Abstract
Plants employ small RNA-mediated posttranscriptional gene silencing as a virus defense mechanism. In response, plant viruses encode proteins that can suppress RNA silencing, but the mode of action of most such proteins is poorly understood. Here, we show that the silencing suppressor protein P0 of two Arabidopsis-infecting poleroviruses interacts by means of a conserved minimal F-box motif with Arabidopsis thaliana orthologs of S-phase kinase-related protein 1 (SKP1), a component of the SCF family of ubiquitin E3 ligases. Point mutations in the F-box-like motif abolished the P0–SKP1 ortholog interaction, diminished virus pathogenicity, and inhibited the silencing suppressor activity of P0. Knockdown of expression of a SKP1 ortholog in Nicotiana benthamiana rendered the plants resistant to polerovirus infection. Together, the results support a model in which P0 acts as an F-box protein that targets an essential component of the host posttranscriptional gene silencing machinery.
Keywords: E3 ubiquitin ligase, RNA silencing, viral pathogenicity, viral suppressor
Posttranscriptional gene silencing (PTGS) in plants is an example of a widespread phenomenon in metazoa in which RNA transcripts are degraded in a sequence-specific manner through the intervention of homologous short (21–24 nt) RNAs known as siRNAs (1, 2). The presence of double-stranded RNA (dsRNA) in the cytoplasm triggers PTGS. siRNAs are generated from the dsRNA by members of the Dicer family of dsRNA-specific endonucleases and are loaded onto an Argonaut-containing multicomponent complex known as RISC (RNA-induced silencing complex), where they act as guide RNAs to mediate degradation of RNA sequences complementary to the siRNAs.
PTGS is important in host defense against viruses, and it is now recognized that many plant viruses encode silencing suppressor proteins, which can counter this defense response (3–5). There is no sequence homology between known silencing suppressor proteins of different virus genera, suggesting that they intervene at different steps in the PTGS pathway. However, with the exception of a group of silencing suppressors exemplified by P19, which binds siRNAs or related molecules and is thought to disrupt PTGS by sequestering these species (6–8), relatively little is known about the mode of action of silencing suppressor proteins.
The Poleroviruses (family Luteoviridae) are a group of plant viruses with a small (≈5.6 kb) plus sense-RNA genome and icosahedral virions that are phloem-limited in their hosts (9). P0, the ≈29-kDa protein encoded by the 5′-proximal gene on polerovirus genomic RNA (Fig. 1A), is a potent silencing suppressor (10). In this work, we show that P0 interacts by means of an F-box-like domain with Arabidopsis thaliana S-phase kinase-related protein 1 (SKP1) orthologs. SKP1 is a core component of the SCF family of E3 ubiquitin ligases and serves to tether the rest of the complex to an F-box protein, which provides specificity in binding to ubiquitin ligase substrate proteins (11, 12). Point mutations in the F-box-like motif of P0 that abolished the P0–SKP1 interaction also abolished the silencing suppressor activity of P0 and diminished viral pathogenicity. Finally, knockdown of SKP1 ortholog levels in Nicotiana benthamiana rendered the plants resistant to Polerovirus infection. Together, these observations support a model in which P0 functions as a virus-coded F-box protein to direct an essential component of the host’s PTGS-based virus defense system to the E3 ubiquitination ligase machinery.
Fig. 1.
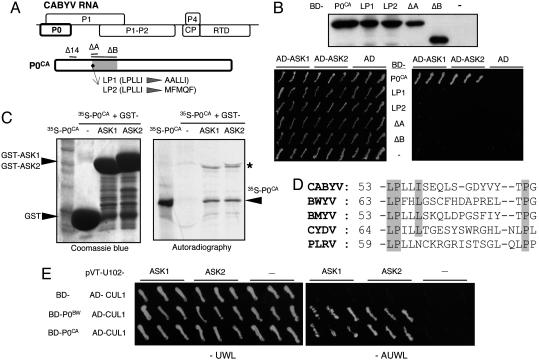
P0CA interacts with ASK1 and ASK2 via an F-box domain. (A) Genetic organization of CABYV RNA. Labeled rectangles symbolize important genes. The genetic organization of BWYV RNA is identical. The blow-up image shows the positions of deletions (above) and point mutations (below) in P0. Shading indicates the notional position of the ≈60-residue F-box domain. (B) Three independent colonies of yeast transformed with bait and prey plasmids expressing the indicated fusion proteins were replicate-streaked on nonselective medium (Left) and on medium selective for a strong two-hybrid interaction (Right). Upper is a Western blot showing accumulation of the WT and mutant P0CA fusion proteins in total yeast protein extracts. (C) Pull-down of [35S]methionine-labeled P0CA by glutathione S-transferase (GST) and GST–ASK1 and GST–ASK2 fusion proteins immobilized on glutathione-Sepharose beads. Proteins immobilized on the beads were visualized by Coomassie blue staining (Left) and 35S-P0CA by autoradiography (Right). An aliquot of the input 35S–P0CA was loaded in the leftmost lane of the gel. The minor bands marked by an asterisk in the two right-hand lanes of the autoradiogram comigrate with the GST–ASK1 and GST–ASK2 bands (Left) and may be 35S–P0CA, which has remained associated with the fusion proteins under denaturing conditions. (D) Sequences of P0 of CABYV, BWYV, Beet mild yellowing virus (RefSeq accession no. "NC 003491), Cereal yellow dwarf virus-RPV (NC 004751), and Potato leafroll virus (NC 001747) near the LPxxI/L motif and a downstream proline conserved in many plant F-box domains. (E) A yeast-bridging assay shows that interaction between P0BW and P0CA and AtCUL1 requires ASK1 or ASK2 as a bridging component. Three independent yeast AH109 colonies triply transformed with plasmids expressing the indicated BD-P0 and AD-CUL1 fusion proteins plus either empty pVT-U102 or pVT–U102 expressing ASK1 or ASK2 were replicate-streaked on a dropout plate under nonstringent (−UWL) and stringent (−AUWL) conditions selecting for interaction between BD–P0 and AD–CUL1. The plates were incubated at 21°C for 7 days.
