Abstract
Amino acid racemases catalyze the stereoinversion of the chiral Cα to produce the d-enantiomers that participate in biological processes, such as cell wall construction in prokaryotes. Within this large protein family, bacterial proline racemases have been extensively studied as a model of enzymes acting with a pyridoxal-phosphate-independent mechanism. Here we report the crystal structure of the proline racemase from the human parasite Trypanosoma cruzi (TcPRACA), a secreted enzyme that triggers host B cell polyclonal activation, which prevents specific humoral immune responses and is crucial for parasite evasion and fate. The enzyme is a homodimer, with each monomer folded in two symmetric α/β subunits separated by a deep crevice. The structure of TcPRACA in complex with a transition-state analog, pyrrole-2-carboxylic acid, reveals the presence of one reaction center per monomer, with two Cys residues optimally located to perform acid/base catalysis through a carbanion stabilization mechanism. Mutation of the catalytic Cys residues abolishes the enzymatic activity but preserves the mitogenic properties of the protein. In contrast, inhibitor binding promotes the closure of the interdomain crevice and completely abrogates B cell proliferation, suggesting that the mitogenic properties of TcPRACA depend on the exposure of transient epitopes in the ligand-free enzyme.
Keywords: B cell mitogen, pyridoxal phosphate-independent proline racemase, epimerases, enzyme–inhibitor complex, titration calorimetry
The vast majority of amino acids found in living cells correspond to the l-stereoisomer at the Cα chiral center. However, d-amino acids are often found as constituents of bacterial cell walls (1, 2) and were identified in archaea and higher eukaryotes (3–5). Amino acid racemases and epimerases, which catalyze the l, d-stereochemistry inversion on free amino acids, have been extensively studied in prokaryotic systems (6). In the d→l sense, cells use d-amino acids to feed the large l-amino acid pool in normal amino acid/protein metabolism; whereas, in the l→d sense, bacteria generate the d-enantiomers widely distributed in bacterial cell walls, in particular d-alanine, d-glutamate, and d, l-diaminopimelate, as peptidoglycan components that function as innate defense against host proteolytic mechanisms (1, 2)
All known racemases catalyze the inversion of the chiral center by deprotonation of the Cα, followed by reprotonation on the opposite face of the planar carbanionic transition-state species. To overcome the high energetic barrier of this reaction [estimated pKa values for the Cα are in the range 21–32 (7, 8)], some racemases evolved to use pyridoxal phosphate (PLP) as cofactor, because formation of an imine PLP–substrate covalent bond greatly acidifies the chiral center by resonance (9). However, a second class of enzymes, which includes proline, aspartate, and glutamate racemases and diaminopimelate epimerase, operates through a two-base mechanism in a cofactor-independent manner (10–13). A foundational paper on the Clostridium sticklandii proline racemase (EC 5.1.1.4) proposed that the enzyme binds one molecule of competitive inhibitor per homodimer (14), based on equilibrium dialysis experiments with the radiolabeled competitive inhibitor pyrrole-2-carboxylic acid (PYC). Isotope incorporation/enzyme kinetics in 2H2O (15) had demonstrated a 1:1 proton transfer (transfer of a proton to and from the same carbon atom) and a two-base mechanism in which no solvent isotope exchange occurs while the active site is occupied, implying as well that the two enzymic bases are monoprotic. Together with the iodoacetamide titration evidence that suggested the presence of one reactive Cys (14), the currently accepted model of proline racemase puts forward a unique active site per dimer, wherein each monomer would contribute its catalytic Cys, generating a highly symmetric two-thiol/thiolate reaction center that could handle equally well both substrate enantiomers. However, direct structural evidence is lacking to validate these hypotheses.
The first eukaryotic proline racemase was identified in the pathogenic parasite Trypanosoma cruzi (16), which produces functional intracellular or secreted versions of the enzyme (17). The secreted version of T. cruzi proline racemase (TcPRACA) is a potent host B cell mitogen that sustains parasite evasion of specific immune responses (16, 18), offering a potential target for the chemotherapy of Chagas disease (19). Somewhat surprisingly, in vitro studies with strong competitive or nonspecific inhibitors of bacterial proline racemases suggested a direct link between the enzymatic and mitogenic properties of TcPRACA (16, 17), although the molecular basis of this correlation remained largely unclear.
In this paper, we investigate the mechanism of catalysis and the mitogenic properties of TcPRACA by using a combination of structural, thermodynamic, mutagenesis, and cellular approaches. The crystal structure of TcPRACA was determined at 2.1-Å resolution in complex with the transition-state analog PYC, revealing that the enzyme possesses two active sites per dimer, each one including two catalytic Cys residues. The structure supports a reaction mechanism operating by stabilization of a carbanionic transition-state species. Crystallographic and calorimetric studies of TcPRACA at different inhibitor concentrations reveal a ligand-promoted closure movement of the monomer, which completely buries the bound ligand from the bulk solvent. Mutagenesis studies of the catalytic residues and B cell proliferation assays of wild-type and mutant proteins reveal that the mitogenic activity of TcPRACA is dissociated from its catalytic activity and may depend on the formation of transiently exposed epitopes in the ligand-free enzyme.
Results
Overall Structure and Topology.
