Abstract
TGF-β has been postulated to play an important role in the development of pancreatic cancers. More than 50% of human pancreatic cancers bear mutations of Sma- and Mad-related protein (Smad) 4, a critical protein required for TGF-β signaling. To evaluate the in vivo function of TGF-β in the development of pancreatic cancers, we generated a transgenic mouse model with pancreas-specific expression of Smad7, a specific inhibitor of TGF-β signaling. Through the use of elastase I promoter, we directed the tissue specific expression of exogenous Smad7. Consistently, the exogenous Smad7 was detected only in the pancreas in the transgenic mice, and, furthermore, phosphorylation of Smad2 was blocked in the pancreatic tissues. At 6 months of age, most transgenic animals developed premalignant ductal lesions in the pancreas, with characteristics of pancreatic intraepithelial neoplasia (PanIN), a precursor to invasive pancreatic cancers. The premalignant lesions of the pancreas were accompanied by accelerated proliferation of the ductal epithelium and acinar cells, as well as increased fibrosis around the ductal lesions. This study not only demonstrated that in vivo inactivation of TGF-β signaling is implicated in the development of early stage of pancreatic cancers, but also provided a promising animal model useful for the investigation and intervention of pancreatic cancers in humans.
Keywords: mouse model, pancreatic cancer, pancreatic intraepithelial neoplasia, fibrosis, Sma- and Mad-related protein 2
Pancreatic cancer represents the fifth leading cause of cancer death in the United States, with a medium survival of 4–6 months. According to a latest estimate, there were 31, 860 new cases of pancreatic cancer and 31, 270 deaths from this disease in 2004 (1). Pancreatic cancer has the worst prognosis of all gastrointestinal cancers, with 5-yr survival rates <5% (2), largely due to a lack of specificity in clinical presentations in the early stage of cancer development and the resistance to conventional cancer therapy. Therefore, it is of paramount importance to develop model systems with early lesions of pancreatic cancers to facilitate the diagnosis and therapy for this deadly disease.
Various genetic changes have contributed to the development of pancreatic cancers. Activation of the Kras proto-oncogene has been found in >90% of pancreatic cancers (3–5). On the other hand, inactivation of various tumor-suppressor genes such as p16INK4a and p53 has been identified in most invasive pancreatic cancers (6). In addition, >50% of pancreatic cancers bear homozygous deletions or inactivating mutations of Sma- and Mad-related protein (Smad) 4 (7). Because Smad4 plays a central role in TGF-β signaling, it has been postulated that disruption of TGF-β signaling plays a critical role in the tumorigenesis of pancreatic cancers (6).
TGF-β superfamily members control cellular growth, differentiation, and apoptosis, as well as embryonic development (8, 9). TGF-β family members regulate gene expression via serine/theronine kinase receptors at the cell surface (10) and a group of intracellular transducers called Smad proteins (11). According to their functional and structural features, Smads are classified as receptor-specific Smads (R-Smads), a common-Smad (Co-Smad or Smad4), and inhibitory Smads (I-Smads) (11–13). TGF-β signaling starts by binding of the ligand with the type II receptor, followed by recruitment of the type I receptor. The activated type I receptor phosphorylates the R-Smads, including Smad2 and Smad3, which then form a heteromeric complex with the Co-Smad, Smad4. The R-Smad/Smad4 complex is translocated into the nucleus where it regulates the transcription of target genes. Smad7 is a member of the I-Smads that is able to antagonize TGF-β signaling by direct interaction with the type I receptor (14).
To provide in vivo evidence that TGF-β signaling is implicated in the development of pancreatic cancer, we generated and analyzed a transgenic mouse model with specific disruption of TGF-β function in the pancreas by Smad7 overexpression. We report here that pancreatic expression of Smad7 is able to induce to development of pancreatic intraepithelial neoplasia (PanIN) lesions in the pancreas.
Results
Generation of Transgenic Mice.
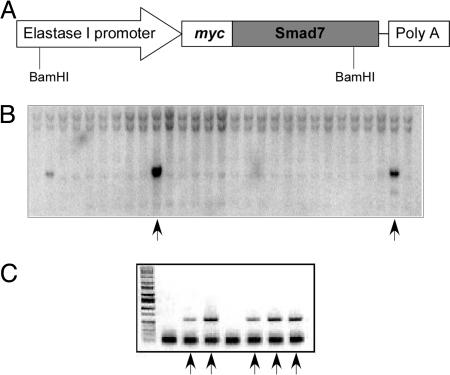
We sought to disrupt the in vivo TGF-β signaling by exogenous expression of Smad7 specifically in the pancreas. Smad7 is an inhibitory Smad protein that is able to bind TGF-β type I receptor and turn off TGF-β signaling in the cells (14). To achieve this goal, we generated a transgenic construct that contains a −205/+8-bp rat elastase I gene promoter/enhancer (15, 16), a myc-tagged Smad7 cDNA, and an SV40 poly(A) tail at the C terminus (Fig. 1A). In the transgenic construct, a myc-tag was constructed at the N terminus of Smad7 transcript to facilitate detection of the transgene expression in the animals. Our previous study showed that this myc-tag did not affect the inhibitory activity of Smad7 on TGF-β and activin signaling (17).
Fig. 1.
Generation of transgenic mouse. (A) A diagram depicting the transgenic construct. A rat elastase I promoter/enhancer (−205/+8bp) was placed upstream of a rat Smad7 cDNA gene. A myc tag was placed at the N-terminal end of the Smad7 transcript. (B) Genotyping of transgenic mice by Southern blotting analysis. Genomic DNA isolated from mouse tails was used in hybridization with a transgene-specific probe in Southern blotting. A representative result is shown here with two positive samples (arrows). (C) Genotyping by PCR. The genomic DNA was used in PCR with primers specific for the transgene. The positive samples are marked with arrows.
