Abstract
The tetrameric K channel ROMK provides an important pathway for K secretion by the mammalian kidney, and the gating of this channel is highly sensitive to changes in cytosolic pH. Although charge–charge interactions have been implicated in pH sensing by this K channel tetramer, the molecular mechanism linking pH sensing and the gating of ion channels is poorly understood. The x-ray crystal structure KirBac1.1, a prokaryotic ortholog of ROMK, has suggested that channel gating involves intermolecular interactions of the N- and C-terminal domains of adjacent subunits. Here we studied channel gating behavior to changes in pH using giant patch clamping of Xenopus laevis oocytes expressing WT or mutant ROMK, and we present evidence that no single charged residue provides the pH sensor. Instead, we show that N–C- and C–C-terminal subunit–subunit interactions form salt bridges, which function to stabilize ROMK in the open state and which are modified by protons. We identify a highly conserved C–C-terminal arginine–glutamate (R-E) ion pair that forms an intermolecular salt bridge and responds to changes in proton concentration. Our results support the intermolecular model for pH gating of inward rectifier K channels.
Keywords: channel gating, inward rectifier, Kir1.1, pH sensing, potassium channel
Small-conductance, inwardly rectifying K+ channels provide one of the major pathways for secretion of K+ across kidney tubule epithelia (1). In the cortical collecting duct these channels mediate regulated K+ secretion important in K+ homeostasis, and in the thick ascending limb of Henle (TAL) they mediate K+ recycling that enables efficient function of the Na-K-2Cl cotransporter responsible for reabsorption of 25% of the filtered load of NaCl. The inward rectifier K+ channel, ROMK (Kir1.1), expressed in Xenopus laevis oocytes, shares many of the biophysical and regulatory properties of these secretory K+ channels (1, 2). N-terminal splice variants of ROMK (ROMK1–3) are differentially expressed along the nephron and impart unique regulatory features to the channel (Fig. 1A and ref. 1). Consistent with the role of ROMK in the TAL, loss-of-function mutations in ROMK (KCNJ1) result in a severe renal salt wasting disorder, Bartter’s syndrome type II (3). In addition, the absence of these small-conductance K+ channels in cortical collecting duct and TAL cells in ROMK (KCNJ1)-deficient mice has established that ROMK encodes these native K+ secretory channels (4).
Fig. 1.
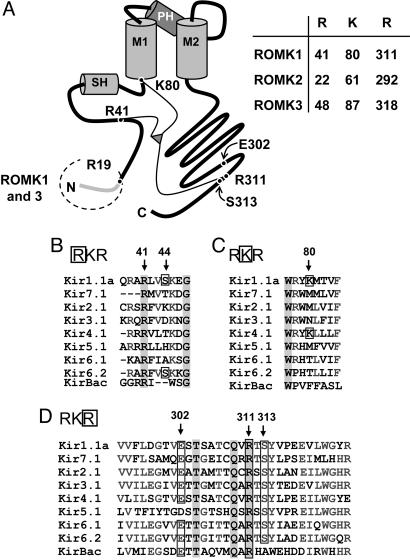
Residues involved in pH sensing by the ROMK channel. (A) Cartoon model of ROMK showing the N terminus, slide helix (SH), first transmembrane domain (M1), pore helix (PH), second-transmembrane domain (M2), and the C terminus. The RKR sites are shown for the three ROMK splice variants. The ROMK1 C-terminal R311 is shown together with the salt bridge partner E302. (B–D) N-terminal (B and C) and C-terminal (D) alignments are shown for members of the Kir inwardly rectifying potassium channel family. Important residues are indicated by residue numbers and arrows. The bacterial Kir ortholog, KirBac1.1, is also shown for comparison.
ROMK channels are posttranslationally regulated by multiple, and often interacting, mechanisms, including phosphorylation by kinases and the binding to both membrane and cytosolic ligands (1). The sensitivity of ROMK channels to cytosolic protons provides a major mechanism for regulation of channel activity (K0.5 ≈ 6.9; refs. 1 and 5–10), and a number of other ROMK channel regulators [e.g., PKA phosphorylation and phosphatidylinositol(4,5)bisphosphate] also function, at least in part, by modifying pH sensitivity. This pH sensing provides an important mechanism for linking renal K excretion with acid–base balance. Although progress has been made in identifying residues involved in pH sensing by ROMK (1), the molecular mechanism by which protons gate this channel is unresolved. Although a triad of basic amino acid residues in the N and C termini of ROMK (RKR triad; Fig. 1) has been proposed to form the proton gating sensor (11), mutational analyses have identified additional residues in diverse locations along the N and C termini of ROMK that modify the pH sensitivity of this channel (1).