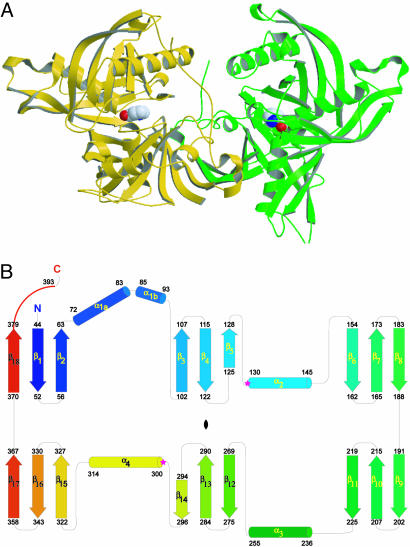
The structure of TcPRACA was determined by single wavelength diffraction methods at 2.1-Å resolution from crystals of selenomethionine-labeled protein (Table 1). The enzyme is a homodimer of approximate dimensions 100 × 60 × 50 Å (Fig. 1A). Within each monomer, the polypeptide chain is organized in two structurally equivalent α/β domains (rms deviation of 2 Å for 112 superposed Cα, 9% sequence identity). Each structural domain consists of three antiparallel three-stranded β-sheets interacting with two α-helices on either side of the sheet (Fig. 1B). An additional α-helix (αN) is visible at the N terminus (positions 33–41) only in the structure of ligand-free monomer at low inhibitor concentrations (Table 1), which is seen in a different, open conformation (see below). One side of the globular monomer contains two interdomain connections, whereas the other side is open to the solvent, with a deep crevice (≈20 Å) extending well into the separating interdomain space where the inhibitor is bound (Fig. 1). A C-terminal extension after the last β-strand (residues 382–393), which interacts with α1- and α3-helices of the same monomer and with α3′ and the β15′–β16′-turn from the second monomer, is located opposite the interdomain connecting hinge and partially occludes the entrance to the crevice.
Table 1.
Data collection, phasing, and refinement statistics
| Data set | SeMet-labeled | PYC complex | PYC hemicomplex |
|---|---|---|---|
| Data resolution, * Å | 70–3 (3.05–3.0) | 69.2–2.1 (2.21–2.1) | 60–2.5 (2.64–2.5) |
| Wavelength, Å | 0.97914 | 1.0072 | 0.934 |
| Measured reflections | 168, 186 | 172, 647 | 65, 088 |
| Multiplicity* | 9.7 (5.1) | 3.7 (3.4) | 2.4 (2.4) |
| Completeness, * % | 99.6 (98.6) | 98.8 (94.1) | 97.6 (97.4) |
| Rsym, *† % | 7.4 (21.9) | 6.9 (12.8) | 7.5 (22.1) |
| I/σ* | 25.8 (6.6) | 15.4 (7.7) | 10 (4) |
| a/b/c, Å | 134.06/91.62/86.03 | 131.15/91.21/85.98 | 136.02/89.38/84.45 |
| β, ° | 123.37 | 126.52 | 125.98 |
| Mean FOM acentric | 0.458 | — | — |
| Phasing power‡ | 2.93 | — | — |
| Resolution, Å | — | 30–2.1 | 30–2.5 |
| Rcryst§ (No. of refs) | — | 0.149 (44, 628) | 0.170 (24, 875) |
| Rfree§ (No. of refs) | — | 0.193 (2, 331) | 0.236 (2, 778) |
| rms bonds, Å | — | 0.019 | 0.018 |
| rms angles, ° | — | 1.72 | 1.76 |
| Protein atoms | — | 5, 376 | 5, 395 |
| Water atoms | — | 358 | 154 |
| Ligand atoms (PYC) | — | 16 | 8 |
FOM, figure of merit; SeMet, selenomethionine; —, not applicable.
*Values in parentheses apply to the high-resolution shell.
†Rsym = ΣhklΣi|Ii(hkl) − I(hkl)|/ΣhklΣiIi(hkl).
‡Anomalous phasing power = 〈[|Fh(calc)|/phase-integrated lack of closure]〉.
§R = Σhkl|F(h)obs − F(h)calc|/Σhkl| F(h)obs|·Rcryst and Rfree were calculated from the working and test reflection sets (refs), respectively.
Fig. 1.
Structure of TcPRACA. (A) Ribbon model of the homodimer with the inhibitor (PYC) shown in Corey–Pauling–Koltun representation. (B) Topology of the TcPRACA monomer showing the two structurally equivalent domains (the pseudodyad axis relating them is depicted at the center). The only exception to the internal symmetry is the difference between the topologically equivalent α1- and α3-helices. Whereas α1 shows a strong kink that results in tight packing of the helix against both the β3–β4–β5-sheet and the β6–β7 turn, α3 is not bent and interacts only with the equivalent β12–β13–β14-sheet. The two catalytic Cys residues are marked with stars at the N termini of α2 and α4.
The dimer interface buries a surface of ≈1, 500 Å2 per monomer. In addition to the C-terminal extension described above, the dimerization surface includes a second, larger contact region between the two monomers: the last β-strand (β18) contributes to the formation of the dimeric species by constituting a full six-stranded antiparallel β-sheet with the equivalent elements of the other monomer (β18′–β1′–β2′). In addition, the loop including the conserved motif Glu-Pro-Arg-Gly at positions 94–97 between α1 and β3 establishes strong contacts with the loop β1′–β2′ of the other monomer.
The overall structure of TcPRACA is similar to those of Haemophilus influenzae diaminopimelate epimerase DapF [Protein Data Bank (PDB) ID codes 1BWZ and 1GQZ] and two proteins of unknown function, Escherichia coli YddE (PDB ID codes 1QYA and 1SDJ) and Brucella melitensis Lr31 (PDB ID code 1TM0). DapF and YddE are homologous (20) and most probably share a similar catalytic mechanism with proline racemase, albeit with a different substrate specificity (10, 21). TcPRACA residues Cys-130 and Cys-300 match the two catalytic Cys residues of DapF, and 60–80% of secondary structural elements are conserved (overall rms deviations of 2.8–3 Å) despite a low sequence identity (Fig. 7, which is published as supporting information on the PNAS web site).
