Abstract
Enteropathogenic Escherichia coli (EPEC) induces a severe watery diarrhea responsible for several hundred thousand infant deaths each year by a process correlated with the loss (effacement) of absorptive microvilli. Effacement is linked to the locus of enterocyte effacement pathogenicity island that encodes an “injection system,” “effector” proteins, and the Intimin outer membrane protein. Here, we reveal that effacement (i) is a two-step process, (ii) requires the cooperative action of three injected effectors (Map, EspF, and Tir) as well as Intimin, and (iii) leads to the retention, not release (into the extracellular milieu), of the detached microvillar material. We also discover that EPEC rapidly inactivates the sodium-d-glucose cotransporter (SGLT-1) by multiple mechanisms. Indeed, the finding that one mechanism occurs more rapidly than microvilli effacement provides a plausible explanation for the rapid onset of severe watery diarrhea, given the crucial role of SGLT-1 in the daily uptake of ≈6 liters of fluids from the normal intestine. The importance of SGLT-1 in the disease process is supported by severe EPEC diarrheal cases being refractory to oral rehydration therapy (dependent on SGLT-1 function). Moreover, the identification of effector activities that alter microvilli structure and SGLT-1 function provides new tools for studying the underlying regulatory processes.
Keywords: Caco-2, diarrhea, effacement, SGLT-1
Enteropathogenic Escherichia coli (EPEC) is the prototypical member of the attaching and effacing family of pathogens that infect a number of mammalian species, including humans (EPEC and enterohemorrhagic Escherichia coli), rabbits, dogs, mice, cattle, and sheep (1, 2). EPEC-mediated disease has been correlated for more than four decades with intimate binding to enterocytes lining the small intestine, loss (effacement) of absorptive microvilli, and recruitment of host cytoskeletal proteins beneath the adherent bacteria: the attaching and effacing lesion (1–3). These processes are mediated by the EPEC locus of enterocyte effacement (LEE) pathogenicity island because transfer of this ≈35-kb region into nonpathogenic E. coli strains confers the ability to induce such changes (4). The LEE encodes transcriptional regulators, a type three secretion system (T3SS), translocators, injected “effectors,” chaperones, and the Intimin outer membrane protein (1, 2, 5). The T3SS/translocators function together to deliver both LEE- and non-LEE-encoded proteins into the host cell.
The T3SS/translocator system, Intimin, and Tir are all essential virulence determinants presumably because of their roles in intimate adherence, a process that requires the T3SS-dependent insertion of Tir into the plasma membrane to act as a receptor for bacterial binding via Tir-Intimin interaction (6–8). However, Tir–Intimin interaction also triggers signaling cascades leading to responses, including phosphorylation of a host phospholipase and recruitment of cytoskeletal proteins beneath the adherent bacteria (5, 9). Intimin can also subvert cellular processes independently of Tir (5). The LEE encodes five additional effectors, namely Map, EspF–EspH, and EspZ (5, 10), although the translocator proteins may also have signaling functions (11). Although relatively little is known about the functions of these molecules in disease, the multifunctional nature of LEE effectors has been established, as has been their ability to act together in redundant, synergistic, and/or antagonistic mechanisms to alter host cellular processes (5,12–19).
A lack of success in identifying the LEE effectors responsible for effacement is indicative of (i) roles for unidentified effectors, (ii) signaling functions for the translocator proteins, and/or (iii) a requirement for multiple effector molecules in the process. In this study, scanning electron microscopy (SEM) analysis of differentiated Caco-2 cells (small intestinal epithelial model) infected with wild-type EPEC versus mutants deleted for one or more LEE-encoded effectors identified a two-step effacement process dependent on the cooperative action of four LEE proteins. The additional discovery that effaced microvilli material is not released into the extracellular milieu led to the development of a biochemical assay to monitor this event and in fact identified a potent diarrheagenic mechanism to possibly explain the rapid onset of severe watery diarrhea (20). The implications of the data to understanding both EPEC pathogenic and host cellular processes are discussed.
Results
Generation of a Biochemical Assay to Monitor EPEC T3SS-Dependent Effacement.