The plasmid construct was linearized and used in microinjection to generate the transgenic mice that were identified by both Southern blot analysis and PCR genotyping (Fig. 1 B and C). From a total of 78 offspring in the C3H background given by the pseudopregnant foster mothers, 5 founders were identified to carry the transgenes. The mice positive for the transgene were crossed with mice of DBA2 strain to generate transgenic mice used in this study. All mice positive for the transgene showed no signs of health problems up to 10 months of age as compared with the WT littermates.
Smad7 Transgene Was Expressed only in the Pancreas.
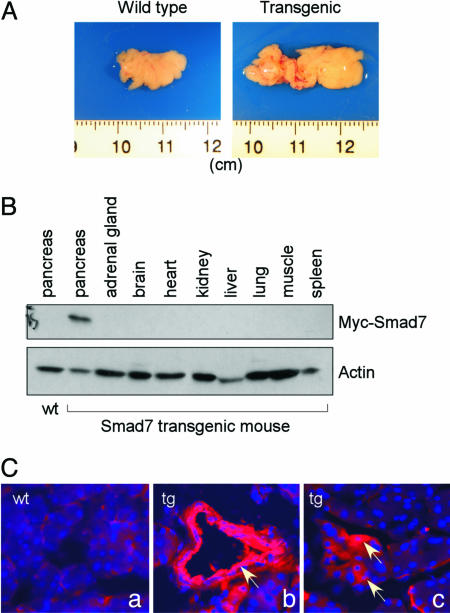
Of the five founders that carried the transgene, only one of them was found to have relatively high expression of the myc-tagged Smad7 by Western blotting analysis with the pancreas isolated from the offsprings of the founders (data not shown). All of the phenotypic and histological analyses were based on the offspring from this founder. At 6 months of age, pancreata were isolated from the transgenic mice as well as their WT littermates. At that age, we found that the pancreas from most transgenic mice was significantly enlarged in comparison with the WT animals (25 of 29 transgenic mice as compared with 7 WT animals). As shown in Fig. 2A, the pancreas from the WT mouse was ≈1.5 cm in length. However, the pancreas from the transgenic animal was ≈2.5 cm in length. This observation was in contrast to those gathered at 2 months of age, when the size of the pancreas from the transgenic mice was indistinguishable from that of the WT (data not shown). To confirm that the expression of the Smad7 gene driven by the elastase I promoter/enhance is limited to the pancreas, we performed Western Blotting analysis using an anti-myc antibody with tissues isolated from one representative transgenic animal at 6 weeks of age. As shown in Fig. 2B, the myc-tagged Smad7 was found only in the pancreas, but not in other tissues including the liver, lung, spleen, kidney, and many other tissues. We next used immunofluorescent staining with an anti-myc antibody to analyze the localization of the exogenously expressed Smad7. As shown in Fig. 2C, no fluorescence signal could be detected in the pancreas from the WT animal. However, Smad7 expression could be easily detected in pancreatic ducts (Fig. 2B), some acinar cells (Fig. 2C), and a few islets (data not shown) in the pancreas from the transgenic mouse. These data, therefore, indicate that Smad7 had a specific expression in the pancreas under the control of the elastase I promoter/enhancer.
Fig. 2.
Targeted expression of Smad7 in the pancreas. (A) Pancreas from the WT (Left) and transgenic mouse (Right) at 6 months of age. Note the enlargement of the pancreas from the transgenic animal. (B) Pancreas-specific expression of the exogenous Smad7 in transgenic mouse. Different tissues were isolated from a transgenic mouse as indicated, and protein preparation from these tissues was used in Western blotting with an anti-myc antibody. (C) Analysis of the myc-tagged Smad7 by immunofluorescent staining. Pancreas sections from WT (Ca) and transgenic mice (Cb and Cc) were used in immunofluorescent staining with an anti-myc antibody followed by an anti-mouse Cy3-coupled secondary antibody. The nuclei were stained with Hoechst 33342. Note the expression of Smad7 in the ductal cells (Cb, indicated by an arrow) and patchy expression in acinar cells (Cc, indicated by arrows).
Overexpression Smad7 in the Pancreas Disrupts TGF-β Signaling in Vivo.
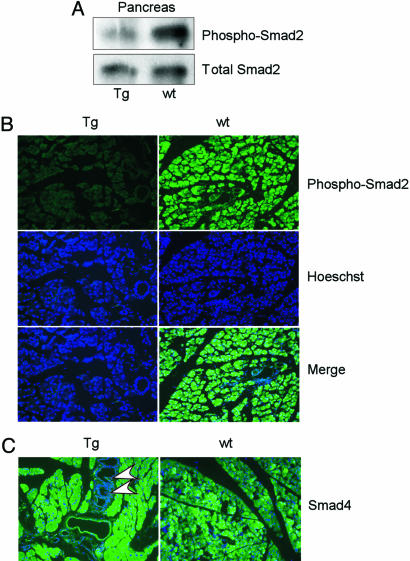
Smad7 is an inhibitory Smad protein that blocks TGF-β signaling through interaction with the TGF-β type I receptor. One of the major signaling events after TGF-β receptor activation is Smad2 phosphorylation (18, 19). We hypothesized that the overexpression of Smad7 in the transgenic animal would attenuate TGF-β signaling through a reduced Smad2 phosphorylation. To address this issue, we performed Western blotting analysis using protein lysates extracted from either transgenic or WT mice. An antibody specific for the phosphorylated Smad2 was used to determine the phosphorylation status of Smad2, and an anti-Smad2 antibody was used to detect the total Smad2 proteins. As shown in Fig. 3A, we found that Smad2 phosphorylation was reduced in the pancreas isolated from the transgenic mice in comparison with the pancreas from the WT littermate. However, the total amount of Smad2 proteins was not changed in both samples. Furthermore, we used immunofluorescent staining to detect phosphorylation of Smad2 with pancreas sections (Fig. 3B). Phosphorylation of Smad2 was clearly observed in the pancreas of the WT mice. However, the fluorescent signal for phospho-Smad2 was almost absent in the pancreas section from the transgenic mice. In addition, we analyzed the expression of Smad4 in the pancreas of the mice by immunostaining. As shown in Fig. 3C, Smad4 was expressed in the pancreas of both the transgenic and WT animals at a similar level, indicating that the Smad7 transgene didn’t affect the expression of Smad4. Taken together, these data demonstrated that overexpression of Smad7 was able to inactivate TGF-β signaling in the pancreas.