The x-ray crystal structure of KirBac1.1 (12), the prokaryotic ortholog of the mammalian inward rectifiers like ROMK, has provided insights into the gating mechanism of these K channels. The N terminus of one subunit interacts with the C terminus of the adjacent subunit (12), and this intermolecular interaction has been suggested to form an important part of the channel gating mechanism (13). Consistent with this view, molecular motions of both the N and C termini of ROMK have been suggested to be involved in pH sensing (14). However, the KirBac1.1 channel crystal structure indicates that the residues in the RKR triad are not in close proximity required to form a single pH sensor. Thus, an alternative mechanism is required to explain pH sensing.
In the present study we assessed the possibility that N- and C-terminal interactions between subunits are critical for pH sensing in ROMK. We show that mutations of the “RKR” residues exhibit complex patterns of pH sensitivity in ROMK isoforms. Other residues that are predicted to alter N–C- or C–Cz-terminal interactions can also modify pH gating. In particular, we identified an intermolecular interaction between the second R in the RKR triad on one subunit and a C-terminal glutamate (E283 in ROMK2) in the adjacent subunit that is critical to pH sensitivity. This R-E pair can be linked by salt bridging in KirBac1.1 (12), and this pair is conserved in all Kir channels (except Kir5.1) and likely provides a fundamental interaction critical to channel gating by ligands. When taken together, these results suggest that there is no single pH sensor in ROMK but rather a complex of N- and C-terminal interactions between subunits that is fundamental to gating, and each interaction can be influenced by protons. Such interactions could explain the often complex pH gating patterns observed in different inward rectifiers (15).
Results and Discussion
Mutation of the N-Terminal Lysine in the Putative RKR pH Sensor Does Not Abolish pH Gating.
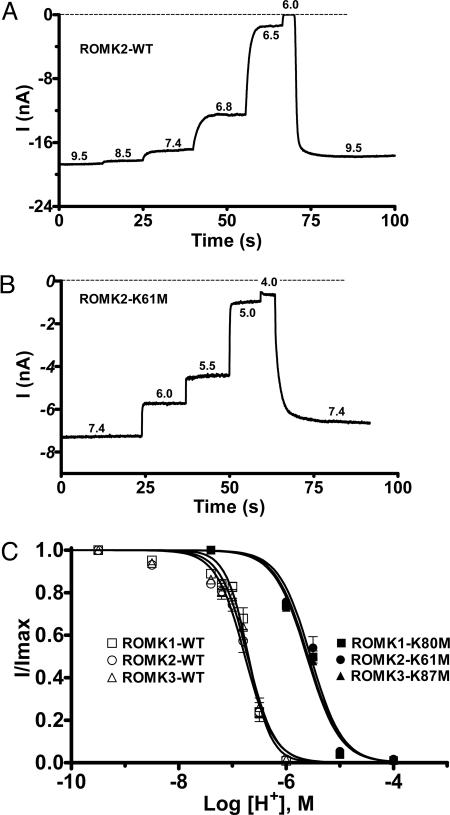
The pH–current relationships for all three ROMK isoforms are shown in Fig. 2. All three channels behaved similarly, with slight activation at pH values >7.4 and complete inhibition at a pH of ≤6.0 (Fig. 2A). Similar observations for ROMK1 and ROMK2 have been reported by others (8, 10). The half-maximal inhibitory pH values, pH0.5, and Hill coefficients of the isoforms were not significantly different (Fig. 2C): ROMK1, pH0.5 = 6.70 ± 0.02 and Hill = 2.1 ± 0.2 (n = 4); ROMK2, pH0.5 = 6.77 ± 0.02 and Hill = 1.6 ± 0.1 (n = 7); ROMK3, pH0.5 = 6.72 ± 0.03 and Hill = 1.7 ± 0.2 (n = 4). Thus, the small alternative splice inserts at the beginning of the NH2 termini in ROMK1 and ROMK3 (Fig. 1) did not alter pH gating.
Fig. 2.
ROMK channel isoforms with the lysine in the RKR triad mutated remain sensitive to pH. (A and B) Representative current traces from WT ROMK2 (A) and ROMK2-K61M (B) from inside-out giant-patches of oocytes showing pH responses to step acidification of the cytosolic face solution. (C) pH dose–response curves are shown for WT ROMK (open symbols) and K-mutant (solid symbols) channels.