The Active Site and Catalytic Mechanism.
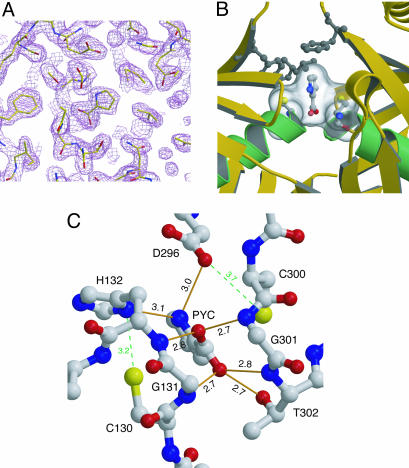
The inhibitor PYC is well defined in the electron density map (Fig. 2A). PYC binds to a completely buried, mostly hydrophobic, cavity in the center of each monomer (Figs. 2 A and B), explaining why the enzyme can only be active on free l/d-prolines. Moreover, no solvent channel is observed to connect the ligand-containing pocket to the bulk solvent, suggesting that the monomer must undergo a major structural rearrangement for substrate access and release (see below). The pyrrole ring can be modeled in either of two possible orientations 180° apart, which cannot be resolved from the diffraction data alone. The proposed orientation was chosen to satisfy the H-bonding capacity of the pyrrole nitrogen atom (N6), making possible hydrogen-bonding contacts with His-132-Nδ1 and Asp-296-Oδ2 (Fig. 2C) and to account for hydrophobic contacts of the ring carbon atoms with aromatic (Phe-102 and Phe-290) and aliphatic (Leu-127) amino acid side chains (Fig. 2B). The carboxylate group of the ligand is strongly fixed in place by five hydrogen bonds (Fig. 2C), suggesting a tetrahedral configuration for the carboxylate oxygens.
Fig. 2.
Protein–inhibitor interactions. (A) Electron density map (2mFo − DFc, contoured at 1σ) showing the inhibitor PYC bound to the active site cleft. (B) PYC and the two catalytic Cys residues are shown in the reaction center in ball-and-sticks representation colored by atom. The solvent-accessible surface area of PYC and both Cys residues is represented as a transparent surface, highlighting the strong interaction. Hydrophobic residues Phe-102, Phe-290, and Leu-127 making contacts with the pyrrole cycle of the inhibitor are shown in gray. (C) Protein–inhibitor hydrogen-bonding interactions. Two protein–protein H bonds proposed to be important for catalysis (Cys-130 SH–His-132 Nδ1 and Cys-300 SH–Asp-296 Oδ2) are highlighted in green.
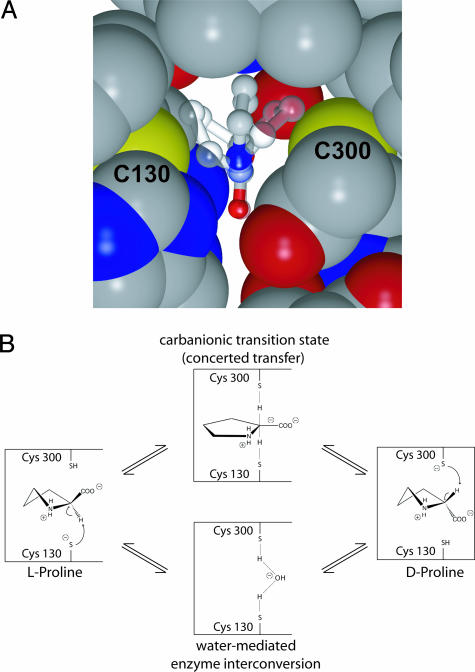
Stereoinversion of the chiral Cα center in proline racemization involves the abstraction of a proton from the carbon atom and reprotonation of the resulting carbanionic intermediate in the opposite stereochemical sense. In the crystal structure of the enzyme-inhibitor complex, the C2 atom of PYC (equivalent to the Cα atom of proline) is in close contact with the SH groups of residues Cys-130 and Cys-300 from equivalent α-helices in each structural domain (Fig. 2B). The sulfur–carbon distances of 3.4–3.5 Å on either side of the pyrrole plane reveal that the two Cys residues sterically favor a planar conformation of the chiral center (Fig. 3A) and are therefore optimally placed to abstract and donate a proton in the two-base mechanism (Fig. 3B) proposed for PLP-independent amino acid racemases (14, 15, 22). In full agreement with the involvement of the two Cys residues in catalysis, the point mutants TcPRACAC130S and TcPRACAC300S show marginal or undetectable racemase activity using l/d-prolines as substrates in polarimetric assays (17) under different buffer and pH conditions (Fig. 8, which is published as supporting information on the PNAS web site).
Fig. 3.
The catalytic mechanism of TcPRACA. (A) Close view of the active site showing the steric hindrance between the protein sulfur atoms (in Corey–Pauling–Koltun representation) and the L- and D-proline substrates (in transparent stick-and-ball representation). The planar inhibitor is also shown. (B) Two-base catalytic mechanism of proline racemases. Cys-130 and Cys-300 are responsible for the deprotonation of L- and D-proline, respectively. The reaction may proceed either through a discrete carbanionic intermediate or by concerted proton transfer and a carbanion-like transition state.
Added to the steric effect of the two catalytic Cys residues on C2 (Fig. 3A), the geometry of the protein–ligand hydrogen-bonding interactions (Fig. 2C) further supports a mechanism of negative charge delocalization of the prochiral carbanion species. The carboxylate group of PYC is engaged in five strong H bonds with protein atoms (donor/acceptor distances of 2.6–2.9 Å), which induce a distortion on the geometry of the carboxylate oxygen atoms from the expected sp2 orbital arrangement toward a tetrahedral configuration.