EPEC interaction with human small intestinal enterocytes is extensively modeled by using differentiated Caco-2 cells because they share many characteristics, including epithelial barrier function and absorptive microvilli (21). Routine evaluation of EPEC-induced effacement by SEM and transmission electron microscopy (4,22–24) is indicative of obstacles in generating more quantitative biochemical-based assays. This premise was supported by the inability to detect significant T3SS-dependent release of three microvillar-associated enzyme activities, including alkaline phosphatase (AP), after EPEC infection of Caco-2 cells (data not shown). This apparent paradox was resolved by confocal microscopic analysis, which consistently revealed the relocation of the majority of AP from the actin-rich brush border to a location above the adherent bacteria (Fig. 1A) in a T3SS-dependent manner (data not shown). This prompted the development of a quantitative assay based on the sodium-d-glucose cotransporter (SGLT-1), because its relocation from the apical surface membrane would prevent the import of glucose, or the nonmetabolizable analog α-methyl-d-glucopyranoside (AMG), because of the requirement of an intracellular sodium gradient (25). The appropriateness of this protein as an effacement marker was supported by its T3SS-dependent relocation, as with AP (Fig. 1A), to a position above the actin-rich brush border (Fig. 1B).
Fig. 1.
EPEC-induced relocation of microvillus material. Caco-2 cells were left uninfected or apically infected with EPEC for 2 h before fixing and staining DNA [blue, bacteria (arrows) and host nuclei], polymerized actin (red), and AP (A) or SGLT-1 (B; green, arrowheads) revealing the relocation of both AP and SGLT-1 from the actin-rich apical surface to a position above the adherent bacteria. Images are composites of sequential sections taken at 2-μm intervals, with Inset (A) showing a magnified view of the boxed region. Note the large intracellular pool of SGLT-1 that constitutes up to two-thirds of the cellular total (35) and the altered distribution after EPEC infection.
Measuring SGLT-1 cellular activity after 30-min synchronized infections with preactivated bacteria (induces T3SS) over a range of multiplicities of infection (moi) supported the use of this assay to monitor effacement. Thus, whereas no significant decrease in SGLT-1 activity was recorded with the espA (translocator defective) mutant, wild-type EPEC induced a dose-dependent decrease, with the highest moi practically abolishing this cellular activity (Fig. 2A) by a process linked to bacteria covering the entire apical surface (Fig. 2B). However, moi of ≈50:1 and ≈200:1 (bacteria/host cell) resulted in bacteria covering only ≈8% and 37% of the cell surface, respectively (Fig. 2B) but inhibited ≈25% and ≈85% of the SGLT-1 activity, respectively (Fig. 2A). This disproportionately high loss of cotransporter function at lower moi is indicative of the existence of an inhibitory mechanism that can rapidly inactivate SGLT-1 at sites distal to the adherent bacteria. Importantly, rapid inactivation of SGLT-1 would undoubtedly play a crucial role in a diarrheal disease because it is responsible for the daily uptake of ≈6 liters of fluid from the normal small intestine (26).
Fig. 2.
Rapid T3SS-dependent inactivation of SGLT-1. (A) Import of the nonmetabolizable glucose analog 14C-labeled AMG [in disintegrations per minute (DPM)] by SGLT-1 is inhibited by EPEC but not the effector delivery mutant espA, in a dose-dependent manner (n = 3; mean ± SE). Statistical significance (∗) between uninfected and indicated infected series (horizontal lines) determined by one-way ANOVA with a post hoc Tukey’s test revealing where significance lies. (B) Percentage of the apical surface covered by EPEC after a 30-min infection at the indicated moi (EPEC/host cell) quantified after SEM evaluation (n = 3; mean ± SE).
SEM analysis was used to investigate whether the disproportionately high loss of SGLT-1 activity at low bacterial dosages was linked to the ability of EPEC to efface microvilli from around the infection site (1, 2). This analysis revealed that microvilli damage was restricted to the area directly below (or in very close proximity to) the adherent bacteria at 30 min, with extensive loss of adjacent microvilli requiring a 1- to 2-h infection (Fig. 3). Thus, it appears that EPEC possesses an effacement-independent mechanism to rapidly inactivate SGLT-1. Furthermore, bacterial “sinking” into the brush border by 30 min, with loss of adjacent microvilli a later event, is indicative of a two-step effacement process (Fig. 3). Notably, centrifugation did not promote uniform extensive adherence because most cells displayed no bacteria, single bacteria, or small microcolonies 5 min after centrifugation (Fig. 3) despite the moi of ≈200:1.
Fig. 3.
Kinetics of EPEC-induced microvilli damage. SEM analysis reveals that centrifugation of preactivated EPEC (moi of ≈200:1) does not promote uniform extensive adherence to differentiated Caco-2 cells because most cells have no bacteria, single bacteria, or small microcolonies 5 min after centrifugation. However, by 30 min, small microcolonies are more evident, with bacteria appearing to sink (arrowheads) into the brush border. Importantly, loss of microvilli from around the infection site occurs at later times, with microvilli appearing to be pulled toward the bacteria (arrows). Extensive loss of microvilli from around the infection site is evident by 2 h with practically all microvilli lost by 4 h.
