Abstract
The E3 ubiquitin (Ub) ligase Itch is a critical regulator of T helper 2 (Th2) cytokine production through its ability to induce Ub-dependent JunB degradation. After T cell receptor engagement, Itch undergoes JNK1-mediated phosphorylation that greatly enhances its enzymatic activity. To investigate how phosphorylation activates an E3 Ub ligase we have identified the JNK1 phosphorylation sites within Itch as S199, S232, and T222, which are located within a Pro-rich region. Phosphorylation of these sites is necessary and sufficient for disrupting an inhibitory interaction between the WW domain of Itch and its catalytic HECT (Homologous to E6-AP C Terminus) domain and induces a conformational change that greatly enhances the catalytic activity of Itch, a HECT E3 ligase found to be directly activated upon its phosphorylation.
Keywords: JNK
Protein phosphorylation, the most common type of posttranslational modification, is involved in many different facets of cellular regulation (1). Ubiquitylation is another common system of posttranslational modification that controls protein stability or protein–protein interactions (2, 3). Addition of ubiquitin (Ub) chains to a target protein can mark it for proteasomal degradation or enable it to interact with other proteins that contain a Ub-binding domain (2). In recent years it has become clear that protein phosphorylation plays an important role in regulation of protein ubiquitylation (4). Protein ubiquitylation depends on a three-component enzyme system, namely, E1, E2, and E3. The E1 ubiquitin-activating enzyme forms a thioester bond between its active Cys and Ub, whereas the E2 Ub-conjugating enzyme (Ubc) transfers this activated Ub moiety to the target protein or onto a growing polyUb chain. E3 Ub ligases facilitate this reaction through different mechanisms (2). One class of E3 ligases, the passive RING (Really Interesting New Gene) finger-containing molecules mostly function as adaptor molecules that allow the E2 to be recruited to the vicinity of the substrate (5). The other class of E3 ligases contain a characteristic HECT (Homologous to E6-AP C Terminus) domain that serves as a catalytic intermediate in the transfer of Ub chains from the E2 to the substrate (2, 3).
Itch is a HECT domain E3 ligase that plays a critical role in the differentiation of T helper 2 (Th2) cells (6). Itch is also an E3 ligase whose catalytic activity is directly stimulated by phosphorylation (4, 7). After activation of T cells through the T cell receptor and the CD28 accessory molecule, Itch is rapidly phosphorylated (7). This modification increases its catalytic activity, leading to enhanced polyubiquitylation of JunB, a critical transcription factor for Th2 cell differentiation (8, 9). Polyubiquitylation accelerates JunB degradation and diminishes activation of one of its critical target genes, encoding the Th2 cytokine IL-4 (7). Decreased IL-4 expression inhibits production of other Th2 cytokines (10). Defects in this pathway due to mutational inactivation of either Itch, MEKK1 or JNK1 enhance production of IL-4 and other Th2 cytokines (6, 7, 11). The mechanism by which JNK1-mediated phosphorylation leads to Itch activation was, however, not understood. Because Itch represents an E3 ligase that is directly activated by protein phosphorylation, we have studied it as a model for understanding the regulatory interface between protein phosphorylation and protein ubiquitylation (4).
HECT-domain-containing E3 ligases, with a few notable exceptions (e.g., E6-AP), contain an N-terminal C2 domain, followed by WW domains, and a C-terminal catalytic HECT domain (2, 3). The C2 domain binds Ca2+ and phospholipids and mediates protein recruitment to intracellular membranes (12). WW domains comprise a β-sheet structure that contains two conserved Trp residues, and mediates protein–protein interactions (13). Targets for WW motifs are usually Pro-rich regions (PRR) (14). HECT domains, unlike RING finger domains, are structurally similar to E2s, including an active Cys residue that transfers the activated Ub from the E2, first onto itself, and then onto its target protein (2, 3). Recent structural studies have revealed a hinge region that provides conformational flexibility to the N- and C-terminal halves of the HECT domain and may also be involved in regulation of its activity (15).
To understand how phosphorylation regulates Itch activity we have mapped the JNK1 phosphorylation sites of Itch to three residues within its PRR: S199, T222 and S232. The nonphosphorylated form of Itch is engaged in an intramolecular interaction between its HECT and WW domains. This self-inhibitory interaction is disrupted by JNK1-mediated phosphorylation of the PRR, which alters the conformation of the WW domain and greatly enhances catalytic activity.
Results
Identification of the JNK1 Phosphorylation Sites.
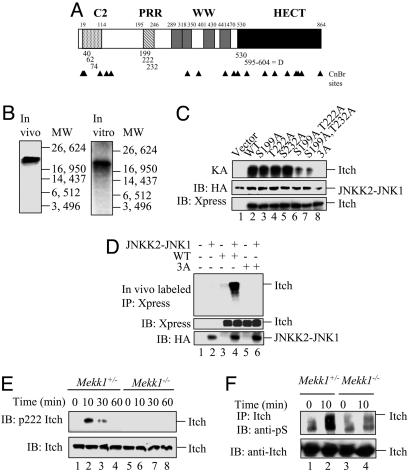
Itch contains seven potential mitogen-activated protein kinase (MAPK) phosphorylation sites (Pro-directed Ser/Thr) and a D domain-like sequence (Fig. 1A), which is required for recognition by different JNK isoforms (16, 17). Like most other HECT domain E3 ligases, Itch contains a C2 domain, four WW motifs, and a HECT domain (2). CNBr peptide mapping (18) was chosen to identify the regions of Itch phosphorylated by JNK1, because it generates three readily identifiable cleavage products containing potential MAPK phosphorylation sites. To phosphorylate Itch by JNK1 in living cells, it was coexpressed in HEK293 cells with a constitutively active JNKK2-JNK1 fusion protein (19). Alternatively, Itch was incubated in vitro with recombinant JNKK2-JNK1 in the presence of γ-labeled ATP. CNBr cleavage of either preparation of JNK1-phosphorylated and 32P-labeled Itch generated a labeled cleavage product 21 kDa in size, corresponding to Itch residues 132–332 (Fig. 1B). This peptide contains three potential MAPK phosphorylation sites, S199, T222, and S232, within a PRR (Fig. 1A). To further define the JNK1 phosphoacceptor sites, Ala-scanning mutagenesis was performed (Table 1) and the Ala substitution mutants used as JNK1 substrates in the in vitro kinase assay. Although the single substitutions S199A, T222A, and S232A did not reduce Itch phosphorylation, a significant reduction in phosphorylation was observed upon combination of these mutations (Fig. 1C). A combination of all three mutations [S199A, T222A, and S232A, i.e., Itch(3A)] completely eliminated Itch phosphorylation. The Itch(3A) substitution mutant was no longer subject to JNK1-mediated phosphorylation in cells (Fig. 1D).
