban M. R., Wang J. Lipids Health Dis. 2005;4:16. doi: 10.1186/1476-511X-4-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Simon J. S., Karnoub M. C., Devlin D. J., Arreaza M. G., Qiu P., Monks S. A., Severino M. E., Deutsch P., Palmisano J., Sachs A. B., et al. Genomics. 2005;86:648–656. doi: 10.1016/j.ygeno.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 13.Vitkup D., Sander C., Church G. M. Genome Biol. 2003;4:R72. doi: 10.1186/gb-2003-4-11-r72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lander E. S. Science. 1996;274:536–539. doi: 10.1126/science.274.5287.536. [DOI] [PubMed] [Google Scholar]
- 15.Chakravarti A. Nat. Genet. 1999;21:56–60. doi: 10.1038/4482. [DOI] [PubMed] [Google Scholar]
- 16.Lohmueller K. E., Pearce C. L., Pike M., Lander E. S., Hirschhorn J. N. Nature Genet. 2003;33:177–182. doi: 10.1038/ng1071. [DOI] [PubMed] [Google Scholar]
- 17.Cohen J. C., Kiss R. S., Pertsemlidis A., Marcel Y. L., McPherson R., Hobbs H. H. Science. 2004;305:869–872. doi: 10.1126/science.1099870. [DOI] [PubMed] [Google Scholar]
- 18.Cohen J., Pertsemlidis A., Kotowski I. K., Graham R., Garcia C. K., Hobbs H. H. Nat. Genet. 2005;37:161–165. doi: 10.1038/ng1509. [DOI] [PubMed] [Google Scholar]
- 19.Frikke-Schmidt R., Nordestgaard B. G., Jensen G. B., Tybjaerg-Hansen A. J. Clin. Invest. 2004;114:1343–1353. doi: 10.1172/JCI20361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boerwinkle E., Visvikis S., Welsh D., Steinmetz J., Hanash S. M., Sing C. F. Am. J. Med. Genet. 1987;27:567–582. doi: 10.1002/ajmg.1320270310. [DOI] [PubMed] [Google Scholar]
- 21.Pritchard J. K. Am. J. Hum. Genet. 2001;69:124–137. doi: 10.1086/321272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang W. Y., Pike N. Med. Hypotheses. 2004;63:748–751. doi: 10.1016/j.mehy.2003.12.057. [DOI] [PubMed] [Google Scholar]
- 23.Bamshad M., Wooding S., Salisbury B. A., Stephens J. C. Nat. Rev. Genet. 2004;5:598–609. doi: 10.1038/nrg1401. [DOI] [PubMed] [Google Scholar]
- 24.Victor R. G., Haley R. W., Willett D. L., Peshock R. M., Vaeth P. C., Leonard D., Basit M., Cooper R. S., Iannacchione V. G., Visscher W. A., et al. Am. J. Cardiol. 2004;93:1473–1480. doi: 10.1016/j.amjcard.2004.02.058. [DOI] [PubMed] [Google Scholar]
- 25.Mostaza J. M., Schulz I., Vega G. L., Grundy S. M. Am. J. Cardiol. 1997;79:1298–1301. doi: 10.1016/s0002-9149(97)00109-4. [DOI] [PubMed] [Google Scholar]
- 26.Wilund K. R., Yu L., Xu F., Hobbs H. H., Cohen J. C. J. Lipid Res. 2004;45:1429–1436. doi: 10.1194/jlr.M400167-JLR200. [DOI] [PubMed] [Google Scholar]
- 27.Rosenberg N. A., Pritchard J. K., Weber J. L., Cann H. M., Kidd K. K., Zhivotovsky L. A., Feldman M. W. Science. 2002;298:2381–2385. doi: 10.1126/science.1078311. [DOI] [PubMed] [Google Scholar]
- 28.Barrett J. C., Fry B., Maller J., Daly M. J. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 29.Gabriel S. B., Schaffner S. F., Nguyen H., Moore J. M., Roy J., Blumenstiel B., Higgins J., DeFelice M., Lochner A., Faggart M., et al. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 30.Qin Z. S., Niu T., Liu J. S. Am. J. Hum. Genet. 2002;71:1242–1247. doi: 10.1086/344207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rost B. Methods Enzymol. 1996;266:525–539. doi: 10.1016/s0076-6879(96)66033-9. [DOI] [PubMed] [Google Scholar]
- 32.Davies J. P., Ioannou Y. A. J. Biol. Chem. 2000;275:24367–24374. doi: 10.1074/jbc.M002184200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.