Role for Multiple LEE-Encoded Effectors in Microvilli Effacement and Rapid SGLT-1 Inactivation.
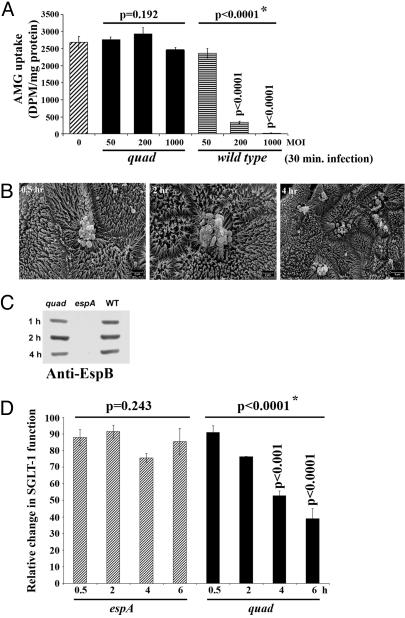
Additional 30-min infections with an available quadruple (quad) mutant (Map-, EspF-, Tir-, and Intimin-deficient) revealed the dependence of both rapid SGLT-1 inactivation and microvilli damage on the absent gene products. Thus, the quad mutant, unlike wild-type EPEC, failed to induce dose-dependent decreases in SGLT-1 activity (Fig. 4A) or significant microvilli damage (Fig. 4B). However, although this mutant was rarely detected on Caco-2 cells (Fig. 4B), it was as proficient as wild-type EPEC, unlike the espA translocator mutant, at delivering the EspB translocator/effector protein into the host cytoplasm (Fig. 4C). Thus, rapid abrogation of SGLT-1 activity and microvilli damage are both linked to the function of one or more of the missing gene products.
Fig. 4.
Microvilli effacement and rapid SGLT-1 inactivation are linked to the cooperative activities of Map, EspF, Tir, and Intimin. Differentiated Caco-2 cells were infected with wild-type EPEC (WT), quad mutant (Map/EspF/Tir/Intimin-deficient) or espA mutant (translocator defective) (moi of ≈200:1, unless otherwise indicated) for the stated times before processing for SGLT-1 activity (A and D), SEM (B), or Western blot analysis (C). Uptake of the nonmetabolizable glucose analog 14C-labeled AMG [in disintegrations per minute (DPM)] after 30-min infections reveal that wild-type EPEC, but not the quad mutant, triggers a dose-dependent loss of SGLT-1 activity (A), although prolonged quad infections induce a progressive effector delivery-dependent loss of SGLT-1 activity (D) (n = 3; mean ± SE). Statistical significance (∗) between uninfected and indicated infected sets (horizontal lines; A) or between indicated sets (D) determined by one-way ANOVA with a post hoc Tukey’s test revealing where significance lies. SEM reveals that the quad mutant rarely binds Caco-2 cells and does not induce significant microvillar damage (B). However, the ability of the quad, but not espA (translocator defective), mutant to deliver the EspB translocator/effector protein into host cells in similar amounts and at a comparable rate with the wild-type strain (C) indicates a role for one or more of the missing gene products in microvilli damage and rapid SGLT-1 inactivation.
Surprisingly, extended infections with the quad, but not espA (translocator defective), mutant triggered a slow progressive loss of SGLT-1 activity, revealing a T3SS-dependent inhibitory mechanism. This mechanism is rather minor because a significant decrease in SGLT-1 function is not evident until 4 h after infection (≈40%) (Fig. 4D), compared with the Map/EspF/Tir/Intimin-mediated inhibition in which ≈85% of SGLT-1 function is abolished during a 30-min infection (Fig. 2A). However, the identification of this additional mechanism provides an assay to define the responsible effector(s) and underlining inhibitory mechanism(s).
Cooperative Roles for Map, EspF, Tir, and Intimin in Rapid Loss of SGLT-1 Activity.