Conformational Changes of TcPRACA upon Inhibitor Binding.
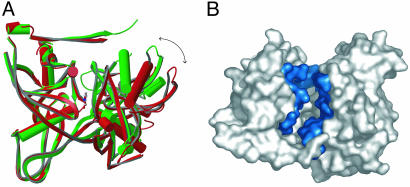
Although we were unable to grow well diffracting crystals of TcPRACA in the absence of ligand, the enzyme was cocrystallized at a lower PYC concentration (0.1 mM) and a complete x-ray diffraction data set could be collected at 2.5-Å resolution. The crystal structure (Table 1) revealed a dimeric enzyme with a single binding site occupied by the inhibitor. Although the PYC-bound monomer is very similar to those in the fully occupied dimeric species, the inhibitor-free monomer shows an open catalytic crevice with a communicating channel connecting the catalytic site to the outside bulk solvent (Fig. 9, which is published as supporting information on the PNAS web site). Optimal structural superposition between the PYC-bound and free monomers requires a rigid-body rotation (17°) of one domain with respect to the other (Fig. 4A). This closure movement induced by inhibitor binding buries 1, 100 Å2 of molecular surface (Fig. 4B) and stabilizes the conformation of the loop containing Cys-130, which is highly mobile in the ligand-free enzyme, with large temperature factors and possible alternative conformations.
Fig. 4.
Conformational changes in TcPRACA upon inhibitor binding. (A) Structural superposition of the crystal structures in the absence (red, open form) and presence (green, closed form) of inhibitor (PYC drawn in ball-and-stick representation). The inhibitor is completely buried within the monomer in the closed form. (B) Solvent-accessible surface area of the open (inhibitor-free) monomer colored on the residues that show a significant increase in solvent exposure when compared with the inhibitor-bound form. The active site is connected to the bulk solvent, allowing substrate access and release.
Thermodynamic Characterization of TcPRACA–PYC Interactions.
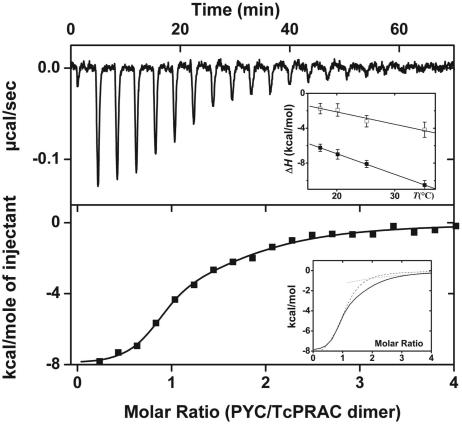
Isothermal titration calorimetry studies (Fig. 5) confirm that the homodimer binds two PYC molecules in solution. The binding isotherms could only be fitted to a model with two interacting sites. Their corresponding binding affinities (K1 = 4.6 × 106 M−1, K2 = 3.0 × 105 M−1) reveal a large negative cooperativity between sites (K1/K2 = 15), which may account for the previously reported stoichiometry of one inhibitor molecule per homodimer in the C. sticklandi enzyme (14). Both association reactions are enthalpy- and entropy-favored, with sizable negative heat capacity changes upon binding (ΔCp), characteristic of large hydrophobic contributions (Fig. 5 Inset). The two binding reactions are of different thermodynamic nature because binding of the first and second PYC molecules are driven by enthalpy (ΔH1°/ΔG1° = 88.5%) and entropy (ΔH2°/ΔG2° = 42.6%), respectively. These results are consistent with the protein undergoing significant conformational changes upon binding in solution. Furthermore, differential scanning calorimetry measurements indicate that TcPRACA exists as a monomer/dimer equilibrium in dilute solution and has a propensity to form upper oligomeric species at higher concentrations, whereas PYC binding is observed to stabilize the dimeric species and to reduce the oligomerization propensity of the protein (data not shown).
Fig. 5.
Calorimetric titration of TcPRACA with PYC at 25°C. (Upper) The heat signal for injections of 10 μl of a solution of 50 mM sodium acetate, pH 5.2, containing 58 μM PYC into a reaction cell containing 1.34 ml of the same buffer with 2.2 μM TcPRACA dimer. The data are shown after substration of the heat of dilution of PYC. (Lower) The integrated heat of injection normalized to the amount of PYC injected (■). The curve through the points represents the best fit to a model with two interacting sites. The parameters defining the fitted curve are K1 = (4.6 ± 0.6) × 106 M−1, ΔH1° = −8.1 ± 0.4 kcal·mol−1, and TΔS1° = 1.1 ± 0.5 kcal·mol−1, and K2 = (3.0 ± 1.6) × 105 M−1, ΔH2° = −3.2 ± 0.8 kcal·mol−1, and TΔS2° = 4.3 ± 1.2 kcal·mol−1. (Upper Inset) The enthalpy changes, ΔH1° (■) and ΔH2° (□), as a function of temperature for the same set of experimental conditions. Straight lines represent linear regression analyses of the data with slope of lines ΔCp, 1 = −237 ± 36 cal·mol−1·K−1 and ΔCp, 2 = −142 ± 30 cal·mol−1·K−1. (Lower Inset) The best fit to the experimental binding isotherm (thick line) and the calculated isotherms for the first (dashed line) and second (dotted line) binding reactions.
Dissociation of TcPRACA Enzymatic and Mitogenic Activities.