Fig. 1.
Mapping the JNK1 phosphorylation sites of Itch. (A) Schematic representation of Itch and its structural motifs. Locations of possible MAPK phosphorylation sites and the D domain are indicated by the numbers below the bar. CNBr cleavage sites are indicated by arrowheads. (B) Identification of a CNBr cleavage product containing JNK1 phosphorylation sites. (Left) Full-length Itch labeled with 32P in HEK293 cells was immunoprecipitated and digested with CNBr. (Right) H6-ItchΔC2 was incubated with recombinant JNKK2-JNK1 in the presence of [γ-32P]ATP and digested with CNBr. Digests were separated on Tris-Tricine gels along with molecular mass markers. (C) Identification of individual JNK1 phosphorylation sites. (Top) WT Itch and Ala substitution mutants thereof affecting all of the potential MAPK phosphorylation sites within the CNBr fragment identified above were incubated with JNKK2-JNK1 and [γ-32P]ATP . (Middle and Bottom) Itch proteins were also immunoblotted (IB) with anti-Xpress antibody (Bottom) and JNKK2-JNK1 was detected with anti-HA antibody (Middle). (D) In vitro JNKK2-JNK1 phosphorylation sites are also sites for JNK1-mediated phosphorylation in intact cells. WT Itch and the Ala substitution mutants were cotransfected into HEK293 cells along with pSRα-JNKK2-JNK1 or an empty vector. (Top) After 24 h, cells were labeled with [32P]orthophosphate, Itch proteins were immunoprecipitated with anti-Xpress, and the gels were separated and autoradiographed. (Middle and Bottom) Part of each lysate was immunoblotted with anti-Xpress (Middle) or anti-HA (Bottom) antibodies. (E) Itch T222 is phosphorylated in response to T cell receptor ligation. Mekk1ΔKD/Δ[supi]KD or Mekk1+/Δ[supi]KD thymocytes were stimulated with anti-CD3 and anti-CD28. At the indicated time points, whole-cell lysates were prepared and analyzed by immunoblotting with anti-pT222 Itch (Upper) or anti-Itch (Lower) antibodies. (F) T cell receptor ligation induces Itch Ser phosphorylation. Mekk1ΔKD/Δ[supi]KD and Mekk1+/Δ[supi]KD thymocytes were stimulated and analyzed as above, except that Itch immunoprecipitates (IP) were immunoblotted with anti-pS (Upper) or anti-Itch (Lower) antibodies.
Table 1.
Itch mutations and their location
| Mutation | Location |
|---|---|
| 199A/D | PRR |
| 222A/D | PRR |
| 232A/D | PRR |
| 199A, 222A | PRR |
| 222A, 232A | PRR |
| 199A, 222A, 232A | PRR |
| 595A, 596A | D domain |
| 595A, 596A, 597A | D domain |
| 595A, 596A, 597A, 599A | D domain |
To confirm whether Itch is phosphorylated on some of these sites in activated T cells, an antibody recognizing phospho-T222 was generated. The phospho-T222 Itch antibody recognizes Itch phosphorylated by JNKK2-JNK1, but did not detect similarly treated Itch T222A substitution mutation (data not shown). After stimulation of Mekk1+/ΔKD thymocytes with anti-CD3 and anti-CD28, induction of T222 phosphorylation was clearly observed (Fig. 1E). The kinetics of T222 phosphorylation were similar to those of Itch phosphorylation detected by 2D PAGE of T cell lysates (7) and endogenous 32P-labeled Itch immunoprecipitated from thymocytes activated with anti-CD3 and anti-CD28 (our unpublished results). T222 phosphorylation was not observed in activated Mekk1ΔKD/ΔKD thymocytes (or Jnk1−/− thymocytes; data not shown), demonstrating that phosphorylation of this site depends on MEKK1 kinase activity (Fig. 1E). Similarly, a clear induction of Itch Ser phosphorylation was seen in Mekk1+/ΔKD but not Mekk1ΔKD/ΔKD thymocytes stimulated by anti-CD3 and anti-CD28 (Fig. 1F). These results demonstrate that Itch is multiply phosphorylated after T cell receptor engagement and that its phosphorylation depends on the JNK kinase kinase MEKK1. Phosphorylation of Itch was not accompanied by changes in its expression level.
JNK1 Binds to Itch Through Its D Domain.