Previous studies had suggested that mutation of the RKR lysine residue to methionine (Fig. 1) produced pH-insensitive channels (8, 10, 14); however, a more complete pH titration to 4 demonstrated that each of the three ROMK isoforms remained pH sensitive, with the N-terminal K→M mutation (Fig. 2 B and C). The pH0.5 and Hill coefficients for these lysine-mutant channels (Fig. 2C) were as follows: ROMK1-K80M, pH0.5 = 5.59 ± 0.03 and Hill 1.4 ± 0.2 (n = 3); ROMK2-K61M, pH0.5 = 5.55 ± 0.04 and Hill 1.5 ± 0.2 (n = 3); ROMK3-K87M, pH0.5 = 5.61 ± 0.03 and Hill 1.4 ± 0.1 (n = 3). On average there is a shift in pH0.5 of ≈1.2 pH units in the acid direction but no significant change in the cooperativity of pH gating (Hill coefficient) with mutation of the N-terminal lysine. Thus, whereas the N-terminal lysine residue contributes to pH sensing, other mechanisms/sites must be contributing to proton titration involved in pH-gating of ROMK channels.
The N Terminus Alters the Response to C-Terminal Arginine Mutations in the Putative RKR pH Sensor.
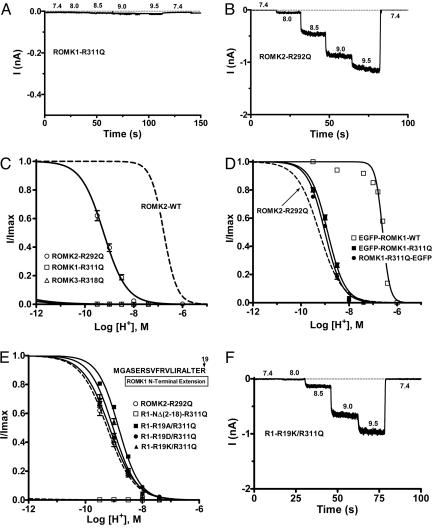
Schulte et al. (11) found that mutation of the C-terminal R311 to glutamine in the ROMK1 “RKR311” (Fig. 1D) resulted in complete loss of channel activity at a pH of 7.4, and activity did not increase with raising the cytosolic-side pH to 10. We confirmed this behavior for the R311Q mutation (Fig. 3 A and C; n = 7). Mutation of the equivalent arginine residue in ROMK3 (R318Q; Fig. 1D) gave a similar result (Fig. 3C). In contrast, although ROMK2-R292Q was inactive at pH 7.4, activity could be restored by increasing cytosolic-face pH (Fig. 3 B and C; n = 6). The pH0.5 for ROMK2-R292Q was alkaline-shifted by >2 pH units, and the Hill coefficient was reduced by ≈1 (pH0.5 = 9.24 ± 0.03 and Hill = 0.9 ± 0.1; n = 4) compared with WT ROMK2 (Fig. 3C). The different response to mutation of the C-terminal arginine of ROMK2 compared with ROMK1 and ROMK3 is likely due to the initial N-terminal extensions in the latter two isoforms (Fig. 1A).
Fig. 3.
Mutation of the C-terminal arginine in the RKR triad disrupts channel activity. (A and B) Representative current traces from giant inside-out patches showing inactive channels at a pH of 7.4. The ROMK2 (B), but not ROMK1 (A), mutant could be activated by increasing internal pH. (C and D) pH dose–response relationships to internal pH for the C-terminal arginine RKR mutant ROMK channels, without (C) or with (D) an EGFP tag. pH sensitivity was rescued in the nonfunctional ROMK1-R311Q channels by the bulky N- or C-terminal EGFP tag. (E and F) Mutation of the N-terminal R19 in ROMK1 rescues the inactive channels in ROMK1-R311Q channels. pH dose–response curves (E) and a representative current trace from an excised giant patch (F) are shown.
The absence of detectable channel activity in ROMK1-R311Q and ROMK3-R318Q (Fig. 3 A–C) could be due either to loss of surface expression of the channel or to a more severe left-shifting of pH0.5 than 9.5, our maximum ability to alkalinize the cytosolic face of a patch. The observation that ROMK2-R292Q channel activity can be restored at pH values >8.0 (Fig. 3B) suggested that these arginine mutant channels may be expressed on the surface but that their apparent inactivity is due to a large shift in pH0.5. To assess this possibility, we determined whether the Flag-tagged ROMK1-R311Q mutant channel is expressed at the oocyte plasma membrane. Giant patch-clamp experiments showed that Flag-tagging ROMK-WT or mutant channels did not alter the responses of the channel to pH; Flag-ROMK1-R311Q channels were inactive, whereas Flag-ROMK1 WT channels exhibited a pH–current response (pH0.5 = 6.73 ± 0.02; n = 4; data not shown) indistinguishable from untagged ROMK1. The Flag-tagged ROMK1-R311Q channel showed clear expression at the oocyte surface, although lower than that from Flag-ROMK1 WT (Fig. 6, which is published as supporting information on the PNAS web site). Surface localization of WT and R311Q mutant ROMK1 proteins in Xenopus oocytes was confirmed by fluorescence confocal microscopy of EGFP-tagged channels (Fig. 7, which is published as supporting information on the PNAS web site). EGFP-ROMK1 and EGFP-ROMK2 channels were functional, and their pH sensitivities were similar to WT ROMK channels (EGFP-ROMK1, pH0.5 = 6.60 ± 0.01, n = 5; EGFP-ROMK2, pH0.5 = 6.69 ± 0.01, n = 4; Fig. 7A). In contrast to Flag-tagged ROMK1-R311Q, fusion of EGFP to the N or C terminus of the ROMK1-R311Q mutant channel “rescued” channel function, resulting in a pH sensitivity that was similar to ROMK2-R292Q (Fig. 3D; EGFP-ROMK1-R311Q, pH0.5 = 8.90 ± 0.02, n = 5; ROMK1-R311Q-EGFP, pH0.5 = 9.00 ± 0.02, n = 4). Comparable results were seen with N-terminal EGFP fusion on ROMK3-R318Q (data not shown). Thus, the addition of the large EGFP tag to either the N or C terminus of ROMK1 or ROMK3 C-terminal arginine mutants resulted in pH sensitivities similar to ROMK2-R292Q.