Fig. 3.
Disruption of TGF-β signaling by Smad7 in the transgenic mouse. (A) Proteins were isolated from pancreata of the WT or transgenic mouse and used in Western blotting with an anti-phosphorylated Smad2 antibody. The same blot was also analyzed by an anti-total-Smad2 antibody. Note the reduction of Smad2 phosphorylation in the pancreas from the transgenic animal. (B) The anti-phosphorylated Smad2 antibody was used in immunofluorescent staining in pancreas sections from WT or transgenic mice. The nuclei were stained with Hoechst 33342. (C) Immunostaining with Smad4 antibody together with Hoechst 33342 staining. The arrowheads indicate PanIN lesions in a 6-month-old transgenic mouse.
In Vivo Disruption of TGF-β Signaling in the Pancreas Induces Premalignant Ductal Lesions.
We next analyzed the histological changes of the pancreas associated with the exogenous expression of Smad7. At 2 months of age, the size and histology of the pancreata from the transgenic mice were indistinguishable from those of the WT animals. However, the pancreas from the transgenic mice at 6 months of age had an obvious change with the characteristics of PanIN lesion that is believed to be a precursor to invasive pancreatic cancers (6, 20).
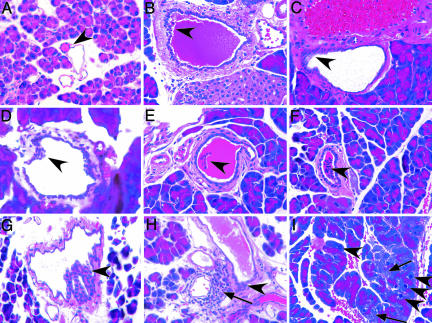
In the pancreas from WT animals, our histological analyses revealed an abundant distribution of acinar tissue with scattered islets and rarely seen ducts (Fig. 4A). The acinar cells that synthesize and secrete digestive enzymes are arranged in grape-like clusters. The pancreatic ducts are tightly lined with flat epithelial cells with uniform round nuclei (Fig. 4A).
Fig. 4.
Histological features of the Smad7 transgenic mouse. (A) Normal intralobular duct of WT pancreas showed single-layer, short cubical duct epithelium (arrowhead) with surrounding acinar cells. (B and C) PanIN-IB lesions with pseudostratified ductal epithelia (arrowhead) in transgenic mouse. (D–F) Various PanIN-IB lesions in intralobular ducts ranging from tufting to papillae formation (arrowhead) in the pancreas of the transgenic mouse. The ducts were surrounded by increasing fibrous stroma. (G) PanIN-II lesion with significant loss of epithelial polarity with moderate cell atypia. The hyperplastic ductal cells formed epithelial papillae (arrowhead). (H) Dilated duct with PanIN-1B lesion showing focal pseudostratified epithelial cells (arrowhead) accompanied by lymph-mononuclear cell infiltration in the adjacent fibrous stroma (arrow). (I) Hyperplastic acinar cells had enlarged nuclei and hyperchromasia (arrowhead) with scattered degeneration (arrow) in transgenic mouse. All pictures are hematoxylin/eosin staining at ×200.
However, hyperplastic lesions of the pancreatic ducts with characteristics of PanIN were observed in all transgenic mice at 6 months of age (20). PanIN lesions are premalignant lesions classified into PanIN-1A, PanIN-1B, PanIN-2, and PanIN-3 based on the degree of cytological and architectural atypia of pancreatic sections (20). PanIN-1A and -1B lesions are represented by a transition from the normal cuboidal epithelial to a columnar phenotype with or without nuclear atypia. PanIN-1B lesions have a papillary or pseudostratified architecture. The PanIN-1B lesions were frequently observed in our transgenic mice with exogenous expression of Smad7. The pseudostratified epithelial changes were frequently seen in intralobal ducts of the transgenic pancreas (Fig. 4 B and C). A spectrum of pseudostratified epithelium, cell tufting, micropapillary, and papillary architecture was frequently seen in the pancreatic ducts in the transgenic mice (Fig. 4 D–F). In these lesions, the ductal epithelial cells were hyperproliferated so that they formed a focally thickening epithelial layer and in some cases protruded into the lumen. However, we could rarely observe typical transitions from the normal cuboidal epithelium to a columnar phenotype, characteristics of the PanIN-1A lesions.
The PanIN-2 lesion is characterized by moderate cytological atypia in the ductal epithelium. We occasionally observed PanIN-2 lesions in the pancreas from the transgenic animals (Fig. 4G). In these lesions, the hyperproliferative ductal cells budded into the lumen with moderate atypia. Accompanying the PanIN lesions, an increased fibrosis could be observed in the regions surrounding the ductal lesion (Fig. 4 B–H). In a few cases, we also observed lymph-mononuclear cell infiltration adjacent to the PanIN lesion (Fig. 4H), indicating a local inflammatory reaction induced by the ductal alteration. Taken together, these observations demonstrated that inactivation of TGF-β in the pancreas is able to lead to hyperproliferation of the ductal epithelium, with accelerated fibrosis around the ductal lesion.