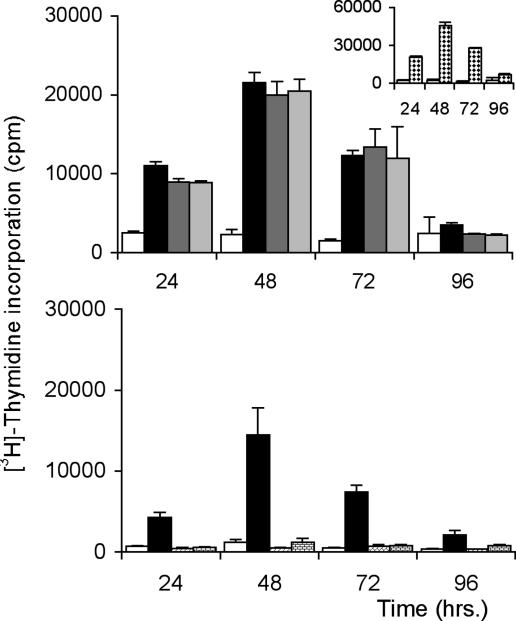
The Cys–Ser amino acid substitutions abolish the catalytic activity, but have little or no effect on the mitogenic activity of B lymphocytes in vitro and in vivo (Fig. 6A). As described before, B cell proliferative activity after wild-type TcPRACA stimulation in vitro is time-dependent and maximized after 48 h of culture, and thymidine uptake is comparable whether TcPRACAC300S or TcPRACAC130S is used. Furthermore, the number of Ig-secreting splenic B cells in mice after i.p. injection of the wild-type or mutant enzymes are enhanced 2- to 4-fold (depending on the class of Ig) compared with naïve mice without particular increase in total spleen cell numbers (Table 2, which is published as supporting information on the PNAS web site). In contrast, in vitro assays of B cell proliferation with TcPRACA either untreated or preincubated with PYC reveals that inhibitor binding renders the enzyme unable to trigger B cell-proliferative activity (Fig. 6B). Based on the structural information, these data strongly suggest that the ligand-induced protein closure movement precludes the interaction of TcPRACA with B cell receptor molecules. Thus, enzyme-triggered polyclonal B cell activation is not directly associated with the enzymatic activity but might depend on conformational epitopes displayed by the enzyme in its open (unliganded) form.
Fig. 6.
Lymphocyte proliferation assays of the wild-type and mutant forms of TcPRACA. (Upper) The mitogenic activity of TcPRACA is preserved after mutation of the catalytic Cys residues. The plot shows the incorporation of DNA precursors by splenic BALB/c (nu−/nu−) cells throughout time. Splenocytes obtained from T cell immunodeficient nude (nu−/nu−) BALB/c mice were cultured in the presence or absence of TcPRACAC300S or TcPRACAC130S proteins and compared with B cell proliferation obtained with the wild-type enzyme or lipopolysaccharide- and ConA-unrelated B and T cell mitogens as controls. Cells were cultured with medium alone (white bars), wild-type TcPRACA (black bars), TcPRACAC130S (dark gray bars), or TcPRACC300S (light gray bars). (Inset) The [3H]thymidine incorporation of splenic nu−/nu− cells after stimulation with lipopolysaccharide B cell mitogen. (Lower) The mitogenic activity of TcPRACA is lost after incubation of the protein with PYC. The plot shows the incorporation of DNA precursors by splenic BALB/c (nu−/nu−) cells throughout time. [3H]Thymidine incorporation is expressed in cpm. Cells were cultured with TcPRACA alone (black bars) and after specific enzyme inhibition performed by previous incubation of the protein with PYC (hatched bars). Controls included cells incubated with medium alone (white bars) or after protein denaturation by heating (dotted bars). [3H]Thymidine incorporation by cells incubated with PYC alone are below 400 cpm (data not shown).
Discussion
In this paper we report the structure of T. cruzi proline racemase (16), an enzyme whose bacterial analog has been extensively studied as a model for PLP-independent mechanism during the last 40 years (14, 15, 22). The overall structure of TcPRACA is similar to that originally found for diaminopimelate epimerase DapF (10). In the two enzymes, the monomer folds into two structurally equivalent domains (23) with undetectable sequence similarity. The symmetry operator relating both domains in TcPRACA is a dyad axis that passes through the reaction center, accounting for the ability of a chiral enzyme to cope with both enantiomeric forms of its substrate (23, 24). The overall structure of two other PLP-independent enzymes, aspartate and glutamate racemase (13, 25), differs from that of TcPRACA and DapF, but they also operate through a similar two-Cys-based acid/base mechanism for catalysis (12, 26). However, to our knowledge, direct structural evidence of a functionally competent enzyme–substrate (or substrate analog) complex was missing for PLP-independent racemases because the only structural data available showed the weak competitive inhibitor d-Gln (Ki = 50 mM) bound to glutamate racemase in a nonfunctional mode (25).
The structure of TcPRACA in complex with the strong competitive inhibitor PYC directly confirms that Cys-130 and Cys-300 are correctly positioned to perform acid/base catalysis on the chiral center of l/d-proline. PYC is strongly fixed in place through H-bonding interactions of its carboxylate group with protein atoms (Fig. 2C). The hypothesis on low-barrier H bonds contributing to stabilize the deprotonated carbanionic transition-state species (7) does not seem to hold in this case. Instead, the geometry of the five H bonds fixing the carboxylate group of PYC indicates a forced tetrahedral configuration for both carboxylate oxygens. This strained architecture suggests an alternative carbanion stabilization mechanism. With the oxygens in a tetrahedral configuration, the resonant double bond π electron of the carboxylates carbon can now establish a C(sp2))–Cα(sp2) π-bond, contributing to the planarity of the chiral Cα and delocalizing its transitory negative charge (7). Furthermore, the close contacts of the catalytic Cys residues with the PYC prochiral Cα center (Fig. 3A) introduce a second stabilizing force for the transition-state species, sterically forcing the tetrahedral chiral center in l/d-proline to a planar configuration. Both Cys residues are assisted by neighbor residues (His-132 and Asp-296) to perform alternate acid and base functions on the α-proton (Fig. 2C). The protonated state of the assistant residues can be thought to cyclically change the effective pKa of the catalytic Cys residues to alternately favor a thiol or thiolate state. Similar Cys-assistant residues (His-186 and Asp-10) have indeed been identified in glutamate racemase (27), where their substitution leads to drastic reductions in kcat and introduces an asymmetric preference for the substrate stereoisomer.