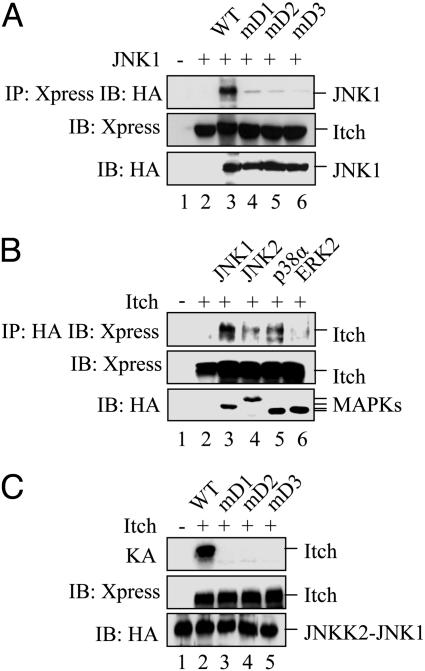
MAPKs interact with their substrates by docking onto D domains (20), a cluster of basic residues followed by a cluster of hydrophobic residues first identified in c-Jun and shown to be essential for its phosphorylation by JNK (16, 21). Itch amino contains such a sequence between residues 595 and 604 (Table 2), similar to the D domain of JIP-1 (22). To analyze its function, the Itch D domain was subjected to Ala substitution mutagenesis (Table 2). WT Itch coimmunoprecipitated with JNK1 when expressed in HEK293 cells, but this interaction was attenuated by each of the D domain mutations (Fig. 2A). Coimmunoprecipitation was also used to analyze the interaction of Itch with different MAPKs. Itch interacted more efficiently with JNK1 than either JNK2, p38α MAPK, or extracellular signal-regulated kinase 2 (Fig. 2B) and illustrates the specificity of the interaction between Itch and JNK1. Because proper MAPK docking is an important prerequisite for substrate phosphorylation (16, 21), it was not surprising that the D domain mutations inhibited Itch phosphorylation by the JNKK2-JNK1 fusion protein as efficiently as the Itch(3A) mutation (Fig. 2C, lane 2 compared with lanes 3–5).
Table 2.
Comparison of the JIP-1 and Itch D domain and its mutant derivatives
| D domain | (R/K)2X1–6ϕXϕ |
|---|---|
| JIP-1 | 155KRPTTLNLFP164 |
| Itch | 595RRRLWVIFPG604 |
| mD1 | 595AARLWVIFPG604 |
| mD2 | 595AAALWVIFPG604 |
| mD3 | 595AAALWAIFPG604 |
Fig. 2.
JNK1 binds the Itch D domain. (A) Itch interacts with JNK1 through its D domain. Xpress-tagged WT Itch and D domain substitution mutants were coexpressed in HEK293 cells with a HA-JNK1 expression vector. (Top) Itch proteins were immunoprecipitated, and association with JNK1 was analyzed by immunoblotting (IB) with anti-HA antibody. (Middle and Bottom) Cell lysates were also immunoblotted with anti-Xpress (Middle) or anti-HA (Bottom) antibodies. (B) Preferential interaction between JNK1 and Itch. HEK293 cells were cotransfected with Xpress-Itch and various HA-tagged MAPK expression vectors. (Top) After immunoprecipitation with anti-HA antibody, the presence of Itch was examined by immunoblotting with anti-Xpress antibody. (Middle and Bottom) Unfractionated lysates were immunoblotted with anti-Xpress (Middle) or anti-HA (Bottom) antibodies. (C) D domain mutations prevent JNK1-mediated phosphorylation. (Top) WT Itch or D domain substitution mutants were incubated with JNKK2-JNK1 and [γ-32P]ATP, and, after gel separation, their phosphorylation was examined by autoradiography. (Middle and Bottom) Reaction mixtures were also analyzed by immunoblotting with anti-HA (Middle) or anti-Xpress (Bottom) antibodies.
Phosphorylation Is Necessary and Sufficient for Itch Activation.
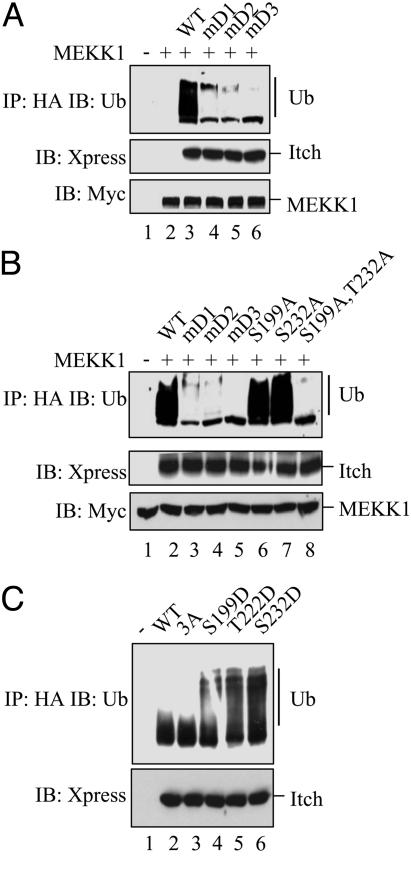
Unlike RING finger E3 ligases, which mostly function as adaptor proteins lacking endogenous catalytic activity, HECT domain E3 ligases possess intrinsic enzymatic activity and exhibit structural similarity to Ubc E2 enzymes (2, 3). We previously demonstrated that JNK1-mediated phosphorylation enhances Itch self-ubiquitylation and JunB ubiquitylation (7). Therefore, we examined the effect of the different D domain mutations on Itch activity. The D domain mutations greatly reduced the level of Itch self-ubiquitylation (Fig. 3A). As shown below (see Fig. 5A), self-ubiquitylation depends on the Itch catalytic domain and is therefore a valid measure of its activity. Although single Ala substitutions of the JNK1 phosphoacceptor residues had no effect on Itch activity, a double substitution of S199A and S232A almost completely inactivated Itch (Fig. 3B). Thus, only those mutations that considerably reduce Itch phosphorylation by JNK1 impact its activity. To further investigate how phosphorylation affects Itch activity, phosphomimic Asp mutations were generated. Even single-substitution mutations, S199D, T222D, or S232D, markedly enhanced Itch activity in the absence of JNK-mediated phosphorylation (Fig. 3C). These findings also explain why elimination of a single phosphorylation site has no effect on Itch activation. The presence of a negative charge at either of these positions within the PRR is sufficient for increasing Itch catalytic activity.
Fig. 3.