Results
P0 interacts with Arabidopsis SKP1-Related (ASK1) and ASK2 in Yeast.
The Poleroviruses Beet western yellows virus (BWYV) and Cucurbit aphid-borne yellows virus (CABYV) infect Arabidopsis. Yeast were transformed with constructs expressing P0 from either BWYV (P0BW) or CABYV (P0CA) fused to the GAL4 DNA binding domain (BD), and the resulting fusion proteins were used as bait in two-hybrid screens of a cDNA library expressing A. thaliana proteins fused to the GAL4 activation domain (AD). With P0BW as bait, none of the 107 double transformants tested grew on selective medium. When P0CA was the bait, six of the 5 × 106 double transformants grew under strong selective conditions. All contained the same prey sequence, corresponding to ASK2 (At5g42190), which is an ortholog of the Saccharomyces cerevisiae Skp1 gene.
SKP1 is a core subunit of the multicomponent SCF (SKP1/Cullin1/F-box/RBX1) E3 ubiquitin ligase. The E3 ubiquitin ligases are a large and diverse group of proteins and protein complexes that can direct ubiquitination of specific target proteins as a signal for their degradation by the 26S proteasome (11, 12). SKP1 orthologs are found in many organisms, including higher plants, where SCF-mediated ubiquitination/proteolysis of target proteins regulates numerous pathways (13). The SKP1 subunit in the SCF complex acts as a specific adapter linking the Cullin1 (CUL1) scaffold protein to one of a large family of F-box proteins, which in turn selectively binds to a target protein. The target is then ubiquitinated by an E2 ubiquitin-conjugating enzyme, which docks at the RBX1 subunit of the SCF complex. Genes for 21 SKP1 orthologs have been identified in Arabidopsis, but ASK2 and the closely similar ASK1 are the most abundant and interact with many F-box proteins (14–16). Although in our original screen no double transformants containing ASK1 were obtained, an interaction with P0CA was observed when AD-ASK1 was expressed in yeast from a vector (pGADT7) with a stronger promoter (Fig. 1B).
The high degree of conservation of SCF components between yeast and plants (15) raised the possibility that a yeast protein could participate in the observed two-hybrid interactions. To determine whether P0CA can associate with ASK1 and ASK2 in the absence of other proteins, glutathione-Sepharose beads loaded with GST–ASK1, GST–ASK2, and GST (expressed in Escherichia coli) were incubated with [35S]methionine-labeled P0CA translation product. After washing, bound proteins were eluted from the beads and analyzed by SDS/PAGE. Coomassie blue staining of the gel revealed the GST, GST–ASK1, and GST–ASK2 in the eluates (Fig. 1C Left), whereas autoradiography detected 35S-labeled P0CA only in the eluates from the beads loaded with GST–ASK1 and GST–ASK2 (Fig. 1C Right). We conclude that association between P0CA and the ASKs is direct.
An F-box-like motif in P0 is necessary for interaction with ASK1 and ASK2. Its interaction with ASK1 and ASK2 suggested that P0CA might contain an F-box, the ≈60-residue domain that is generally situated near the N terminus of an F-box protein (11). The F-box consensus has no strictly invariant residues and contains gaps so that it is often difficult to reliably identify an F-box from the sequence alone. However, inspection of the P0CA sequence detected the short motif LPLLI (residues 53–57; Fig. 1D), which matches the start of the F-box consensus sequence (LPxxI/L), the most highly conserved part of the domain in plant F-box proteins (15). A similarly positioned motif is present in the P0s of other poleroviruses (Fig. 1D), even though overall sequence identity among different P0s is low (9).