In previous work, we have shown that the mitogenic property of TcPRACA toward host B cells was fully compromised by prior incubation of the protein with specific and nonspecific inhibitors (16, 17), suggesting an intriguing link between the mitogenic and catalytic activities of the protein. We demonstrate here that this association is indirect given that the substitution of either catalytic residue (Cys-130 or Cys-300) abrogates the enzymatic activity but still preserves the full capacity of the protein to trigger B cell proliferation, as demonstrated by in vitro and in vivo assays.
In contrast, the structural analysis of TcPRACA revealed that the enzyme undergoes significant conformational changes upon inhibitor binding, switching from an open state in the ligand-free protein to a closed state in the enzyme-inhibitor complex. As a result of this closure movement, a large accessible surface area in the free enzyme (≈1, 100 Å2) is occluded from the bulk solvent in the complex (Fig. 4B). Consistent with calorimetric measurements in solution, this conformational flexibility is directly correlated with the mitogenic activity of the protein (Fig. 6B), strongly suggesting a plausible model to account for B cell triggering, as suggested for other B cell activators (28). Thus, the B cell mitogenic effects of TcPRACA would depend on exposed conformational epitopes in the ligand-free protein, which become partially or totally unavailable upon formation of the enzyme-inhibitor complex. In agreement with this model, incubation of TcPRACA with different concentrations of substrates (l/d-proline) decreases but does not eliminate the mitogenic activity of the protein, whereas formation of the more stable enzyme-inhibitor complex completely abrogates B cell proliferation. Because TcPRACA is involved in immune evasion and parasite infectivity, the present findings set a firm basis for further studies of B cell receptor–protein interactions that may uncover opportunities for therapeutic intervention in Chagas disease.
Methods
Protein Production and Purification.
The protein TcPRACA was produced in E. coli BL21 (DE3) (Invitrogen) and purified with immobilized metal affinity chromatography on nickel columns as described in ref. 17. The active peak was further submitted to gel filtration chromatography at 2.5 ml/min in a Superdex200 26/60 column (Amersham Pharmacia Biosciences) previously equilibrated in 200 mM sodium acetate, pH 5.2. The protein was concentrated to 6–7 mg/ml in 25 mM sodium acetate, pH 5.2. The selenomethionine-labeled protein was produced in B834(DE3) E. coli cells (Novagen) as described in ref. 29. Recombinant protein expression and purification were achieved as described before for unlabeled TcPRACA.
Site-directed mutagenesis to obtain the point mutants TcPRACAC130S and TcPRACAC300S was performed as described in ref. 17 or by using a QuikChange XL kit (Stratagene). The numbering convention used in this paper starts with an added Met before position 32 of the full-length ORF. The mutants were produced as described above for the wild-type enzyme and behave as the nonmodified enzyme in SDS/PAGE, immunoblotting with polyclonal anti-TcPRACA antibodies, dynamic light scattering, and PYC-binding as assayed by isothermal titration calorimetry (data not shown).
Crystallization.
Initial crystallization conditions of TcPRACA were found by using a hanging-drop-based sparse-matrix screening strategy. In the absence of PYC, crystals were typically smaller and diffracted poorly. The best crystals were obtained by mixing 2–3 μl of the protein solution (preincubated with different concentrations of PYC) with an equal volume of crystallization buffer (0.1 M ammonium acetate/50 mM tri-sodium citrate dihydrate, pH 5.6/15% wt/vol polyethylene glycol 4000), equilibrated over 1 ml of the same buffer. These crystals are monoclinic, space group C2, and contain a homodimer in the asymmetric unit.
Crystallographic Studies.
x-ray diffraction data sets were collected from frozen single crystals at the European Synchrotron Radiation Facility (Grenoble, France, beamlines BM14 and ID29) and processed with the programs mosflm, scala, and truncate from the ccp4 program suite (30) (Table 1). The structure was solved by using a single-wavelength anomalous diffraction experiment from a highly redundant 3-Å data set of the selenomethionine-labeled protein. Twenty-two of 24 Se sites were found by direct methods with the program shake‘n′bake (31) and refined with the program sharp (32). Electron density modification was performed with solomon (33); chain tracing and model building were done with the program o (34). Initial refinement was performed with cns (35) by using simulated annealing in torsional space. Iterative model rebuilding and refinement was further performed by using o and cns, and, at the final stages, refinement was completed with the program refmac5 (30), using a translation/libration/screw model against the native data set at 2.1-Å resolution. In the final model, the first 41 residues of the protein and the last 13 (monomer A) or 17 (monomer B), which include the affinity tag, are not visible in the electron density map. The structure of a second crystal of TcPRACA, incubated at low PYC concentration (0.1 mM PYC) (PYC hemicomplex data set in Table 1), was solved by molecular replacement and refined as described above. In this structure, one of the monomers shows nine extra residues in the N terminus, which define an additional α-helix.
B Cell Mitogenic Activity Assays.