In addition to PanIN lesions, ductal stasis and dilation were observed in most pancreas sections (Fig. 4 B and E), suggesting that pancreatic ducts were either functionally impaired or partially blocked. Patchy hyperplasia and degeneration could be observed among the acinar cells (Fig. 4I), consistent with the scattered expression pattern of the Smad7 transgene in the acinar cells (Fig. 2Cc).
Epithelial Features of the PanIN Lesions in the Transgenic Mouse.
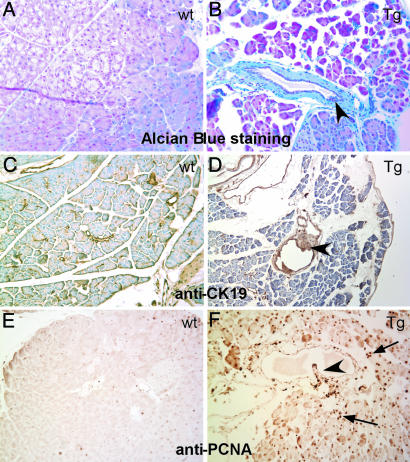
To better define the PanIN lesions in the transgenic mice, we applied Alcian blue staining and immunohistochemistry analysis. Alcian blue is one of the most widely used cationic dyes to detect glycosaminoglycans that are abundantly synthesized by the pancreatic ductal epithelium. We observed that Alcian blue stain was positive in the ductal epithelium overlying the pancreatic ducts in both WT and transgenic mice (Fig. 5 A and B). An intense Alcian blue staining was observed in PanIN lesions in the transgenic mice, indicating the epithelial nature of PanINs in the transgenic mice. To further confirm this finding, we performed immunohistochemistry studies to detect expression of cytokeratin-19 (CK-19), a specific epithelial cell marker for pancreatic ducts. As expected, intense CK-19 signals were detected in normal pancreatic ducts as well as in PanIN lesions (Fig. 5 C and D). Taken together, these data demonstrated that the PanIN lesions seen in our transgenic mice were originated from pancreatic ductal epithelium.
Fig. 5.
Characterization of PanIN lesions in the transgenic mouse. (A and B) Alcian blue staining visualized mucin content in a PanIN ductal lesion (arrowhead) in transgenic mouse, and such stain is absent in its WT counterpart. (C and D) Immunohistochemistry for CK19, PanIN lesions (arrowhead) showed the hyperplastic epithelial cells expressing CK19. (E and F) Immunohistochemistry labeling of PCNA demonstrated the high percentage of proliferating cells in PanIN lesion (arrowhead) as well as acinar cells (arrow) in transgenic mouse but only scarce staining in WT pancreas.
Disruption of TGF-β Signaling Promotes Proliferation of Ductal and Acinar Cells.
TGF-β signaling is thought to be a major tumor suppressing pathway due to its anti-proliferative activity (8, 9). We hypothesized that inactivation of TGF-β signaling by Smad7 in the pancreas would promote cell proliferation. We assessed this issue by detecting proliferating cell nuclear antigen (PCNA) expression. As shown in Fig. 5E, the basal level of PCNA expression was low in the pancreas of WT animals, and the strong PCNA-positive ductal cells were seldom observed. A few of the PCNA-positive acinar cell were scattered through the exocrine portion of the pancreas (Fig. 5E). In the transgenic mice, the ductal cells with PanIN lesions were found to express high levels of PCNA (Fig. 5F). The PCNA expression level in acinar cells in the transgenic mice was elevated as well. Therefore, it seemed that overexpression of Smad7 in the pancreas can induce cellular escape from TGF-β-mediated antiproliferative activities.
Discussion
In this report, we demonstrated that disruption of TGF-β signaling is able to induce PanIN formation in the mouse. Through a rat elastase I promoter/enhancer, we targeted pancreas-specific expression of Smad7, an inhibitory Smad that antagonizes TGF-β signaling by association with the TGF-β type I receptor (14). Our immunoblotting analysis confirmed that the exogenous Smad7 was expressed only in the pancreas, but not in other tissues of the animal. The inhibition of TGF-β signaling was confirmed by a reduction of Smad2 phosphorylation in the pancreas. At 6 months of age, most transgenic animals with Smad7 expression developed premalignant lesions in the pancreas with the characteristics of PanIN. Such premalignant ductal lesions of the pancreas were accompanied by accelerated fibrosis surrounding the ducts. Therefore, this study demonstrated that in vivo inactivation of TGF-β signaling is implicated in the development of the early stage of pancreatic cancers.
TGF-β is characterized as an antiproliferative cytokine, especially in the early stage of cancer development (8, 9). Most cancer cells escape from the growth inhibitory activity of TGF-β by genetic mutations of the members involved in TGF-β signaling. In pancreatic cancers, it has been reported that >50% of the cases have Smad4 mutations, thus making the cells insensitive to TGF-β (7). It was also reported that the expression of Smad7 is frequently elevated in pancreatic cancers (21). However, it has not been proven in the past that disruption of TGF-β function is directly associated with the development of pancreatic cancers. By using a nonpancreatic-specific promoter, it was found that overexpression of a dominant negative TGF-β type II receptor (DNR) was able to affect the growth and differentiation of the acinar cells in the pancreas, but without typical PanIN lesions (22). In our study, Smad7 was targeted to express in the pancreas by a rat elastase I promoter/enhancer, and the transgenic animals had characteristic PanIN lesions. The phenotypic difference between our report and the study using DNR is likely associated with the promoters used in driving gene expression in the pancreas. The widely expressed metallothionein 1 (MT1) promoter was used to guide DNR expression, and many lines of transgenic studies indicated that MT1 promoter is mainly limited to acinar expression in the pancreas (23, 24). Therefore, overexpression of DNR under the MT1 promoter is mainly associated with lesions of acinar cells, but not of ductal epithelium. Our study used the −205/+8-bp region of rat elastase I promoter/enhance. In our immunohistochemistry studies, we found that the exogenous Smad7 was expressed in ductal epithelium and acinar cells (Fig. 2C) as well as islet (data not shown). This finding is consistent with our observation that Smad7 overexpression in the transgenic mouse is associated with PanIN lesions in the ductal epithelium accompanied by an enhanced proliferation of the acinar cells. In addition, we observed premalignant lesions in the pancreas only in 6-month-old transgenic mice, but not in 2-month-old animals. Therefore, it is likely that other genetic alterations accumulated over time are needed for the formation of these premalignant changes.