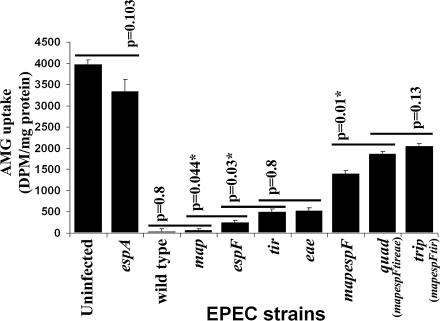
Infections with mutants deleted for individual effectors revealed only minor roles for EspF, Tir, and Intimin in inhibiting SGLT-1 activity, because cells infected with the espF, tir, and eae mutants retained ≈6%, ≈13%, and ≈13% of the cellular activity, respectively, compared with ≈1% for map-infected and wild-type EPEC-infected cells (Fig. 5). Similar values for tir and eae mutants (≈13%) is indicative of yet another Tir subversive activity dependent on previous Tir–Intimin interaction (5). This premise was supported by finding that the triple (trip; mapespFtir) and quad mutants, which differ only in Intimin expression, inhibited SGLT-1 activity to the same extent (≈50%) (Fig. 5). Moreover, tir and eae mutants inhibiting a much greater percentage of SGLT-1 activity compared with the trip/quad mutants implicated participating roles for Map and/or EspF. Indeed, redundant cooperative roles were uncovered by showing that a mapespF double mutant reduced SGLT-1 activity to ≈35% of total levels compared with ≈1% and ≈6% for map and espF mutants (Fig. 5). Thus, the rapid Map/EspF/Tir/Intimin-related inhibition of SGLT-1 activity involves the cooperative action of all four proteins.
Fig. 5.
Map, EspF, Tir, and Intimin function together to inhibit SGLT-1 function. Differentiated Caco-2 cells were infected for 6 h with wild-type EPEC, the espA translocator mutant, or strains deleted for one or more of the map, espF, tir, and eae (Intimin) genes before assessing SGLT-1 activity by measuring uptake of the nonmetabolizable glucose analog 14C-labeled AMG [in disintegrations per minute (DPM)]. This reveals (i) minor roles for EspF, Tir, and Intimin, (ii) redundant cooperative roles for Map and EspF, (iii) a role for Tir–Intimin interaction, and (iv) that all four proteins function together to inhibit SGLT-1 function (n = 3; mean ± SE). Statistical significance (∗) between indicated pairs (horizontal lines) determined by Student’s t test. Note the ≈50% loss of SGLT-1 activity with quad/trip mutant (differ only in Intimin expression) is due to a slow Map/EspF/Tir/Intimin-independent mechanism (Fig. 4D).
Uncoupling the Two-Step Effacement Process.
SEM analysis was used to investigate the relationship between the mapespF-mediated partial loss of SGLT-1 activity (Fig. 5) and microvilli damage. This analysis revealed similar binding kinetics to wild-type EPEC (Figs. 3 and 6A). However, bacterial sinking into the brush border of Caco-2 cells (Fig. 6A) or human gastrointestinal biopsy material (Fig. 6B) without inducing extensive damage to adjacent microvilli revealed essential roles for Map and EspF in extensive microvilli damage and that effacement was a two-step process. However, complementing this defect on Caco-2, but not biopsy material (data not shown), by plasmid expression of Map in the double mutant revealed important signaling differences between in vitro and in vivo differentiated cells. Moreover, the mapespF mutant defect in triggering extensive microvilli damage compared to the quad defects in this step, in addition to bacterial sinking into the brush border (Fig. 4), implies that Tir/Intimin function together in the bacterial sinking process.
Fig. 6.
Uncoupling the two-step effacement process. Centrifugation of the preactivated mapespF double mutant onto differentiated Caco-2 cells, like EPEC (Fig. 3), does not promote uniform extensive adherence despite the initial moi of ≈200:1 (EPEC/host cells). (A) Although adherence is once again linked to bacteria sinking into the brush border, the double mutant, unlike wild-type EPEC (Fig. 3), does not trigger extensive microvilli damage. (B) A similar defect is reproduced with human gastrointestinal biopsy tissue. Thus, effacement is a two-step effacement process in which Map and EspF function to induce the extensive loss of microvilli.
Discussion
One of the most perplexing and long-standing questions in EPEC pathogenesis relates to the mechanism by which this pathogen uses its T3SS system to trigger the loss of absorptive microvilli (effacement), a process linked to disease for more than four decades (1, 2). In this paper, we reveal, by using the Caco-2 small intestinal epithelial model system, that effacement is a two-step process that requires the activity of at least three injected effector proteins (Map, EspF, and Tir) and the Intimin outer membrane protein. The demonstrated redundant and cooperative nature of these molecules explains the previous lack of success in detecting roles for individual effectors in effacement using this model system. Although infection of biopsy material has indicated possible roles for these same proteins in effacement (22), its capacity to unearth the described underlying complexity is limited by the fact that (i) strains deficient for Tir or Intimin expression bind very poorly to biopsy material and induce little microvilli damage and (ii) a T3SS-defective mutant induces some microvilli damage (22). A suggestive role for EspH in effacement (22) highlights the possibility that other effectors may also participate in this process. However, the identified role of four multifunctional proteins (5) in effacement not only illustrates the complexity of pathogen–host interactions but also future obstacles to defining the contribution of specific effector functions and/or subverted cellular pathways in disease.