Based on these observations, it is possible that the N-terminal extensions on ROMK1 and ROMK3 isoforms of 19 or 26 residues (Fig. 1A), respectively, may have contributed to the pH behavior of these channels resulting from mutations of their C-terminal lysine residues. Such a possibility would support the importance of N- and C-terminal interactions in gating. The last seven residues of the N-terminal extensions of ROMK1 and ROMK3 are identical (“IRALTER19”), and this region has been shown to alter interactions of ROMK with the ABC protein SUR2B (16). Using a similar mutational strategy, we found that arginine 19 (R19; Fig. 1A) in ROMK1 (equivalent to R26 in ROMK3) was the single residue that caused the divergent pH sensitivities of ROMK1-R311Q vs. ROMK2-R292Q (Fig. 3 E and F). Mutation of R19 to alanine (A), aspartate (D), or lysine (K) resulted in rescue of the ROMK1-R311Q mutant. This result identifies R19 in ROMK1 (or R26 in ROMK3) as a modifier of pH sensing and provides further evidence supporting the importance of N- and C-terminal interactions in pH gating.
Inactivity of ROMK1-R311Q Channels Is Due to Severe Left-Shifting of the pH0.5.
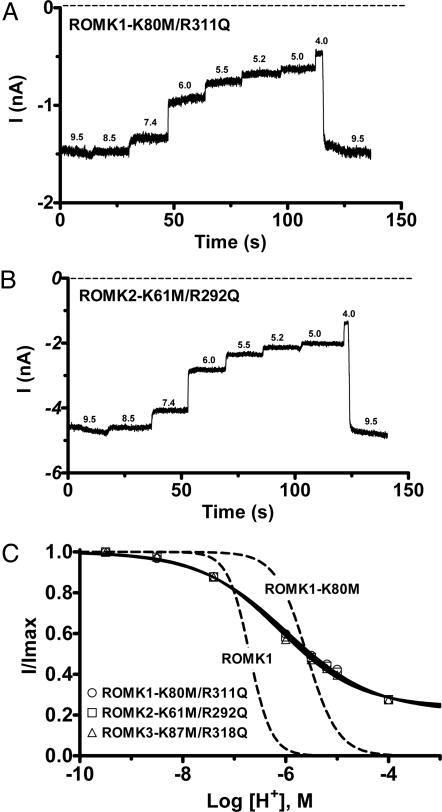
Because ROMK1-R311Q and ROMK3-R318Q channels are expressed at the oocyte membrane, it is possible that their inactivity is due to a large (>2 pH units) right shift in pH sensitivity (i.e., channels are more sensitive to pH). Given that mutation of lysine to methionine (e.g., ROMK1-K80M; Fig. 2C) decreases the sensitivity of ROMK channels to inhibition by internal protons and shifts pH0.5 ≈1 pH unit in the acidic direction (Fig. 3), we reasoned that N-terminal K→M mutations might also rescue pH sensing by C-terminal R→Q mutant channels. Fig. 4 shows that the K→M mutations rescued the inactive ROMK1-R311Q and ROMK3-R318Q channels and returned the pH0.5 of the three mutant channels back to the physiological range (Fig. 4C): ROMK1-K80M/R311Q (pH0.5 = 5.98 ± 0.08, n = 6), ROMK2-K61M/R292Q (pH0.5 = 6.06 ± 0.03, n = 12), and ROMK3-K87M/R318Q (pH0.5 = 6.14 ± 0.06, n = 6). However, pH 4 inhibited the double mutant channels by only ≈80% (Fig. 4C), and the Hill coefficients were greatly reduced: ROMK1-K80M/R311Q, 0.51 ± 0.03; ROMK2-K61M-R292Q, 0.54 ± 0.02; ROMK3-K87M/R318Q, 0.56 ± 0.03. Thus, the pH-sensitive gate closing the channel appears defective in the RKR N- and C-terminal double mutants.