The cellular origin of ductal pancreatic carcinoma has been a controversial issue (25, 26). Studies by Grippo et al. (27) using the elastase promoter driving Kras expression in the pancreas demonstrated acinar-to-ductal metaplasia in aged mice, but no ductal lesions in young animals. However, our studies with Smad7 transgene revealed clear PanIN lesions, without obvious acinar dysplasia surrounding the ductal changes, indicating that the premalignant changes of the ductal epithelium are not likely derived from acinar cells. However, it is possible that the premaligant ductal cells are originated from islet cells or multipotent precursor cells due to transdifferentiation. This possibility is supported by our observation that the PanIN lesions are frequently neighbored by islets (data not shown). In addition, this issue can be addressed in the future by analysis using lineage-specific markers such as Pdx-1.
It is noteworthy that Smad7 is also able to block activin signaling by interacting with the activin type I receptor (17). Activin ligands and receptors are expressed in the developing pancreas (28), and activin shares a similar antiproliferative effect as TGF-β in most cell types (29). Overexpression of Smad7 in the transgenic mouse, therefore, may also antagonize the activin function in the pancreas. Expression of dominant negative activin type II receptors in the pancreas was associated with islet hypoplasia and impaired differentiation of both endocrine and exocrine cells, but without PanIN lesions (30, 31). In our study, overexpression of Smad7 in the pancreas is able to induce characteristic PanIN lesions distinct from the phenotypes of transgenic mice that express either dominant negative TGF-β receptors or dominant negative activin receptors. At present, we could not rule out the possibility that our observation with Smad7 overexpression might be caused by a combined antagonization of both TGF-β and activin signaling in the pancreas. To a certain extent, Smad7 overexpression in our model is equivalent to deletion of Smad4 in human pancreatic carcinomas in which the loss of Smad4 is expected to abrogate the signaling pathways of both TGF-β and activin. In addition, it is noteworthy that Smad7 may exert its functions independent of Smad2/Smad3 activation upon TGF-β signaling, such as activation of p38 mitogen-activated protein kinase (MAPK) via TGF-β-activated kinase 1 (TAK1) and MAPK kinase 3 (MKK3) (32). Interestingly, activin is also reported to activate p38 MAPK to stimulate expression of pro-endocrine gene neurogenin 3 in pancreatic endocrine cells (33). Therefore, it needs to be determined in the future whether deregulation of p38 MAPK by Smad7 overexpression is implicated in the formation of PanIN lesions in the pancreas.
In humans, PanIN has been considered a direct noninvasive neoplastic precursor to pancreatic cancers (6). A stepwise molecular progression model for the carcinogenesis in the pancreas has been postulated based on the stages of PanIN and the associated genetic mutations from human studies (6, 34). The early event in PanIN is always associated with mutations of Kras (3–5) and telomere shortening (35). The loss of p16INK4a commonly occurs at the intermediate stage. Mutations of p53 and Smad4 have been considered as late events that occur in PanINs with increasing severity (6, 34, 36). The involvement of these mutations in the development of pancreatic cancers has been extensively investigated in mouse models (25). Consistent with the molecular progression model of pancreatic cancer formation, only activating Kras mutation has been shown to be able to directly induce full spectrum PanIN lesions and low-frequency progression to invasive and metastatic adenocarcinoma (37), although the tumorigenic effect of the Kras mutation in pancreatic cancer development is variable depending on the targeting promoters used in the mouse models (27, 38). Inactivation of p16INK4a alone was not able to produce any neoplastic lesion in the pancreas. However, the activated Kras mutation is able to cooperate with p16INK4a deficiency to induce progressive and metastatic pancreatic carcinoma, indicating a role of Kras in tumor initiation and p16INK4a in tumor promotion (39). The tumor-promoting activity of p16INK4a is further supported by the finding that c-myc expression in the pancreas can induce cancer formation only in the presence of p16INK4a deficiency (40). In addition, the tumor promotion function of p53 was evidenced by the finding that p53 deficiency is able to accelerate tumor development in the TGF-α transgenic mouse (41).
The in vivo function of Smad4 in pancreatic cancer formation using mouse models is limited by the findings that homozygous Smad4-deficient mice die before day 7.5 of embryogenesis (42, 43). Heterozygous Smad4-deficient mice, however, fail to yield any form of pancreatic pathology (42, 43). Our findings that inhibition of TGF-β signaling by Smad7 is able to induce PanIN lesions in the pancreas would argue for a unique function of TGF-β in the early initiation of pancreatic cancer, in addition to its tumor promotion activity postulated from human mutation studies (6, 34, 36). In other words, our results would put TGF-β into the same category as Kras mutations that are believed to the major initiating events during carcinogenesis in the pancreas. This proposed tumor initiation activity of TGF-β can be tested, for example, by crossing the Smad7 transgenic mouse with the p16INK4a-deficient mouse to determine whether or not TGF-β disruption can cooperate with loss of p16INK4a to recapitulate the full spectrum of pancreatic cancer progression.