The ability of a plasmid expressing Map to complement the mapespF effacement defect on Caco-2, but not biopsy material, was suggestive of important signaling differences between in vitro and in vivo differentiated cells. This premise is supported by the detection of putative roles for individual effectors in effacement with biopsy material (22), unlike the Caco-2 model system (data not shown). This difference may provide an explanation as to why eae and tir mutants (i) show no effacement defect on Caco-2 cells (5), (ii) bind very poorly to biopsy material and induce little microvilli damage (22), and (iii) fail to bind or efface in natural animal infections (7, 24) if one assumes that Tir/Intimin-interaction is required to unleash the effacing activities of Map/EspF. Indeed, the dependence of specific effector functions on Intimin interaction with Tir or non-Tir molecules has been demonstrated (5, 8, 12, 15, 27). Moreover, we have shown that efficient effector delivery can occur independently of adherence (Fig. 4C), with this likely occurring in vivo because tir and eae mutants induce T3SS-dependent inflammatory responses (7, 24). Thus, the minor microvilli damage on biopsy material presumably reflects low EspF/Map activity in the absence of Tir or Intimin. In contrast, signaling differences within Caco-2 cells allow Map and EspF to function in a redundant manner and independently of Tir–Intimin.
Roles for Tir/Intimin in bacterial sinking into the brush border and Map/EspF in extensive microvilli damage identifies these molecules as novel tools to study the cellular pathways regulating these processes. Indeed, the absence of filamentous actin in the detached microvillar material (Fig. 1) suggests that the actin-rich microvilli structure is retracted or depolymerized before microvilli detachment. The ready detection of retained microvillar material by confocal (Fig. 1), but not electron microscopy, reveals a limitation of the current electron microscopy processing protocol in this respect and is suggestive of a mechanism to retain microvillar material. This prediction is supported by SEM, which frequently detects microvilli being “pulled” toward the adherent bacteria (Fig. 3). This retention process is apparently not restricted to the model system because EPEC infection of biopsy material resulted in the release of lower-than-expected levels of the microvilli marker AP (28, 29). Retention, even transiently, of microvilli material could play an important role in pathogenesis by perhaps delaying detection of EPEC by the host defense system and/or providing growth-promoting activities.
Importantly, discovering that microvillar material is in fact retained, not released, opened the way to develop assays to monitor the detachment process. Our choice of SGLT-1 proved to be excellent because (i) EPEC inhibited SGLT-1 activity in a T3SS-dependent manner, and (ii) SGLT-1 is the major water pump in the small intestine responsible for the daily uptake of ≈70% (≈6 liters) of fluid from the normal intestine (26). Indeed, the finding that EPEC rapidly inhibits the activity of the apical Na+/H+ exchanger NHE3 is consistent with our data (30) and is suggestive of a common inhibitory mechanism. Crucially, the capacity of EPEC to rapidly abolish SGLT-1 activity by a mechanism unlinked to microvilli effacement provides a potent diarrheagenic mechanism that could account for the rapid onset of severe watery diarrhea (20). SGLT-1 inactivation during EPEC infection is supported by severe diarrheal cases being refractory to oral rehydration therapy (1), a process that relies on the sodium/glucose cotransporter to correct electrolyte imbalance and promote fluid uptake (31). Indeed, the responsiveness of milder diarrheal cases to oral rehydration therapy (1) indicates that disease symptoms are related to the degree of SGLT-1 inactivation, a process that would depend on many factors, such as bacterial inoculum size, “infectivity” state (see below), host age, health status, and host/bacterial genotypes. Although EPEC undoubtedly subverts other cellular responses that can contribute to diarrhea, such as disruption of barrier function and immune response-triggered secretory responses (1, 5), these would be relatively late events compared with the rapid adherence-associated loss of SGLT-1 activity and thus likely to be secondary or additive mechanisms.
The finding that centrifugation does not promote uniform extensive bacterial adherence (Figs. 3 and 6A), with the kinetics of SGLT-1 inactivation only delayed by ≈60–90 min in the absence of preactivation and centrifugation (data not shown), illustrates that this in vitro data is very relevant to the in vivo situation. This premise is reinforced by the fact that experimental in vivo animal infection studies also (i) involve high bacterial inoculums with extensive binding to enterocytes (3, 7, 24, 32), and (ii) probably involve preactivated bacteria because signals within the gastrointestinal tract induce the T3SS (33), as well as a “hyperinfective,” presumably, preactivated state (32).