Fig. 4.
Mutation of the N-terminal lysine in the RKR triad reactivates the inactive ROMK1-R311Q and ROMK3-R318Q channels. (A and B) Representative current traces from excised inside-out giant patches showing pH responses to step acidification of the cytosolic face solution. (C) pH dose–response curves are shown for ROMK K-R double mutant channels. Fits to ROMK1 and ROMK1-K80M from Fig. 2 are shown for comparison (dashed lines).
The C-Terminal Arginine in the RXR Forms an R-E Ion Pair Between Adjacent Subunits That Is Involved in pH Sensing.
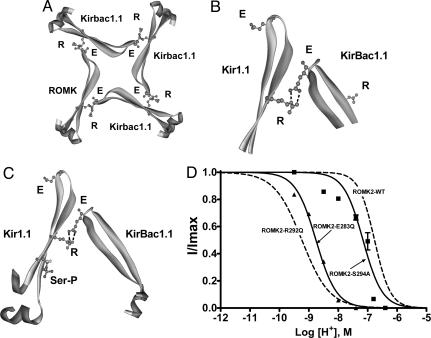
The recently solved x-ray structure of the bacterial channel KirBac1.1 (12) has structural similarity with the crystal structure of the mammalian Kir3.1 channel (13, 17) and is believed to be representative of other Kir channel family members (13). In the KirBac1.1, the analogous C-terminal arginine of “RXR” forms a salt bridge with a C-terminal glutamate in an adjacent subunit (Fig. 5 A and B), and this salt bridge is conserved in all known Kir channels (Fig. 1D) except Kir5.1. The latter channel forms functional channels only when coexpressed with Kir4.1/2, and this heteromultimer exhibits pH sensitivity (18–20). This E-R ion pair has recently been found to contribute to the open-state stability of the Kir6.2 channel (21). For most ion pairs exposed to solvent, changes in the pH of the surrounding solution can lead to either making or breaking of these salt bridges (22). Acid pH breaks a critical salt bridge between transmembrane domains and activates the bacterial colicin ion channel (23), and mutation of a salt bridge between subunits of the AMPA receptor decreases dimer stability and accelerates deactivation (24). Thus, we reasoned that if subunit–subunit interactions are important to pH sensing of ROMK, then disruption of this R-E ion pair should alter channel pH0.5. Consistent with this possibility, mutation of the glutamate partner of the R-E ion pair (ROMK2-E283Q) shifted the pH dependence of channel gating in the alkali direction with a pH0.5 = 8.76 ± 0.02 (n = 3; Fig. 5D). The magnitude of the shift in pH0.5 from ROMK2-WT was similar in ROMK2-R292Q and ROMK2-E283Q (≈0.48 pH units).
Fig. 5.
Contributions of the E-R salt bridge and phosphorylation to channel activity are independent and additive. (A) Tetrameric ring of Kir channel subunits viewed from above showing the proposed C–C-terminal E-R salt bridge interaction between the glutamate (E283) and the arginine (R292) on an adjacent subunit. Approximately 20 residues on either side of the proposed E-R salt bridge are shown. The model of Kir1.1 was generated by sequence homology to KirBac1.1 and based on the x-ray structure data set of KirBac1.1 (1N7P) using Swiss-Model (http://swissmodel.expasy.org/swiss-model.html). The other three subunits are KirBac1.1. (B) Magnified E-R salt bridge interaction from A. (C) The E-R salt bridge is shown in relation to the modeled phosphorylated serine residue on the Kir1.1 subunit. The serine (S294 in ROMK2) was substituted for a phosphorylated serine; therefore, this figure should be interpreted as a cartoon. Noting this, the cartoon agrees with the experimental data that phosphorylation at the serine is independent of, and does not appear to affect, the E-R salt bridge interaction. (D) pH dose–response relationships for the C-terminal R, E, and S mutants.