In summary, our studies demonstrated that disruption of TGF-β is able to induce early PanIN lesions in the mouse pancreas. This animal model would greatly aid in understanding the tumor initiation and/or tumor promotion function of TGF-β blockage, a long sought culprit in pancreatic cancer formation due to the extensive mutations of Smad4 and other TGF-β-signaling components in pancreatic cancers. Because this animal model is associated with early pancreatic premalignant lesions, it would serve as a unique tool to facilitate mechanistic studies of early stage pancreatic cancers as well as designs of strategies for early detection and early therapy for the preinvasive lesions of pancreatic cancers.
Materials and Methods
Generation of Transgenic Mice.
The transgene was constructed by standard recombinant DNA techniques by fusing a rat elastase I promoter/enhancer fragment (−205/+8 bp) with a myc-tagged rat Smad7 cDNA, as shown in Fig. 1. The elastase I enhancer/promoter plasmid was a gift kindly provided by R. Macdonald and G. Swift (Texas Southwestern Medical Center, Dallas). It has been found that this region of the elastase I gene is able to direct specific gene expression in both the endocrine and exocrine pancreas (15, 16). The linearized transgene was used in microinjection into fertilized mouse eggs at the Indiana University Transgenic/Knockout facility according to National Institutes of Health (NIH) animal guidelines. All transgenic mice were generated with the C3H strain and maintained with the DBA2 strain.
Characterization of Transgenic Mice.
Genomic DNA was extracted from a 2-mm tail biopsy with a Genomic DNA Extraction Kit (Promega). Mice were genotyped by both Southern blotting analysis and PCR. For Southern blotting, the genomic DNA were digested with BamHI, separated on agarose gel, and transferred to the Hybond N membrane (Amersham). The membranes were subjected to hybridization with a 32P-labeled DNA probe specific for the transgene, followed by autoradiography. Samples yielding a 3-kb hybridized band were considered positive for the transgene. For PCR genotyping, 100 ng of aliquot of the genomic DNA was used in a PCR mixture containing the following primer pairs: 5′-CTTTGTACTTTCATGTCACCTGTGC-3′ and 5′-CGCCGGACGAGCGCAGATCGTT-3′. The positive samples yielded a 620-bp product.
Tissue Harvest, Protein Extraction, and Western Blot Analysis.
Animals were euthanized with a lethal dose of CO2 per institutional guidelines. Pancreata and other tissues/organs were removed and immediately washed with cold 1× PBS, homogenized, and solubilized in cold RIPA buffer (50 mM Tris·HCl, pH 7.4/1% Nonidet P-40/0.25% sodium deoxycholate/150 mM NaCl/1 mM EDTA) containing complete proteinase inhibitor mixture and β-glycerophosphate (Sigma). Proteins from the various tissues were resolved by SDS/PAGE, and transferred to poly(vinylidene difluoride) (PVDF) membranes (Millipore). The myc-tagged Smad7, actin, phosphorylated Smad2, and total Smad2 were detected by Western blotting by using mouse anti-myc antibody (Roche Applied Science, Indianapolis), rabbit anti-actin antibody (Santa Cruz Biotechnology), rabbit anti-phosphorylated-Smad2 antibody (Cell Signaling Technology, Beverly, MA), and mouse anti-Smad2 antibody (Santa Cruz Biotechnology), respectively.
Histological Analysis.
Mouse pancreas specimens were fixed in a 4% paraformaldehyde solution in PBS overnight at 4°C and embedded in paraffin by using standard techniques. The paraffin-embedded sections (5 μm) were stained with hematoxylin/eosin (H&E) to determine the histopathological features. For Alcian blue staining, the paraffin sections were deparaffinized and rehydrated with standard protocols. The sections were then stained in Alcian blue/acetic acid (pH 2.5) solution for 10 min at room temperature, followed by extensive wash in tap water. The nuclei were counterstained in 0.1% Fast Red solution for 5 min at room temperature.
Immunofluorescence Staining and Immunohistochemistry.
For cryosectioning, pancreata were washed twice in cold PBS, and fixed in 4% paraformaldehyde in PBS overnight at 4°C. The samples were dehydrated in 30% sucrose in PBS and embedded in Tissue-Tek OCT (Sakura USA, Torrance, CA). For immunofluorescence staining, the cryostat sections of 10 μm thickness were air-dried and soaked with 1× PBS followed with 1 h of blocking in 1% normal serum. The slides were incubated at 4°C with primary antibody overnight, followed by incubation with a fluorescence-conjugated secondary antibody for 2 h at room temperature. The nuclei were stained with Hoechst 33342. For paraffin-embedded sections, immunohistochemistry was done on 5-μm tissue sections by using the Hiostostain-Plus kit according to the manufacturer’s instructions (Invitrogen). Before staining, antigen retrieval procedures were performed by incubation with 0.5% pepsin (Sigma) in 5 mM HCl for 20 min at 37°C. The primary antibodies used were as follows: mouse anti-myc antibody (1:200; Roche Applied Science), rabbit anti-phosphorylated-Smad2 antibody (1:1, 000; Cell Signaling Technology), mouse anti-PCNA antibody (1:3, 000; Sigma), mouse anti-cytokeratin-19 (TROMA III) (1:10; Developmental Studies Hybridoma Bank, Iowa City, IA), and rabbit anti-Smad4 antibody (1:500; Santa Cruz Biotechnology). Indirect immunofluorescence was performed with goat anti-mouse Cy3-coupled or goat anti-rabbit Cy5-coupled secondary antibodies at 1:500 dilution (Jackson ImmunoResearch).