In conclusion, the dependence of the virulence-associated loss of microvilli on four LEE-encoded effectors not only illustrates the complexity of EPEC-host interactions but also the future challenge of deciphering the contribution of individual effector functions or subverted cellular process in disease. Identifying effector functions linked with (i) microvilli retraction, (ii) microvilli detachment, and (iii) rapid SGLT-1 inactivation (unlinked to microvilli effacement) reveals the responsible bacterial proteins as new tools to probe the cellular mechanisms regulating these host processes. Discovering that detached microvillar material is retained not only reveals limitations of current electron microscopic procedures but also opens the way to develop quantitative effacement assays. The presented data also identified an important difference between in vitro and in vivo differentiated cells and provides important insights into the infection process. Crucially, identifying that EPEC can rapidly inactivate the major water pump of the small intestine (SGLT-1) reveals a potent diarrheagenic mechanism to possibly explain the rapid onset of severe watery diarrhea (20).
Materials and Methods
Bacterial Strains and Plasmids.
EPEC E2348/69, map, espF, tir, and eae (Intimin) strains, as well as the construction of the Map-expressing plasmid, have been described or referenced within refs. 8, 12, and 27. Multiple mutants were generated by the sequential deletion of individual genes by allelic exchange by using pCVD442-based suicide vectors, as described or referenced within refs. 8, 12, and 27. Deletion of each gene was confirmed by PCR and Western blot analyses, with the latter also serving to confirm the expression and/or secretion of the Esp translocators, as well as nondeleted Map, EspF, Tir, and/or Intimin proteins.
Cultured Cells and Infection Protocol.
Caco-2 cells (American Type Culture Collection no. HTB-37) were cultured and infected as described in ref. 15 on 1.1-cm2 porous membrane filters (0.4-μm pore size; Corning Life Sciences) at a density of ≈5 × 105 cells per filter with media replaced every 2 days for use 20 days after seeding. Bacteria were pregrown in DMEM (Invitrogen) for 3 h without shaking to induce the effector delivery system. Culture OD600 (1 OD600 = ≈1 × 109 bacteria per ml) was measured and adjusted to the required bacterial moi before adding 0.5 ml to the cell monolayers, centrifuged (500 × g; 5 min), and incubated (37°C; 5% CO2) for the required time.
AMG Uptake Assay.
Measurement of SGLT-1-specific uptake of AMG was a modification of that described in ref. 34. The infected monolayers were washed four times with 37°C medium B (34) before incubating for 1 h at 37°C with medium B containing bactericidal levels of gentamycin (100 μg/ml final concentration). The apical medium was replaced with medium B containing 0.1 mM AMG for 10 min to block nonspecific AMG-binding sites before replacing with 0.1 mM AMG containing 0.15 μCi/ml 14C-labeled AMG (Amersham Biosciences; 1 Ci = 37Gbq) for 10 min. Duplicate experiments were performed in the presence of the SGLT-1-specific inhibitor phlorizin (0.2 mM final concentration; Sigma). AMG uptake was stopped by adding ice-cold medium B/0.2 mM phlorizin and washing five times with this solution before lysing in ice-cold 100 mM NaOH. Radioactivity levels (in disintegrations per minute) were measured in Ultima Gold scintillation mixture (PerkinElmer Life and Analytical Sciences) by using a Tricarb 2700TR scintillation analyzer (Packard). SGLT-1-mediated AMG uptake was the difference in radioactivity between phlorizin-treated and untreated cells.
Microscopy and Western Blot Analyses.
Standard techniques were used as described (15). Antibodies used were against intestinal AP (ab7322; Abcam, Cambridge, MA), SGLT-1 (Chemicon), and EspF and EspB (laboratory of B.K.) for detection with goat anti-rabbit fluorescein isothiocyanate-conjugated (Jackson ImmunoResearch) or AP-conjugated (Zymed) antibodies. Polymerized actin was labeled with tetramethylrhodamine isothiocyanate-conjugated phalloidin and DNA with 4′,6-diamidino-2-phenylindole (Vectashield mounting media; Vector Laboratories) (15). Western blot analysis (15) was with 20 μg of saponin released (0.1% wt/vol) host “cytoplasmic” protein fraction.
In Vitro Organ Culture.