In ROMK, the C-terminal arginine is also part of a consensus PKA site (“RTS”; Fig. 1D), and the associated serine residue can be phosphorylated by PKA (25). Hence, mutation of the arginine residue would not only affect the R-E ion pair interaction but would also prevent phosphorylation of the serine residue. Fig. 5D shows that mutation of this serine shifts the pH0.5 for proton inhibition in the alkali direction by ≈0.5 pH units (ROMK2-S294A, pH0.5 = 7.16 ± 0.06 and n = 3, vs. ROMK2 WT, pH0.5 = 6.77 ± 0.02 and n = 7; P < 0.01). The serine phosphorylation state could modulate the pKa of the R-E ion pair interaction or may form a phosphate salt bridge interaction between subunits. However, the alkaline shift with ROMK2-S294A appears to be additive with ROMK2-E283Q to result in the pH0.5 shift with ROMK2-R292Q (Fig. 5D; ≈2-pH-unit shift in ROMK2-R292Q: pH0.5 = 9.24 ± 0.03, n = 4). This finding suggests that the phosphorylation of the serine is not mediating its effects through modifying the strength of the E-R salt bridge, consistent with the orientation of the phosphate group on the serine residue shown in Fig. 5C. In addition, the KirBac1.1 channel crystal structure indicates that the conserved R-E salt bridge and the position occupied by lysine-80 are not in close proximity to form a single pH sensor. Thus, both phosphorylation and the R-E salt bridge appear to be mediating their effects on pH sensing by increasing channel stability in the open state.
In summary, we propose that N–C and C–C intersubunit salt bridge interactions are involved in stabilizing the ROMK channel in the open state and that these interactions involve salt bridges that can be modified by pH. This model may be applicable to pH sensing by other channels and solute transporters. However, other types of charge–charge interactions could also contribute to pH sensing in channels. For example, a charge screening mechanism has also been proposed for the effects of cytoplasmic pH and polyvalent cations on the activity state of the TRPM7 or Mg2+-inhibited cation current channel (26). For the TRPM7 channel, cation screening of membrane phosphatidylinositol(4,5)bisphosphate (PIP2) charges appears to be important for polyvalent cation-mediated regulation of channel activity. Similarly, PIP2 greatly influences the open state of ROMK, and this effect can be modulated by internal pH (6). Thus, multiple types of salt bridge interactions may be contributing to the very complex behavior of channels to changes in cytosolic pH.
Materials and Methods
Molecular Biology.
cDNA encoding ROMK1, ROMK2, and ROMK3 sequences were individually subcloned into the pSPORT-1 vector (Invitrogen). To determine surface expression, EGFP or the FLAG tag (DYKAFDNL) was ligated in-frame with ROMK1, ROMK2, and ROMK3 WT channels at either the N or C terminus as indicated. In each case, a 10-glycine linker was present between EGFP and ROMK. These EGFP-tagged constructs and the WT channels were used as templates for QuikChange site-directed mutagenesis (Stratagene). Plasmids containing channel constructs were verified by sequencing (Keck Facility of Yale University), linearized, and used to generate full-length cRNA transcripts by using mMESSAGE mMACHINE T7 High Yield Transcription kit (Ambion, Austin, TX). cRNA was diluted in water, and 50 nl of solution containing 5 ng of cRNA was injected into oocytes.
Preparation and Injection of Oocytes.
All animal protocols were approved by the Yale Institutional Animal Care and Use Committee. Standard methods were used to express ROMK WT and mutant cRNAs in X. laevis oocytes (7). Briefly, frogs were anesthetized in 0.02% 3-aminobenzoic acid ethyl ester for 3–7 min (tricaine; titrated to pH 7.2 with 5 mM Hepes using NaOH). Oocytes were removed by partial ovariectomy, separated into clumps of ≈10 by using forceps, and then enzymatically defolliculated by treatment with 2 mg/ml collagenase (type I, Worthington) in a Ca2+-free solution (96 mM NaCl, 2 mM KCl, 1 mM MgCl2, 5 mM Hepes, adjusted to pH 7.4 with NaOH) while being gently agitated for 1 h. Oocytes were then washed 12 times in ND-96 solution (96 mM NaCl, 2 mM KCl, 1.8 mM CaCl2, 1 mM MgCl2, 5 mM Hepes, adjusted to pH 7.4 with NaOH) and subsequently placed in ND-96 solution containing 500 units/ml penicillin and 500 μg/ml streptomycin (Invitrogen, Carlsbad, CA). Immediately after isolation, healthy-stage V–VI oocytes were injected with 50 nl of water (negative control) or cRNA by using an air injector (Inject+Matic, Geneva). Oocytes were incubated at 18°C for 24–72 h before experiments.
Giant Patch-Clamp.