In vitro proliferation was done by using freshly recovered splenocytes from BALB/c nu/nu− mice purchased from Charles River Breeding Laboratories seeded at a concentration of 2 × 105 cells per well and incubated with increasing concentrations of recombinant TcPRACA or mutated proteins (25–100 μg/ml), or with lipopolysaccharide–B cell mitogen (5 μg/ml) for 24, 48, 72 and 96 h in 5% FCS–RPMI medium 1640. Analysis of proliferative activity in the presence of 50 μg/ml of enzymatically active TcPRACA was compared with the proliferation obtained by using the same amounts of enzyme preincubated for 10 min at 37°C with 1 mM PYC. A 16-h [3H]thymidine pulse at 1–4 μCi (1 Ci = 37 GBq) per well was performed before harvesting the cultures. [3H]Thymidine uptake was determined in a β-plate liquid scintillation counter (LKB–Wallac). All points were done in triplicate, and the corresponding SD was calculated. Results are representative of five independent experiments. In vivo assays were performed after 0, 5, and 7 days of i.p. injection of wild-type or mutant TcPRACA (50 μg per mouse) proteins. The total number of cells or Ig-producing B cells was determined as described in ref. 36.
Microcalorimetry.
The association reaction of TcPRACA with PYC was quantified by isothermal titration calorimetry using an isothermal titration microcalorimeter from Microcal (Amherst, MA) and the protocols described in ref. 37. Briefly, TcPRACA stock solution dialyzed overnight at 4°C against 50 mM sodium acetate buffer, pH 5.2, was diluted (1.7–21.8 μM) with the same buffer. The protein samples were titrated in the calorimeter cell (1.4 ml) by successive 10-μl injections of 40–800 μM PYC in the same buffer until a final inhibitor/enzyme molar ratio of 4:1. Heat signals were corrected for the heat of dilution of PYC and normalized to the concentration of PYC added. Normalization and deconvolution of the binding isotherms was carried out with the program origin 7 (38) provided by the manufacturer. Thermodynamic parameters are the mean of three experiments; error estimations are the SDs from mean values. The molar change in heat capacity (ΔCp) was determined according to the equation
where ΔH°(TR) is the molar enthalpy change at TR, the reference temperature, taken as 298.15 K.
Supplementary Material
Acknowledgments
We thank S. Petres and L. de Mendonça Lima for assistance with protein purification and A. Waters, J. C. Barale, Y. Janin, and P. E. Bost for helpful discussions. This work was supported by grants from the Institut Pasteur and Centre National de la Recherche Scientifique. M.G. is a fellow at the Minister̀e de la Recherche et de la Technologie of France.
Abbreviations
- PLP
pyridoxal phosphate
- PYC
pyrrole-2-carboxylic acid
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 1W61 and 1W62).
References
- 1.Thompson R. J., Bouwer H. G., Portnoy D. A., Frankel F. R. Infect. Immun. 1998;66:3552–3561. doi: 10.1128/iai.66.8.3552-3561.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Caceres N. E., Harris N. B., Wellehan J. F., Feng Z., Kapur V., Barletta R. G. J. Bacteriol. 1997;179:5046–5055. doi: 10.1128/jb.179.16.5046-5055.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee J. A., Long Z., Nimura N., Iwatsubo T., Imai K., Homma H. Arch. Biochem. Biophys. 2001;385:242–249. doi: 10.1006/abbi.2000.2163. [DOI] [PubMed] [Google Scholar]
- 4.Long Z., Lee J. A., Okamoto T., Sekine M., Nimura N., Imai K., Yohda M., Maruyama T., Sumi M., Kamo N., et al. Biochem. Biophys. Res. Commun. 2001;281:317–321. doi: 10.1006/bbrc.2001.4353. [DOI] [PubMed] [Google Scholar]
- 5.Hashimoto K., Fukushima T., Shimizu E., Okada S., Komatsu N., Okamura N., Koike K., Koizumi H., Kumakiri C., Imai K., Iyo M. Prog. Neuropsychopharmacol. Biol. Psychiatry. 2004;28:385–388. doi: 10.1016/j.pnpbp.2003.11.009. [DOI] [PubMed] [Google Scholar]
- 6.Tanner M. E., Kenyion G. L. In: Comprehensive Biological Catalysis: A Mechanistic Reference. Sinnott M., editor. Vol. II. San Diego: Academic; 1997. pp. 7–42. [Google Scholar]
- 7.Cleland W. W. In: Comprehensive Biological Catalysis: A Mechanistic Reference. Sinnott M., editor. Vol. II. San Diego: Academic; 1998. pp. 1–6. [Google Scholar]
- 8.Tanner M. E. Acc. Chem. Res. 2002;35:237–246. doi: 10.1021/ar000056y. [DOI] [PubMed] [Google Scholar]
- 9.Sun S., Toney M. D. Biochemistry. 1999;38:4058–4065. doi: 10.1021/bi982924t. [DOI] [PubMed] [Google Scholar]