Proximal small intestinal mucosal biopsies (fourth part duodenum) were obtained with fully informed parental consent and local ethical committee approval by using grasp forceps during routine endoscopic (EG/EC-41 pediatric endoscope; Fujinon, Tokyo, Japan) investigation of intestinal disorders. Macroscopically normal tissue samples from five patients (aged 63, 93, 142, 150, and 209 months) were infected for 8 h and analyzed as described (23).
Acknowledgments
We acknowledge excellent technical support from Sabine Quitard. Images were obtained at the Medical Research Council (United Kingdom) through supported facilities at the University of Bristol (Cell Imaging Facility, Department of Biochemistry) and the University of Newcastle (Biomedical Electron Microscopy Unit). We thank Profs. Nigel Robinson (University of Newcastle), Nick Simmons (University of Newcastle), and Barry Holland (Université Paris Sud) for their help with this manuscript. B.K. was funded by Wellcome Trust as a Senior Fellow in Basic Biomedical Sciences.
Abbreviations
- AMG
α-methyl-d-glucopyranoside
- AP
alkaline phosphatase
- EPEC
enteropathogenic Escherichia coli
- LEE
locus of enterocyte effacement
- moi
multiplicity of infection
- SEM
scanning electron microscopy
- SGLT-1
sodium-d-glucose cotransporter
- T3SS
type three secretion system.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Nataro J. P., Kaper J. B. Clin. Microbiol. Rev. 1998;11:142–201. doi: 10.1128/cmr.11.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen H. D., Frankel G. FEMS Microbiol. Rev. 2005;29:83–98. doi: 10.1016/j.femsre.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 3.Fagundes-Neto U., Scaletsky I. C. Sao Paulo Med. J. 2000;118:21–29. doi: 10.1590/S1516-31802000000100006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McDaniel T. K., Kaper J. B. Mol. Microbiol. 1997;23:399–407. doi: 10.1046/j.1365-2958.1997.2311591.x. [DOI] [PubMed] [Google Scholar]
- 5.Dean P., Maresca M., Kenny B. Curr. Opin. Microbiol. 2005;8:28–34. doi: 10.1016/j.mib.2004.12.010. [DOI] [PubMed] [Google Scholar]
- 6.Marches O., Ledger T. N., Boury M., Ohara M., Tu X., Goffaux F., Mainil J., Rosenshine I., Sugai M., De Rycke J., Oswald E. Mol. Microbiol. 2003;50:1553–1567. doi: 10.1046/j.1365-2958.2003.03821.x. [DOI] [PubMed] [Google Scholar]
- 7.Deng W., Vallance B. A., Li Y., Puente J. L., Finlay B. B. Mol. Microbiol. 2003;48:95–115. doi: 10.1046/j.1365-2958.2003.03429.x. [DOI] [PubMed] [Google Scholar]
- 8.Kenny B., DeVinney R., Stein M., Reinscheid D. J., Frey E. A., Finlay B. B. Cell. 1997;91:511–520. doi: 10.1016/s0092-8674(00)80437-7. [DOI] [PubMed] [Google Scholar]
- 9.Kenny B. Microbiology. 2002;148:1967–1978. doi: 10.1099/00221287-148-7-1967. [DOI] [PubMed] [Google Scholar]
- 10.Kanack K. J., Crawford J. A., Tatsuno I., Karmali M. A., Kaper J. B. Infect. Immun. 2005;73:4327–4337. doi: 10.1128/IAI.73.7.4327-4337.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kodama T., Akeda Y., Kono G., Takahashi A., Imura K., Iida T., Honda T. Cell Microbiol. 2002;4:213–222. doi: 10.1046/j.1462-5822.2002.00176.x. [DOI] [PubMed] [Google Scholar]
- 12.Kenny B., Ellis S., Leard A. D., Warawa J., Mellor H., Jepson M. A. Mol. Microbiol. 2002;44:1095–1107. doi: 10.1046/j.1365-2958.2002.02952.x. [DOI] [PubMed] [Google Scholar]
- 13.Jepson M. A., Pellegrin S., Peto L., Banbury D. N., Leard A. D., Mellor H., Kenny B. Cell Microbiol. 2003;5:773–783. doi: 10.1046/j.1462-5822.2003.00315.x. [DOI] [PubMed] [Google Scholar]
- 14.Nagai T., Abe A., Sasakawa C. J. Biol. Chem. 2005;280:2998–3011. doi: 10.1074/jbc.M411550200. [DOI] [PubMed] [Google Scholar]