Oocytes were immersed in a hyperosmotic solution for 1–2 min (200 mM N-methyl-d-glucamine/2 mM KCl/1 mM MgCl2/10 mM EGTA/10 mM Hepes, adjusted to pH 7.4 with HCl), and the vitelline membranes were removed by using forceps (Dumont no. 5). Electrodes were pulled from borosilicate glass capillaries (Sutter Instruments, Novato, CA) on a Narishige PP-83 and polished on a MF-83 microforge (Narishige, Tokyo). Electrodes had tip resistances of 0.3–0.6 MΩ when filled with pipette solution (150 mM KCl/1 mM MgCl2/1 mM CaCl2/5 mM Mes/Tris, adjusted to pH 7.4 with either Mes or Tris buffer). Mg2+-free bath solutions were used in all experiments and perfused by multibarrel quick-exchange solution system (model SF-77B, Warner Instruments, Hamden, CT). Bath solutions at pH 6.0–9.5 contained 150 mM KCl, 2 mM EDTA, 10 mM Mes (2-morpholinoethanesulfonic acid), and 10 mM Tris [Tris(hydroxymethyl)aminomethane] and were adjusted to varied pH values with Mes or Tris. Bath solutions at pH 4.0–6.0 contained 150 mM KCl, 2 mM EDTA, 10 mM imidazole, and 10 mM phthalic acid and were adjusted to various pH values with HCl (27). Macroscopic currents were recorded in the inside-out giant patch configuration by using a EPC-7 amplifier (Heka Elektronik, Lambrecht/Pfalz, Germany) and filtered through at 1 KHz through an 8-pole low-pass Bessel filter (Warner Instruments). Data were acquired by means of a DigiData 1200 Series analogue-to-digital converter (Axon Instruments, Union City, CA) driven by commercial software (pclamp clampex 8.20, Axon Instruments). Macroscopic currents were analyzed off-line with clampfit 8.20 and prism 4.03 software (GraphPad, San Diego). Throughout experimental procedures, water-injected controls were used to assess background endogenous current levels and served as a comparison with oocytes expressing exogenous channels. All chemicals were purchased from Sigma-Aldrich unless otherwise indicated.
Analysis of pH Sensitivity.
Many of the mutant channels have increased sensitivity to internal protons and are not fully active even at pH 9.5 (the most alkaline we could go without destroying the integrity of the membrane-glass seal). Hence, to normalize partially active experimental data, we first fit pH–current data using Eq. 1 to determine the theoretical maximum current (Imax). Channel current data were then normalized to this maximal value (I/Imax) and refit by using the same equation. For data sets where Imin was greater than zero, Imin was allowed to float; otherwise, Imin and Imax were constrained to 0 and 1, respectively, during the fit to the normalized data.
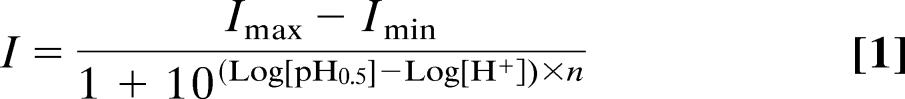 |
In Eq. 1, I is either the raw macroscopic current or the normalized I/Imax current. Imin and Imax are minimum and maximum macroscopic currents, respectively. pH0.5 is the log of the half-maximum inhibitory concentration of protons, and n is the Hill coefficient. In the normalized graphs the solid lines on the dose–response curves are least-squares fits to mean data using Eq. 1. All data are presented as means ± SEM and were fit by using prism 4.02 for Windows (GraphPad).
Additional Details.
See Supporting Methods, which is published as supporting information on the PNAS web site.
Supplementary Material
Acknowledgments
These studies were supported by National Institutes of Health Grants DK17433 and DK054999 (to S.C.H). Q.L. is supported by American Heart Association Heritage Fellowship 0425865T.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Hebert S. C., Desir G., Giebisch G., Wang W. Physiol. Rev. 2005;85:319–371. doi: 10.1152/physrev.00051.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Palmer L. G., Choe H., Frindt G. Am. J. Physiol. 1997;273:F404–F410. doi: 10.1152/ajprenal.1997.273.3.F404. [DOI] [PubMed] [Google Scholar]
- 3.Lorenz J. N., Baird N. R., Judd L. M., Noonan W. T., Andringa A., Doetschman T., Manning P. A., Liu L. H., Miller M. L., Shull G. E. J. Biol. Chem. 2002;277:37871–37880. doi: 10.1074/jbc.M205627200. [DOI] [PubMed] [Google Scholar]