The JNK1 phosphoacceptor sites and the D domain are required for JNK1-mediated Itch activation. (A) D domain mutations prevent Itch activation by JNK1. Xpress-tagged WT Itch or D domain mutants were coexpressed in HEK293 cells with Myc-tagged MEKK1. (Top) Itch proteins were immunoprecipitated with anti-Xpress antibody and subjected to self-ubiquitylation in vitro. The extent of Itch ubiquitylation was examined by immunoblotting of gel-separated reaction mixtures with anti-Ub antibody. (Middle and Bottom) Itch and MEKK1 levels were examined by immunoblotting of unfractionated cell lysates with anti-Xpress (Middle) or anti-Myc (Bottom) antibodies. (B) JNK1-mediated phosphorylation is required for Itch activation. Xpress-tagged WT Itch, D domain (mD1, mD2, and mD3), and JNK1 phosphoacceptor site (S199A and T222A) mutants were expressed in HEK293 cells along with Myc-tagged MEKK1. (Top) Xpress-tagged Itch proteins were immunoprecipitated and subjected to self-ubiquitylation in vitro, and gels were separated and ubiquitylation was analyzed by immunoblotting with anti-Ub antibody. (Middle and Bottom) Unfractionated cell lysates were immunoblotted with anti-Xpress (Middle) or anti-Myc (Bottom) antibodies. (C) Phosphomimic substitution mutations activate Itch. Xpress-tagged WT Itch and mutants that either abolish (S199A, T222A, and S232A) or mimic (S199D, T222D, or S232D) its JNK1 phosphoacceptor sites were expressed in HEK293 cells. After immunoprecipitation with anti-Xpress, self-ubiquitylation activity of the Itch proteins was examined as described for A.
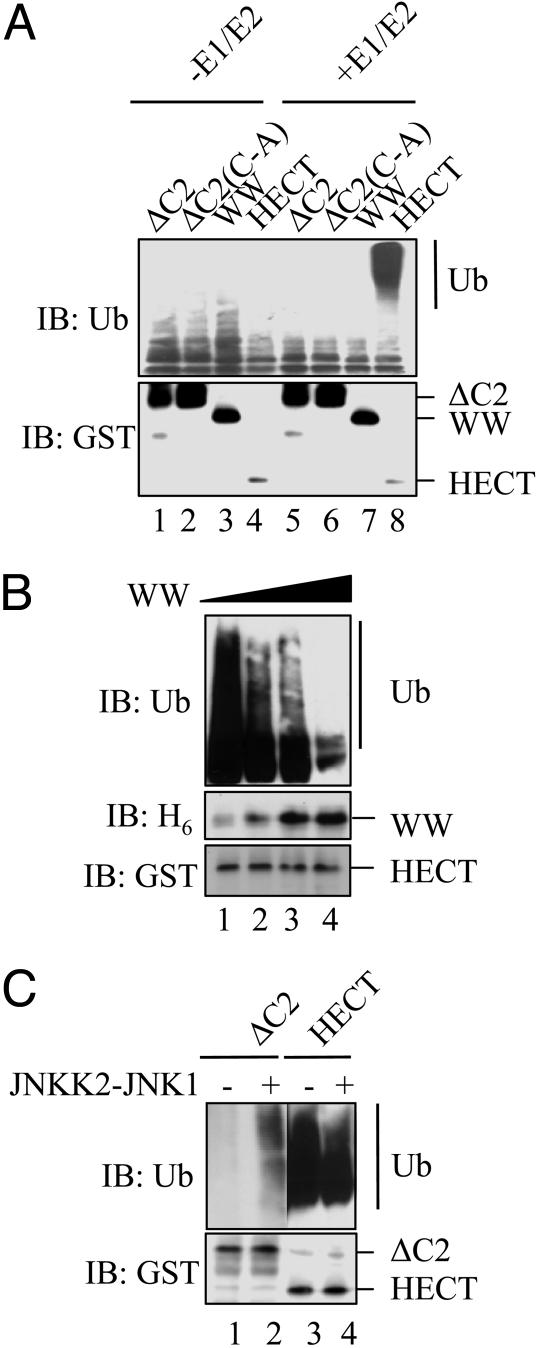
Fig. 5.
The Itch HECT domain is inhibited by N-terminal sequences. (A) Self-ubiquitylation activity of different Itch fragments was tested in the presence (lanes 5–8) or absence (lanes 1–4) of E1 and E2 (Ubc7). HECT(C-A) is a catalytically inactive mutant used to demonstrate the dependence of polyubiquitylation on Itch activity. Reaction mixtures were analyzed by immunoblotting with anti-Ub (Upper) or anti-GST (Lower) antibodies. (B) The enzymatic activity of the Itch HECT domain is inhibited by the WW domain. GST-ItchHECT was incubated with increasing quantities of H6-ItchWW (0, 0.25, 0.5, and 1 μg) together with E1 and E2. Reaction mixtures were analyzed by immunoblotting with anti-Ub (Top), anti-H6 (Middle), or anti-GST (Bottom) antibodies. (C) Suppression of HECT activity is relieved by JNKK2-JNK1-induced phosphorylation. GST-ItchΔC2 or GST-ItchHECT were incubated in an ubiquitylation assay with (lanes 2 and 4) or without (lanes 1 and 3) JNKK2-JNK1. ItchΔC2 and HECT domain ubiquitylation was analyzed as described above.
Phosphorylation Modulates Intramolecular Interactions.