- 15.Dean P., Kenny B. Mol. Microbiol. 2004;54:665–675. doi: 10.1111/j.1365-2958.2004.04308.x. [DOI] [PubMed] [Google Scholar]
- 16.Matsuzawa T., Kuwae A., Yoshida S., Sasakawa C., Abe A. EMBO J. 2004;23:3570–3582. doi: 10.1038/sj.emboj.7600359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shaw R. K., Smollett K., Cleary J., Garmendia J., Straatman-Iwanowska A., Frankel G., Knutton S. Infect. Immun. 2005;73:4385–4390. doi: 10.1128/IAI.73.7.4385-4390.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hardwidge P. R., Deng W., Vallance B. A., Rodriguez-Escudero I., Cid V. J., Molina M., Finlay B. B. Infect. Immun. 2005;73:2586–2594. doi: 10.1128/IAI.73.5.2586-2594.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tomson F. L., Viswanathan V. K., Kanack K. J., Kanteti R. P., Straub K. V., Menet M., Kaper J. B., Hecht G. Mol. Microbiol. 2005;56:447–464. doi: 10.1111/j.1365-2958.2005.04571.x. [DOI] [PubMed] [Google Scholar]
- 20.Donnenberg M. S., Tacket C. O., James S. P., Losonsky G., Nataro J. P., Wasserman S. S., Kaper J. B., Levine M. M. J. Clin. Invest. 1993;92:1412–1417. doi: 10.1172/JCI116717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Delie F., Rubas W. Crit. Rev. Ther. Drug Carrier Syst. 1997;14:221–286. [PubMed] [Google Scholar]
- 22.Shaw R. K., Cleary J., Murphy M. S., Frankel G., Knutton S. Infect. Immun. 2005;73:1243–1251. doi: 10.1128/IAI.73.2.1243-1251.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hicks S., Frankel G., Kaper J. B., Dougan G., Phillips A. D. Infect. Immun. 1998;66:1570–1578. doi: 10.1128/iai.66.4.1570-1578.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marches O., Nougayrede J. P., Boullier S., Mainil J., Charlier G., Raymond I., Pohl P., Boury M., De Rycke J., Milon A., Oswald E. Infect. Immun. 2000;68:2171–2182. doi: 10.1128/iai.68.4.2171-2182.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bissonnette P., Gagne H., Coady M. J., Benabdallah K., Lapointe J. Y., Berteloot A. Am. J. Physiol. 1996;270:G833–G843. doi: 10.1152/ajpgi.1996.270.5.G833. [DOI] [PubMed] [Google Scholar]
- 26.Meinild A., Klaerke D. A., Loo D. D., Wright E. M., Zeuthen T. J. Physiol. 1998;508(Pt 1):15–21. doi: 10.1111/j.1469-7793.1998.015br.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Warawa J., Finlay B. B., Kenny B. Infect. Immun. 1999;67:5538–5540. doi: 10.1128/iai.67.10.5538-5540.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Batt R. M., Hart C. A., McLean L., Saunders J. R. Gut. 1987;28:1283–1290. doi: 10.1136/gut.28.10.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Embaye H., Hart C. A., Getty B., Fletcher J. N., Saunders J. R., Batt R. M. Gut. 1992;33:1184–1189. doi: 10.1136/gut.33.9.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hecht G., Hodges K., Gill R. K., Kear F., Tyagi S., Malakooti J., Ramaswamy K., Dudeja P. K. Am. J. Physiol. 2004;287:G370–G378. doi: 10.1152/ajpgi.00432.2003. [DOI] [PubMed] [Google Scholar]
- 31.Casburn-Jones A. C., Farthing M. J. Gut. 2004;53:296–305. doi: 10.1136/gut.2003.022103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wiles S., Dougan G., Frankel G. Cell Microbiol. 2005;7:1163–1172. doi: 10.1111/j.1462-5822.2005.00544.x. [DOI] [PubMed] [Google Scholar]
- 33.Kenny B., Abe A., Stein M., Finlay B. B. Infect. Immun. 1997;65:2606–2612. doi: 10.1128/iai.65.7.2606-2612.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Delezay O., Verrier B., Mabrouk K., van Rietschoten J., Fantini J., Mauchamp J., Gerard C. J. Cell Physiol. 1995;163:120–128. doi: 10.1002/jcp.1041630114. [DOI] [PubMed] [Google Scholar]
- 35.Kipp H., Khoursandi S., Scharlau D., Kinne R. K. Am. J. Physiol. 2003;285:C737–C749. doi: 10.1152/ajpcell.00041.2003. [DOI] [PubMed] [Google Scholar]