- 4.Lu M., Wang T., Yan Q., Yang X., Dong K., Knepper M. A., Wang W., Giebisch G., Shull G. E., Hebert S. C. J. Biol. Chem. 2002;277:37881–37887. doi: 10.1074/jbc.M206644200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leipziger J., MacGregor G. G., Cooper G. J., Xu J., Hebert S. C., Giebisch G. Am. J. Physiol. 2000;279:F919–F926. doi: 10.1152/ajprenal.2000.279.5.F919. [DOI] [PubMed] [Google Scholar]
- 6.Leung Y. M., Zeng W. Z., Liou H. H., Solaro C. R., Huang C. L. J. Biol. Chem. 2000;275:10182–10189. doi: 10.1074/jbc.275.14.10182. [DOI] [PubMed] [Google Scholar]
- 7.McNicholas C. M., MacGregor G. G., Islas L. D., Yang Y., Hebert S. C., Giebisch G. Am. J. Physiol. 1998;275:F972–F981. doi: 10.1152/ajprenal.1998.275.6.F972. [DOI] [PubMed] [Google Scholar]
- 8.Choe H., Zhou H., Palmer L. G., Sackin H. Am. J. Physiol. 1997;273:F516–F529. doi: 10.1152/ajprenal.1997.273.4.F516. [DOI] [PubMed] [Google Scholar]
- 9.Tsai T. D., Shuck M. E., Thompson D. P., Bienkowski M. J., Lee K. S. Am. J. Physiol. 1995;268:C1173–C1178. doi: 10.1152/ajpcell.1995.268.5.C1173. [DOI] [PubMed] [Google Scholar]
- 10.Fakler B., Schultz J. H., Yang J., Schulte U., Brändle U., Zenner H. P., Jan L. Y., Ruppersberg J. P. EMBO J. 1996;15:4093–4099. [PMC free article] [PubMed] [Google Scholar]
- 11.Schulte U., Hahn H., Konrad M., Jeck N., Derst C., Wild K., Weidemann S., Ruppersberg J. P., Fakler B., Ludwig J. Proc. Natl. Acad. Sci. USA. 1999;96:15298–15303. doi: 10.1073/pnas.96.26.15298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuo A., Gulbis J. M., Antcliff J. F., Rahman T., Lowe E. D., Zimmer J., Cuthbertson J., Ashcroft F. M., Ezaki T., Doyle D. A. Science. 2003;300:1922–1926. doi: 10.1126/science.1085028. [DOI] [PubMed] [Google Scholar]
- 13.Gulbis J. M., Doyle D. A. Curr. Opin. Struct. Biol. 2004;14:440–446. doi: 10.1016/j.sbi.2004.06.008. [DOI] [PubMed] [Google Scholar]
- 14.Schulte U., Hahn H., Wiesinger H., Ruppersberg J. P., Fakler B. J. Biol. Chem. 1998;273:34575–34579. doi: 10.1074/jbc.273.51.34575. [DOI] [PubMed] [Google Scholar]
- 15.Wu J., Cui N., Piao H., Wang Y., Xu H., Mao J., Jiang C. J. Physiol. 2002;543:495–504. doi: 10.1113/jphysiol.2002.025247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dong K., Xu J., Vanoye C. G., Welch R., MacGregor G. G., Giebisch G., Hebert S. C. J. Biol. Chem. 2001;276:44347–44353. doi: 10.1074/jbc.M108072200. [DOI] [PubMed] [Google Scholar]
- 17.Nishida M., MacKinnon R. Cell. 2002;111:957–965. doi: 10.1016/s0092-8674(02)01227-8. [DOI] [PubMed] [Google Scholar]
- 18.Casamassima M., D’Adamo M. C., Pessia M., Tucker S. J. J. Biol. Chem. 2003;278:43533–43540. doi: 10.1074/jbc.M306596200. [DOI] [PubMed] [Google Scholar]
- 19.Lourdel S., Paulais M., Cluzeaud F., Bens M., Tanemoto M., Kurachi Y., Vandewalle A., Teulon J. J. Physiol. 2002;538:391–404. doi: 10.1113/jphysiol.2001.012961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tanemoto M., Kittaka N., Inanobe A., Kurachi Y. J. Physiol. 2000;525:587–592. doi: 10.1111/j.1469-7793.2000.00587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin Y. W., Jia T., Weinsoft A. M., Shyng S. L. J. Gen. Physiol. 2003;122:225–237. doi: 10.1085/jgp.200308822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Simonson T., Carlsson J., Case D. A. J. Am. Chem. Soc. 2004;126:4167–4180. doi: 10.1021/ja039788m. [DOI] [PubMed] [Google Scholar]
- 23.Musse A. A., Merrill A. R. J. Biol. Chem. 2003;278:24491–24499. doi: 10.1074/jbc.M302371200. [DOI] [PubMed] [Google Scholar]
- 24.Horning M. S., Mayer M. L. Neuron. 2004;41:379–388. doi: 10.1016/s0896-6273(04)00018-2. [DOI] [PubMed] [Google Scholar]
- 25.Xu Z. C., Yang Y., Hebert S. C. J. Biol. Chem. 1996;271:9313–9319. doi: 10.1074/jbc.271.16.9313. [DOI] [PubMed] [Google Scholar]
- 26.Kozak J. A., Matsushita M., Nairn A. C., Cahalan M. D. J. Gen. Physiol. 2005;126:499–514. doi: 10.1085/jgp.200509324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sabirov R. Z., Okada Y., Oiki S. Pflügers Arch. Eur. J. Physiol. 1997;433:428–434. doi: 10.1007/s004240050296. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.