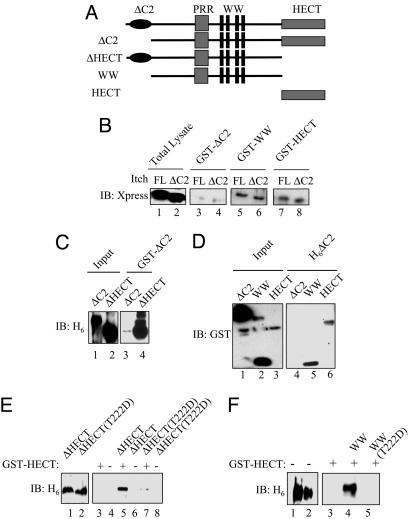
The results described above revealed that phosphorylation of Itch at residues located outside of the catalytic HECT domain exerts a strong effect on its enzymatic activity. Because Itch is a monomeric protein based on gel filtration analysis (our unpublished results), these results suggest that its activity may be regulated through intramolecular interactions. We therefore tested the ability of the different Itch functional domains to interact with each other and how these interactions are affected by phosphorylation. Fragments encompassing separate and multiple Itch domains were fused to GST or a hexahistidine (H6) tag, expressed in Escherichia coli and purified (Fig. 4A). The ability of GST-ItchΔC2, GST-ItchWW, or GST-ItchHECT to pull-down either full-length Itch or ItchΔC2 expressed in HEK293 cells was tested. GST-ItchWW and GST-ItchHECT interacted equally well with either full-length Itch or Itch ΔC2 (Fig. 4B). By contrast, full-length Itch or ItchΔC2 interacted much less efficiently with GST-ItchΔC2 and suggests that different regions of Itch can interact with each other and that this interaction involves mainly the WW and HECT domains. Furthermore, GST-ItchΔC2 appears to be in a conformation that interacts poorly with either full-length Itch or ItchΔC2, even though these proteins contain intact WW and HECT domains. This finding is consistent with our inability to detect oligomers of Itch by gel filtration.
Fig. 4.
Intramolecular interactions between different Itch domains. (A) Schematic representation of Itch constructs used in these experiments. (B) HEK293 cells were transfected with the indicated Xpress-Itch vectors and lysed. Lysates were incubated with recombinant GST-ItchΔC2, GST-ItchWW, or GST-ItchHECT and immobilized onto glutathione agarose beads, and bound proteins were analyzed by immunoblotting (IB) with anti-Xpress antibody. (C) Recombinant H6-ItchΔC2 or H6-ItchΔHECT was incubated with GST-ItchΔC2 and immobilized onto glutathione agarose, and bound proteins were analyzed by immunoblotting with anti-H6 antibody. (D) GST-ItchΔC2, GST-ItchWW, or GST-ItchHECT was incubated with H6-ItchΔC2 or H6-ItchΔHECT bound to Ni2+ agarose. After washing, the beads were spun down, and bound proteins were analyzed by immunoblotting with anti-GST antibody. (E) A phosphomimic mutation reduces the interaction between the WW and HECT domains. GST-ItchHECT was bound to glutathione agarose and incubated with H6-ItchΔHECT or H6-ItchΔHECT(T222D) produced in E. coli. GST pull-downs were performed and analyzed as above. (F) An Itch phosphomimic mutation inhibits the interaction between the WW and HECT domains. GST-ItchHECT was bound to glutathione agarose and incubated with H6-ItchWW or H6-ItchWW(T222D). GST pull-downs were performed and analyzed as described above.
To further investigate this hypothesis, an entirely in vitro interaction assay was performed. GST-ItchΔC2 interacted poorly with H6-ItchΔC2 but strongly with H6-ItchΔHECT (Fig. 4C) and suggests that the WW motifs present in H6-ItchΔHECT interact with the HECT domain in GST-ItchΔC2 and that H6-ItchΔC2 is in a conformation not available for an interaction with another ItchΔC2 molecule. Similarly, when the in vitro interaction assay was performed in reverse, H6-ItchΔC2 interacted strongly with either GST-ItchWW or GST-ItchHECT but weakly with GST-ItchΔC2 (Fig. 4D). The most parsimonial interpretation of these results is that full-length Itch or ItchΔC2 are engaged in intramolecular interactions between the HECT domain and the central WW domain (defined as the region containing the PRR and WW motifs) and are therefore unable to interact with another Itch molecule that contains the WW and HECT domains. However, when presented with an excess of isolated HECT or WW domains, these intramolecular interactions can be replaced by intermolecular interactions, a behavior governed by the law of mass action.
The ability of H6-ItchΔHECT and GST-ItchHECT to directly interact was tested by GST pull-down (Fig. 4E). H6-ItchΔHECT, which contains the PRR and WW motifs, was bound by GST-ItchHECT. The phosphomimic T222D mutation inserted into H6-ItchΔHECT abolished this interaction (Fig. 4E). The interaction between H6-ItchWW and GST-ItchHECT was similarly analyzed, and H6-ItchWW was efficiently retained by GST-ItchHECT, but the H6-ItchWW(T222D) mutant was not bound by GST-ItchHECT (Fig. 4F). These results strongly support a model based on intramolecular association between the WW domain and the HECT domain that is relieved by JNK1-mediated phosphorylation of residues within the PRR of Itch. Most likely, phosphorylation of the PRR alters the conformation of the central WW domain such that its affinity for the HECT domain is reduced. Although the Itch HECT domain is engaged by the WW domain, its N-terminal half, which contains the JNK docking site (D domain), remains available for the binding of JNK1 (Fig. 2B). These results are consistent with the recent identification of a flexible hinge region that separates the two halves of the HECT domain (15).
Itch Activity Is Negatively Regulated by Its WW Domain.
The enzymatic activity of GST-ItchΔC2, GST-ItchΔC2(C-A), GST-ItchWW, or GST-ItchHECT was tested in vitro in the presence or absence of E1 and E2 (Ubc7). In the absence of E1 and E2, Itch self-ubiquitylation was negligible, but, in the presence of E1 and E2, GST-ItchHECT exhibited strong self-ubiquitylation, an activity that was much higher than the one exhibited by the other GST-Itch fusion proteins (Fig. 5A). Such an activity was not exhibited by GST-ItchΔC2(C-A), in which the catalytic Cys residue was replaced with an Ala, confirming that self-ubiquitylation is indeed a reflection of Itch’s own catalytic activity. These results also indicate that sequences present N-terminal to the HECT domain, presumably the WW domain, suppress the catalytic activity of the HECT domain.