- 10.Cirilli M., Zheng R., Scapin G., Blanchard J. S. Biochemistry. 1998;37:16452–16458. doi: 10.1021/bi982138o. [DOI] [PubMed] [Google Scholar]
- 11.Finlay T. H., Adams E. J. Biol. Chem. 1970;245:5248–5260. [PubMed] [Google Scholar]
- 12.Gallo K. A., Tanner M. E., Knowles J. R. Biochemistry. 1993;32:3991–3997. doi: 10.1021/bi00066a020. [DOI] [PubMed] [Google Scholar]
- 13.Liu L., Iwata K., Kita A., Kawarabayasi Y., Yohda M., Miki K. J. Mol. Biol. 2002;319:479–489. doi: 10.1016/S0022-2836(02)00296-6. [DOI] [PubMed] [Google Scholar]
- 14.Rudnick G., Abeles R. H. Biochemistry. 1975;14:4515–4522. doi: 10.1021/bi00691a028. [DOI] [PubMed] [Google Scholar]
- 15.Cardinale G. J., Abeles R. H. Biochemistry. 1968;7:3970–3978. doi: 10.1021/bi00851a026. [DOI] [PubMed] [Google Scholar]
- 16.Reina-San-Martin B., Degrave W., Rougeot C., Cosson A., Chamond N., Cordeiro-Da-Silva A., Arala-Chaves M., Coutinho A., Minoprio P. Nat. Med. 2000;6:890–897. doi: 10.1038/78651. [DOI] [PubMed] [Google Scholar]
- 17.Chamond N., Gregoire C., Coatnoan N., Rougeot C., Freitas-Junior L. H., da Silveira J. F., Degrave W. M., Minoprio P. J. Biol. Chem. 2003;278:15484–15494. doi: 10.1074/jbc.m210830200. [DOI] [PubMed] [Google Scholar]
- 18.Reina-San-Martin B., Cosson A., Minoprio P. Parasitol. Today. 2000;16:62–67. doi: 10.1016/s0169-4758(99)01591-4. [DOI] [PubMed] [Google Scholar]
- 19.Chamond N., Goytia M., Coatnoan N., Barale J. C., Cosson A., Degrave W. M., Minoprio P. Mol. Microbiol. 2005;58:46–60. doi: 10.1111/j.1365-2958.2005.04808.x. [DOI] [PubMed] [Google Scholar]
- 20.Grassick A., Sulzenbacher G., Roig-Zamboni V., Campanacci V., Cambillau C., Bourne Y. Proteins. 2004;55:764–767. doi: 10.1002/prot.20025. [DOI] [PubMed] [Google Scholar]
- 21.Lloyd A. J., Huyton T., Turkenburg J., Roper D. I. Acta Crystallogr. D. 2004;60:397–400. doi: 10.1107/S0907444903027999. [DOI] [PubMed] [Google Scholar]
- 22.Albery W. J., Knowles J. R. Biochemistry. 1986;25:2572–2577. doi: 10.1021/bi00357a043. [DOI] [PubMed] [Google Scholar]
- 23.Liu L., Iwata K., Yohda M., Miki K. FEBS Lett. 2002;528:114–118. doi: 10.1016/s0014-5793(02)03264-7. [DOI] [PubMed] [Google Scholar]
- 24.Lamzin V. S., Dauter Z., Wilson K. S. Curr. Opin. Struct. Biol. 1995;5:830–836. doi: 10.1016/0959-440x(95)80018-2. [DOI] [PubMed] [Google Scholar]
- 25.Hwang K. Y., Cho C. S., Kim S. S., Sung H. C., Yu Y. G., Cho Y. Nat. Struct. Biol. 1999;6:422–426. doi: 10.1038/8223. [DOI] [PubMed] [Google Scholar]
- 26.Tanner M. E., Gallo K. A., Knowles J. R. Biochemistry. 1993;32:3998–4006. doi: 10.1021/bi00066a021. [DOI] [PubMed] [Google Scholar]
- 27.Glavas S., Tanner M. E. Biochemistry. 2001;40:6199–6204. doi: 10.1021/bi002703z. [DOI] [PubMed] [Google Scholar]
- 28.Karpusas M., Cachero T. G., Qian F., Boriack-Sjodin A., Mullen C., Strauch K., Hsu Y. M., Kalled S. L. J. Mol. Biol. 2002;315:1145–1154. doi: 10.1006/jmbi.2001.5296. [DOI] [PubMed] [Google Scholar]
- 29.Wingfield P. T. In: Current Protocols in Protein Science. Coligan J. E., Dunn B. M., Ploegh H. L., Speicher D. W., Wingfield P. T., editors. Vol. 1. New York: Wiley; 2000. pp. 5.3.9–5.3.14. [Google Scholar]
- 30.Collaborative Computational Project Number 4. Acta Crystallogr. D. 1994;50:760–763. [Google Scholar]
- 31.Weeks C. M., Miller R. Acta. Crystallogr. D. 1999;55:492–500. doi: 10.1107/s0907444998012633. [DOI] [PubMed] [Google Scholar]
- 32.Bricogne G., Vonrhein C., Flensburg C., Schiltz M., Paciorek W. Acta Crystallogr. D. 2003;59:2023–2030. doi: 10.1107/s0907444903017694. [DOI] [PubMed] [Google Scholar]
- 33.Abrahams J. P., Leslie A. G. W. Acta Crystallogr. D. 1996;52:30–42. doi: 10.1107/S0907444995008754. [DOI] [PubMed] [Google Scholar]
- 34.Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. Acta Crystallogr. A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 35.Brunger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J.-S., Kuszewski J., Nilges M., Pannu N. S., et al. Acta Crystallogr. D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 36.Minoprio P. In: Molecular Mechanisms of Pathogenesis in Chagas Disease. Kelly J. M., editor. New York: Kluwer Academic/Plenum; 2003. pp. 101–110. [Google Scholar]
- 37.Schaeffer F., Matuschek M., Guglielmi G., Miras I., Alzari P. M., Beguin P. Biochemistry. 2002;41:2106–2114. doi: 10.1021/bi011853m. [DOI] [PubMed] [Google Scholar]
- 38.Wiseman T., Williston S., Brandts J. F., Lin L. N. Anal. Biochem. 1989;179:131–137. doi: 10.1016/0003-2697(89)90213-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.