Acknowledgments
We thank Drs. R. Macdonald and G. Swift for the rat elastase promoter plasmid. The mouse anti-cytokeratin 19 (TROMA III) antibody developed by Dr. Rolf Kemler was obtained from the Developmental Studies Hybridoma Bank under the auspices of the National Institute of Child Health and Human Development and maintained by The University of Iowa, Department of Biological Sciences. This work was supported by research grants from the American Cancer Society (PRG-00-273-01-MGO), the National Institute of Diabetes and Digestive and Kidney Diseases (R01 DK55991), the Chinese Academy of Sciences (Bairen Plan), and the National Natural Science Foundation of China (30470870 to Y.C.).
Abbreviations
- PanIN
pancreatic intraepithelial neoplasia
- Smad
Sma- and Mad-related protein
- PCNA
proliferating cell nuclear antigen.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Brand R., Mahr C. Curr. Gastroenterol. Rep. 2005;7:122–127. doi: 10.1007/s11894-005-0050-9. [DOI] [PubMed] [Google Scholar]
- 2.Warshaw A. L., Fernandez-del Castillo C. N. Engl. J. Med. 1992;326:455–465. doi: 10.1056/NEJM199202133260706. [DOI] [PubMed] [Google Scholar]
- 3.Klimstra D. S., Longnecker D. S. Am. J. Pathol. 1994;145:1547–1550. [PMC free article] [PubMed] [Google Scholar]
- 4.Moskaluk C. A., Hruban R. H., Kern S. E. Cancer Res. 1997;57:2140–2143. [PubMed] [Google Scholar]
- 5.Rozenblum E., Schutte M., Goggins M., Hahn S. A., Panzer S., Zahurak M., Goodman S. N., Sohn T. A., Hruban R. H., Yeo C. J., Kern S. E. Cancer Res. 1997;57:1731–1734. [PubMed] [Google Scholar]
- 6.Hansel D. E., Kern S. E., Hruban R. H. Annu. Rev. Genomics Hum. Genet. 2003;4:237–256. doi: 10.1146/annurev.genom.4.070802.110341. [DOI] [PubMed] [Google Scholar]
- 7.Hahn S. A., Schutte M., Hoque A. T., Moskaluk C. A., da Costa L. T., Rozenblum E., Weinstein C. L., Fischer A., Yeo C. J., Hruban R. H., Kern S. E. Science. 1996;271:350–353. doi: 10.1126/science.271.5247.350. [DOI] [PubMed] [Google Scholar]
- 8.Siegel P. M., Massague J. Nat. Rev. Cancer. 2003;3:807–821. doi: 10.1038/nrc1208. [DOI] [PubMed] [Google Scholar]
- 9.Derynck R., Akhurst R. J., Balmain A. Nat. Genet. 2001;29:117–129. doi: 10.1038/ng1001-117. [DOI] [PubMed] [Google Scholar]
- 10.Wrana J. L., Attisano L., Carcamo J., Zentella A., Doody J., Laiho M., Wang X. F., Massague J. Cell. 1992;71:1003–1014. doi: 10.1016/0092-8674(92)90395-s. [DOI] [PubMed] [Google Scholar]
- 11.Massague J. Annu. Rev. Biochem. 1998;67:753–791. doi: 10.1146/annurev.biochem.67.1.753. [DOI] [PubMed] [Google Scholar]
- 12.Heldin C. H., Miyazono K., ten Dijke P. Nature. 1997;390:465–471. doi: 10.1038/37284. [DOI] [PubMed] [Google Scholar]
- 13.Miyazono K., Kusanagi K., Inoue H. J. Cell. Physiol. 2001;187:265–276. doi: 10.1002/jcp.1080. [DOI] [PubMed] [Google Scholar]
- 14.Hayashi H., Abdollah S., Qiu Y., Cai J., Xu Y. Y., Grinnell B. W., Richardson M. A., Topper J. N., Gimbrone M. A., Jr, Wrana J. L., Falb D. Cell. 1997;89:1165–1173. doi: 10.1016/s0092-8674(00)80303-7. [DOI] [PubMed] [Google Scholar]
- 15.Ornitz D. M., Palmiter R. D., Hammer R. E., Brinster R. L., Swift G. H., MacDonald R. J. Nature. 1985;313:600–602. doi: 10.1038/313600a0. [DOI] [PubMed] [Google Scholar]
- 16.Kruse F., Rose S. D., Swift G. H., Hammer R. E., MacDonald R. J. Genes Dev. 1993;7:774–786. doi: 10.1101/gad.7.5.774. [DOI] [PubMed] [Google Scholar]
- 17.Liu X., Nagarajan R. P., Vale W., Chen Y. FEBS Lett. 2002;519:93–98. doi: 10.1016/s0014-5793(02)02718-7. [DOI] [PubMed] [Google Scholar]
- 18.Abdollah S., Macias-Silva M., Tsukazaki T., Hayashi H., Attisano L., Wrana J. L. J. Biol. Chem. 1997;272:27678–27685. doi: 10.1074/jbc.272.44.27678. [DOI] [PubMed] [Google Scholar]
- 19.Souchelnytskyi S., Tamaki K., Engstrom U., Wernstedt C., ten Dijke P., Heldin C. H. J. Biol. Chem. 1997;272:28107–28115. doi: 10.1074/jbc.272.44.28107. [DOI] [PubMed] [Google Scholar]
- 20.Kern S., Hruban R., Hollingsworth M. A., Brand R., Adrian T. E., Jaffee E., Tempero M. A. Cancer Res. 2001;61:4923–4932. [PubMed] [Google Scholar]
- 21.Kleeff J., Ishiwata T., Maruyama H., Friess H., Truong P., Buchler M. W., Falb D., Korc M. Oncogene. 1999;18:5363–5372. doi: 10.1038/sj.onc.1202909. [DOI] [PubMed] [Google Scholar]