This inhibition may depend on the ability of the HECT domain to associate with the central WW domain, which contains the WW motifs and the PRR. To test this hypothesis, self-ubiquitylation by the HECT domain was measured in the presence of increasing concentrations of the GST-ItchWW fusion protein. Indeed, high concentrations of the ItchWW domain inhibited self-ubiquitylation of the ItchHECT domain (Fig. 5B). Given that the ItchΔC2 construct contains the PRR, which can be phosphorylated by activated JNK1 but has low enzymatic activity compared with the isolated ItchHECT domain, we tested whether JNK1-mediated phosphorylation enhances the activity of ItchΔC2. Indeed, coincubation of ItchΔC2 with active JNKK2-JNK1 fusion protein enhanced its self-ubiquitylation, whereas JNKK2-JNK1 had no effect on the already high self-ubiquitylation activity of the isolated HECT domain (Fig. 5C).
JNK1-Induced Phosphorylation Alters the Structure of Itch and Renders It Sensitive to Proteolysis.
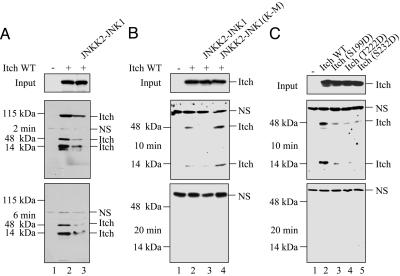
To test whether Itch undergoes a conformational change after JNK1-mediated phosphorylation, we examined the effect of phosphorylation on its sensitivity to proteolysis. N-terminally Xpress-tagged, full-length Itch was rapidly degraded by trypsin, generating two partial proteolysis products after 2 min of digestion (Fig. 6A). Under these conditions, JNK1-phosphorylated Itch was digested faster than nonphosphorylated Itch. Despite the accelerated digestion of the phosphorylated full-length Itch molecule, the amounts of the 46 and 16 kDa partial digestion products also were decreased after JNK1-mediated phosphorylation (Fig. 6A). These results suggest that JNK1-mediated phosphorylation increases the protease sensitivity of these subfragments, which are located at the N-terminal half of Itch (because they contain the Xpress tag). After 6 min, full-length Itch was no longer detected by the anti-Xpress antibody whether it was phosphorylated or not, but JNK1-mediated phosphorylation still resulted in a more rapid disappearance of the two partial digestion products, which contain the N-terminal Xpress tag (Fig. 3A). Thus, JNK1-mediated phosphorylation alters the structure of the N-terminal half of Itch, increasing its overall sensitivity to tryptic digestion.
Fig. 6.
Itch phosphorylation increases its susceptibility to proteolysis. (A) JNK1-mediated phosphorylation increases susceptibility to proteolysis. HEK293 cells were cotransfected with Xpress-tagged Itch expression vectors and JNKK2-JNK1. After 24 h, lysates were made. (Top) A portion of each lysate (Input) was analyzed by immunoblotting with anti-Xpress antibody to determine Itch expression. (Middle and Bottom) Another portion of each lysate was digested with trypsin for 2 min (Middle) or 6 min (Bottom) and analyzed by immunoblotting with anti-Xpress antibody. (B) (Top) HEK293 cells were cotransfected with Xpress-tagged Itch and either the JNKK2-JNK1 or JNKK2-JNK1(K-M) expression vector. (Middle and Bottom) After 24 h, lysates were made and analyzed as above, except that trypsin digestion was for 10 min (Middle) or 20 min (Bottom) min. (C) Phosphomimic Itch mutations alter sensitivity to proteolysis. HEK293 cells were transfected with WT Itch or phosphomimic substitution mutants. After 24 h, lysates were prepared, and Itch expression levels (Top) and sensitivity to trypsin proteolysis after 10 min (Middle) or 20 min (Bottom) of digestion was analyzed as described above.
To confirm that enhanced Itch proteolysis was due to JNK-mediated phosphorylation, HEK293 cells were cotransfected with either JNKK2-JNK1 or JNKK2-JNK1(K-M) (where K is Lys and M is Met) expression vectors. Itch cotransfected with the kinase inactive construct was considerably more resistant to proteolysis than Itch cotransfected with JNKK2-JNK1 (Fig. 6B). Similarly, the Itch phosphomimic mutants (S199D, T222D, or S232D) were more sensitive to tryptic digestion than WT Itch (Fig. 6C), demonstrating that the sensitivity of the N-terminal half of Itch containing the PRR and the WW motifs to tryptic digestion is influenced by JNK1-mediated phosphorylation. The most likely interpretation of these results is that Itch undergoes a conformational change after JNK1-mediated phosphorylation at the PRR region that renders it more sensitive to proteolysis in vitro. Despite being present in a more “open” conformation, we found no evidence that activated Itch is turned over more rapidly in intact cells (Fig. 1E).
Discussion
Although initially thought to be constitutively active, it has become increasingly clear that HECT domain E3 ligases are subject to regulation (4). Nedd4-2 is a HECT E3 ligase that is negatively regulated by Sgk1-mediated phosphorylation (23). However, phosphorylation does not appear to alter the catalytic activity of the HECT domain; rather, it reduces the ability of Nedd4-2 to associate with its substrate, the epithelial Na+ channel. The domain of Nedd4-2 responsible for substrate binding and how it is affected by phosphorylation has not been identified. Smurf2, a HECT E3 ligase involved in TGF-β signaling, was shown to be regulated by a mechanism involving an interaction with phosphorylated Smad7, which in turn facilitates the recruitment of Ubc7 (24). In addition, the activity of the recently described HECT E3 ligase Mule/ARF-BP1 is negatively regulated by its binding partner ARF (25), and we speculate that phosphorylation may modulate the substrate specificity of this enzyme, thereby explaining its diametrically opposed, substrate-dependent effects on cell survival (25–27). Therefore, understanding the mechanisms through which protein phosphorylation and other modifications control the activity of HECT E3 ligases has become of great importance in cellular regulation.
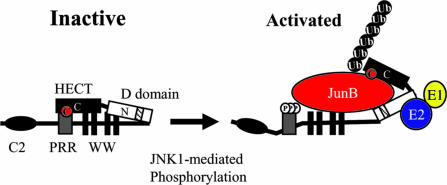
Here we describe a mechanistic analysis of the regulation of a HECT E3 ligase by phosphorylation. Our results suggest that the central region of Itch, which contains its PRR and WW motifs, interacts with its HECT domain to inhibit the activity of the HECT domain. Upon JNK1-mediated phosphorylation of sites located within the PRR, the intramolecular interaction between the central WW domain of Itch (defined as the PRR plus the WW motifs) and the HECT domain is weakened, most likely as a result of electrostatic repulsions that alter the structure of the WW domain (Fig. 7). This conformational change results in a dramatic increase in the catalytic activity of the HECT domain because it relieves the inhibitory interaction between the HECT domain and the WW domain. A similar increase in catalytic activity can be brought about by severing the HECT domain from the rest of the molecule or by introduction of phosphomimic mutations into the PRR. In addition to activating the HECT domain, phosphomimic substitutions also induce the same conformational change as brought about by JNK1-mediated phosphorylation and, thus, weaken the interaction between the WW and HECT domains. A similar mechanism may be involved in the activation of other HECT domain E3 ligases.
Fig. 7.
Itch activity is regulated by a phosphorylation-induced conformational change. In its unphosphorylated state, the activity of the Itch HECT domain, composed of N- and C-terminal halves separated by a hinge, is inhibited through an interaction with the WW domain, composed of the PRR and WW motifs. JNK1 binds the D domain (checked bar within the N-terminal half of the HECT domain) and phosphorylates Itch on three sites within the PRR and alters the conformation of the WW domain and weakens the interaction between it and the HECT domain, thereby increasing the catalytic activity of the HECT domain. The WW motifs of Itch may also mediate JunB recruitment.
Materials and Methods
Transfections, Antibodies, and Plasmids.
Transient transfections were performed by using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions, with 6-cm2 dishes and 8 μg of total DNA per transfection. The antibodies used were anti-Xpress (Santa Cruz Biotechnology), anti-hemagglutinin (anti-HA) (Roche), anti-Myc (Santa Cruz Biotechnology), anti-GST (Pharmingen), anti-Ub (Zymed), anti-Itch (generated by Y. C. Liu, La Jolla Institute for Allergy and Immunology), anti-phospho-T222 Itch (a gift from Chemicon International), anti-CD3ε, and anti-CD28 (both from Pharmingen). pCDNA3-HA-MEKK1, pRSα-HA-JNKK2-JNK1, pCMV5-Myc-MEKK1, and pEF-Xpress-Itch were constructed as described in ref. 6. Itch mutations (Table 1) were constructed by using a QuikChange mutagenesis kit (Stratagene). The bacterial expression plasmids pRSET-ItchΔC2, pRSET-ItchWW, and pRSET-ItchΔHECT were constructed by standard cloning procedures (28) from pEF-Itch Xpress cDNA.
Ubiquitylation Assays.
H6-tagged Ub, E1, and GST-Ubc7 (E2) were prepared and used in ubiquitylation assays as described in ref. 7. Reaction mixtures contained 100 nM E1, 0.5 μM Ubc7, 5 μM Ub, 2 mM ATP, and immunopurified Itch or recombinant Itch fragments. To examine Itch self-ubiquitylation, reactions were incubated for 2 h at room temperature and analyzed by immunoblotting with anti-Ub.
Pull-Down Assays.
GST-ItchΔC2, GST-ItchWW, or GST-ItchHECT were bound to glutathione agarose beads (Sigma) and incubated for 2 h with lysates of transfected HEK293 cells. Beads were washed three times in GST wash buffer (250 mM NaCl/10 mM Tris, pH 7.5/1 mM DTT) and analyzed by immunoblotting. For H6 pull-down assays, H6-ItchΔC2 or H6-ItchΔHECT was bound to Ni2+ agarose beads (Qiagen, Valencia, CA), incubated with GST-Itch fusion proteins, and analyzed as described above.
Kinase Assays, in Vivo Metabolic Labeling, and CNBr Mapping.
Kinase assays are described in ref. 7. In vivo metabolic labeling of transfected HEK293 cells and thymocytes using [32P]orthophosphate (Amersham Pharmacia Biosciences) was performed as described in refs. 18 and 29. CNBr mapping including peptide separation on Tris-Tricine gels was as described in refs. 18 and 29.
Thymocyte Preparation and Stimulation and Partial Proteolysis.
Thymocytes were isolated from Mekk1ΔKD/ΔKD and Mekk1+/ΔKD mice as described in ref. 7. Thymocytes were stimulated with anti-CD3 and anti-CD28 over a 60-min time course, and whole-cell extracts were prepared (7). HEK293 cell lysates were digested by Trypsin (Sigma) under conditions of limited proteolysis as described in refs. 30 and 31. Digestions were stopped by boiling the reactions in SDS/PAGE loading buffer.
Acknowledgments
We thank Chemicon International for the generous gift of anti-pItch T222. This work was supported by National Institutes of Health Grants ES04151 and ES06376 (to M.K.). M.K. is an American Cancer Society Research Professor.