- 22.Bottinger E. P., Jakubczak J. L., Roberts I. S., Mumy M., Hemmati P., Bagnall K., Merlino G., Wakefield L. M. EMBO J. 1997;16:2621–2633. doi: 10.1093/emboj/16.10.2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sandgren E. P., Luetteke N. C., Palmiter R. D., Brinster R. L., Lee D. C. Cell. 1990;61:1121–1135. doi: 10.1016/0092-8674(90)90075-p. [DOI] [PubMed] [Google Scholar]
- 24.Palmiter R. D., Sandgren E. P., Koeller D. M., Brinster R. L. Mol. Cell. Biol. 1993;13:5266–5275. doi: 10.1128/mcb.13.9.5266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leach S. D. Cancer Cell. 2004;5:7–11. doi: 10.1016/s1535-6108(03)00337-4. [DOI] [PubMed] [Google Scholar]
- 26.Pour P. M., Pandey K. K., Batra S. K. Mol. Cancer. 2003;2:13. doi: 10.1186/1476-4598-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grippo P. J., Nowlin P. S., Demeure M. J., Longnecker D. S., Sandgren E. P. Cancer Res. 2003;63:2016–2019. [PubMed] [Google Scholar]
- 28.Dichmann D. S., Miller C. P., Jensen J., Scott Heller R., Serup P. Dev. Dyn. 2003;226:663–674. doi: 10.1002/dvdy.10270. [DOI] [PubMed] [Google Scholar]
- 29.Vale W., Wiater E., Gray P., Harrison C., Bilezikjian L., Choe S. Ann. N.Y. Acad. Sci. 2004;1038:142–147. doi: 10.1196/annals.1315.023. [DOI] [PubMed] [Google Scholar]
- 30.Yamaoka T., Idehara C., Yano M., Matsushita T., Yamada T., Ii S., Moritani M., Hata J., Sugino H., Noji S., Itakura M. J. Clin. Invest. 1998;102:294–301. doi: 10.1172/JCI2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shiozaki S., Tajima T., Zhang Y. Q., Furukawa M., Nakazato Y., Kojima I. Biochim. Biophys. Acta. 1999;1450:1–11. doi: 10.1016/s0167-4889(99)00022-1. [DOI] [PubMed] [Google Scholar]
- 32.Edlund S., Bu S., Schuster N., Aspenstrom P., Heuchel R., Heldin N. E., ten Dijke P., Heldin C. H., Landstrom M. Mol. Biol. Cell. 2003;14:529–544. doi: 10.1091/mbc.02-03-0037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ogihara T., Watada H., Kanno R., Ikeda F., Nomiyama T., Tanaka Y., Nakao A., German M. S., Kojima I., Kawamori R. J. Biol. Chem. 2003;278:21693–21700. doi: 10.1074/jbc.M302684200. [DOI] [PubMed] [Google Scholar]
- 34.Maitra A., Adsay N. V., Argani P., Iacobuzio-Donahue C., De Marzo A., Cameron J. L., Yeo C. J., Hruban R. H. Mod. Pathol. 2003;16:902–912. doi: 10.1097/01.MP.0000086072.56290.FB. [DOI] [PubMed] [Google Scholar]
- 35.van Heek N. T., Meeker A. K., Kern S. E., Yeo C. J., Lillemoe K. D., Cameron J. L., Offerhaus G. J., Hicks J. L., Wilentz R. E., Goggins M. G., De Marzo A. M., Hruban R. H., Maitra A. Am. J. Pathol. 2002;161:1541–1547. doi: 10.1016/S0002-9440(10)64432-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yeo T. P., Hruban R. H., Leach S. D., Wilentz R. E., Sohn T. A., Kern S. E., Iacobuzio-Donahue C. A., Maitra A., Goggins M., Canto M. I., et al. Curr. Probl. Cancer. 2002;26:176–275. doi: 10.1067/mcn.2002.129579. [DOI] [PubMed] [Google Scholar]
- 37.Hingorani S. R., Petricoin E. F., Maitra A., Rajapakse V., King C., Jacobetz M. A., Ross S., Conrads T. P., Veenstra T. D., Hitt B. A., et al. Cancer Cell. 2003;4:437–450. doi: 10.1016/s1535-6108(03)00309-x. [DOI] [PubMed] [Google Scholar]
- 38.Brembeck F. H., Schreiber F. S., Deramaudt T. B., Craig L., Rhoades B., Swain G., Grippo P., Stoffers D. A., Silberg D. G., Rustgi A. K. Cancer Res. 2003;63:2005–2009. [PubMed] [Google Scholar]
- 39.Aguirre A. J., Bardeesy N., Sinha M., Lopez L., Tuveson D. A., Horner J., Redston M. S., DePinho R. A. Genes Dev. 2003;17:3112–3126. doi: 10.1101/gad.1158703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lewis B. C., Klimstra D. S., Varmus H. E. Genes Dev. 2003;17:3127–3138. doi: 10.1101/gad.1140403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wagner M., Greten F. R., Weber C. K., Koschnick S., Mattfeldt T., Deppert W., Kern H., Adler G., Schmid R. M. Genes Dev. 2001;15:286–293. doi: 10.1101/gad.184701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sirard C., de la Pompa J. L., Elia A., Itie A., Mirtsos C., Cheung A., Hahn S., Wakeham A., Schwartz L., Kern S. E., et al. Genes Dev. 1998;12:107–119. doi: 10.1101/gad.12.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang X., Li C., Xu X., Deng C. Proc. Natl. Acad. Sci. USA. 1998;95:3667–3672. doi: 10.1073/pnas.95.7.3667. [DOI] [PMC free article] [PubMed] [Google Scholar]