Abbreviations
- H6
hexahistidine
- PRR
proline-rich region
- Th2
T helper 2
- Ub
ubiquitin
- MAPK
mitogen-activated protein kinase
- HA
hemagglutinin.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Pawson T., Scott J. D. Trends Biochem. Sci. 2005;30:286–290. doi: 10.1016/j.tibs.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 2.Liu Y. C. Annu. Rev. Immunol. 2004;22:81–127. doi: 10.1146/annurev.immunol.22.012703.104813. [DOI] [PubMed] [Google Scholar]
- 3.Ingham R. J., Gish G., Pawson T. Oncogene. 2004;23:1972–1984. doi: 10.1038/sj.onc.1207436. [DOI] [PubMed] [Google Scholar]
- 4.Zhong Q., Gao W., Du F., Wang X. Cell. 2005;121:1085–1095. doi: 10.1016/j.cell.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 5.Joazeiro C. A., Wing S. S., Huang H., Leverson J. D., Hunter T., Liu Y. C. Science. 1999;286:309–312. doi: 10.1126/science.286.5438.309. [DOI] [PubMed] [Google Scholar]
- 6.Fang D., Elly C., Gao B., Fang N., Altman Y., Joazeiro C., Hunter T., Copeland N., Jenkins N., Liu Y. C. Nat. Immunol. 2002;3:281–287. doi: 10.1038/ni763. [DOI] [PubMed] [Google Scholar]
- 7.Gao M., Labuda T., Xia Y., Gallagher E., Fang D., Liu Y. C., Karin M. Science. 2004;306:271–275. doi: 10.1126/science.1099414. [DOI] [PubMed] [Google Scholar]
- 8.Li B., Tournier C., Davis R. J., Flavell R. A. EMBO J. 1999;18:420–432. doi: 10.1093/emboj/18.2.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hartenstein B., Teurich S., Hess J., Schenkel J., Schorpp-Kistner M., Angel P. EMBO J. 2002;21:6321–6329. doi: 10.1093/emboj/cdf648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lavender P., Cousins D., Lee T. Chem. Immunol. 2000;78:16–29. doi: 10.1159/000058813. [DOI] [PubMed] [Google Scholar]
- 11.Dong C., Yang D. D., Tournier C., Whitmarsh A. J., Xu J., Davis R. J., Flavell R. A. Nature. 2000;405:91–94. doi: 10.1038/35011091. [DOI] [PubMed] [Google Scholar]
- 12.Newton A. C. Curr. Biol. 1995;5:973–976. doi: 10.1016/s0960-9822(95)00191-6. [DOI] [PubMed] [Google Scholar]
- 13.Sudol M., Chen H. I., Bougeret C., Einbond A., Bork P. FEBS Lett. 1995;369:67–71. doi: 10.1016/0014-5793(95)00550-s. [DOI] [PubMed] [Google Scholar]
- 14.Sudol M. Trends Biochem. Sci. 1996;21:161–163. [PubMed] [Google Scholar]
- 15.Verdecia M. A., Joazeiro C. A., Wells N. J., Ferrer J. L., Bowman M. E., Hunter T., Noel J. P. Mol. Cell. 2003;11:249–259. doi: 10.1016/s1097-2765(02)00774-8. [DOI] [PubMed] [Google Scholar]
- 16.Kallunki T., Deng T., Hibi M., Karin M. Cell. 1996;87:929–939. doi: 10.1016/s0092-8674(00)81999-6. [DOI] [PubMed] [Google Scholar]
- 17.Huang T. T., Wuerzberger-Davis S. M., Seufzer B. J., Shumway S. D., Kurama T., Boothman D. A., Miyamoto S. J. Biol. Chem. 2000;275:9501–9509. doi: 10.1074/jbc.275.13.9501. [DOI] [PubMed] [Google Scholar]
- 18.Hunter T., Sefton B. Methods Enzymol. 1991;200:149–152. doi: 10.1016/0076-6879(91)01014-s. [DOI] [PubMed] [Google Scholar]
- 19.Zheng C., Xiang J., Hunter T., Lin A. J. Biol. Chem. 1999;274:28966–28971. doi: 10.1074/jbc.274.41.28966. [DOI] [PubMed] [Google Scholar]
- 20.Tanoue T., Maeda R., Adachi M., Nishida E. EMBO J. 2001;20:466–479. doi: 10.1093/emboj/20.3.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goldsmith E. J., Cobb M. H., Chang C. I. Methods Mol. Biol. 2004;250:127–144. doi: 10.1385/1-59259-671-1:127. [DOI] [PubMed] [Google Scholar]
- 22.Dickens M., Rogers J. S., Cavanagh J., Raitano A., Xia Z., Halpern J. R., Greenberg M. E., Sawyers C. L., Davis R. J. Science. 1997;277:693–696. doi: 10.1126/science.277.5326.693. [DOI] [PubMed] [Google Scholar]
- 23.Debonneville C., Flores S. Y., Kamynina E., Plant P. J., Tauxe C., Thomas M. A., Munster C., Chraibi A., Pratt J. H., Horisberger J. D., et al. EMBO J. 2001;20:7052–7059. doi: 10.1093/emboj/20.24.7052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ogunjimi A. A., Briant D. J., Pece-Barbara N., Le Roy C., Di Guglielmo G. M., Kavsak P., Rasmussen R. K., Seet B. T., Sicheri F., Wrana J. L. Mol. Cell. 2005;19:297–308. doi: 10.1016/j.molcel.2005.06.028. [DOI] [PubMed] [Google Scholar]
- 25.Chen D., Kon N., Li M., Zhang W., Qin J., Gu W. Cell. 2005;121:1071–1083. doi: 10.1016/j.cell.2005.03.037. [DOI] [PubMed] [Google Scholar]
- 26.Warr M. R., Acoca S., Liu Z., Germain M., Watson M., Blanchette M., Wing S. S., Shore G. C. FEBS Lett. 2005;579:5603–5608. doi: 10.1016/j.febslet.2005.09.028. [DOI] [PubMed] [Google Scholar]
- 27.Shmueli A., Oren M. Cell. 2005;121:963–965. doi: 10.1016/j.cell.2005.06.018. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook J., Russell D. W. Molecular Cloning: A Laboratory Manual. 3rd Ed. Woodbury, NY: Cold Spring Harbor Lab. Press; 2001. [Google Scholar]
- 29.DiDonato J., Mercurio F., Rosette C., Wu-Li J., Suyang H., Ghosh S., Karin M. Mol. Cell. Biol. 1996;16:1295–1304. doi: 10.1128/mcb.16.4.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Buchberger A., Theyssen H., Schroder H., McCarty J. S., Virgallita G., Milkereit P., Reinstein J., Bukau B. J. Biol. Chem. 1995;270:16903–16910. doi: 10.1074/jbc.270.28.16903. [DOI] [PubMed] [Google Scholar]
- 31.Buchberger A., Valencia A., McMacken R., Sander C., Bukau B. EMBO J. 1994;13:1687–1695. doi: 10.1002/j.1460-2075.1994.tb06433.x. [DOI] [PMC free article] [PubMed] [Google Scholar]