To investigate the significance of the LPLLI sequence, mutant forms of P0CA were tested for interaction with ASK1 and ASK2 in the two-hybrid system. In mutants ΔA and ΔB, 10 or 62 residues were deleted starting with the LPLLI sequence. In mutants LP1 and LP2, the wild-type (WT) sequence was replaced by AALLI and MFMQF, respectively (Fig. 1A). The LP2 mutation does not change the amino acid sequence of P1, which is encoded by an overlapping ORF in viral RNA (Fig. 1A). All of the mutant proteins were stable in yeast but none reacted with ASK1 or ASK2 in the two-hybrid assay (Fig. 1B).
P0BW Interacts with ASK1 and ASK2.
P0BW also contains an F-box-like motif (LPFHL; Fig. 1D), but no yeast double transformants containing ASK1 or ASK2 were obtained during the two-hybrid screen with P0BW (see above) or when double-transformant yeast expressing BD-P0BW and either AD-ASK1 or AD-ASK2 were incubated at 28°C (see Fig. 5, which is published as supporting information on the PNAS web site). Incubation of the plates at 21°C, however, permitted growth of the double transformants on selective media (Fig. 5). The BD-P0CA/AD-ASK1 and BD-P0CA/AD-ASK2 double transformants also grew well at the lower temperature (Fig. 5). A P0BWLP1 mutant (LPFHL replaced by AAFHL) did not interact with ASK1 or ASK2 in yeast at either 21°C or 28°C (data not shown).
ASK1 and ASK2 Can Incorporate P0 into a Trimeric Complex with CUL1.
To determine whether P0BW and P0CA can form trimeric complexes with A. thaliana CUL1 and an ASK, bridge assays were carried out with BD-P0 and AD-AtCUL1 fusion proteins expressed together in yeast along with ASK1 or ASK2. Growth under selective conditions occurred when either ASK1 or ASK2 was provided as a bridging component but not when the ASKs were omitted (Fig. 1E). Thus, the association between ASK and P0 does not interfere with docking of the complex with CUL1.
P0CA and ASK1 Interact in Planta.
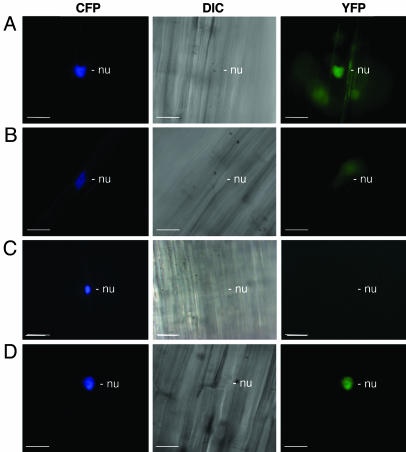
The interaction between P0CA and ASK1 in planta was investigated by using bimolecular fluorescence complementation (17, 18). In this assay, the yellow fluorescence protein (YFP) is expressed as N-terminal (YN) and C-terminal (YC) nonfluorescent fragments. Restoration of YFP fluorescence occurs when the two fragments are brought into proximity by an interaction between two proteins that have been fused to the YN and YC fragments, respectively. Plasmids expressing P0CA–YN and YC–ASK1 were cobombarded into epidermal cells of etiolated mustard seedlings. To identify transformed cells, the bombardment mix also contained a plasmid expressing the cyan fluorescent protein (CFP) fused to the parsley common plant regulatory protein 2 (CPRF2), which localizes to the nucleus (17). A YFP signal was observed in 26 of 41 transformed cells examined. The fluorescence was most intense in the nucleus (Fig. 2A), as observed for a control interaction between YN–ASK1 and YC–EID1 (empfindlicher im dunkelroten Licht), a pair of proteins known to interact in the nucleus (17) (Fig. 2D). In the case of the P0CA–YN/YC–ASK1 interaction, a faint YFP signal was also present in the cytoplasm (Fig. 2A). A similar result was obtained when the fusion partners were reversed: cobombardment with P0CA–YC and YN–ASK1 gave rise to YFP fluorescent nuclei in 34 of 56 transformed cells examined (data not shown).
Fig. 2.
Visualization of the interaction between P0 and ASK1 in planta using bimolecular fluorescence complementation. Three-day-old dark-grown mustard seedlings were transformed by particle bombardment with combinations of plasmids expressing different YN- and YC-fusion proteins. To identify transformed cells, a plasmid expressing the CFP fused to the parsley common plant regulatory protein 2 (CPRF2), which localizes to the nucleus (nu), was included during bombardment. Images were recorded 5 h after bombardment by using CFP-specific and YFP-specific filters. Shown are typical cells bombarded with plasmids expressing P0CA–YN and YC–ASK1 (A), P0CALP1–YN and YC–ASK1 (B), P0CA–YN and YC (C), and YN–ASK1 and YC–EID1 (D), a pair of proteins known to interact (17). A differential interference contrast (DIC) image is shown between each pair of fluorescent images. Regions of diffuse YFP fluorescence in A and B that are not confined to a single cell are background. (All images are at the same magnification; scale bar: 40 μm.)
To confirm the role of the F-box motif in the interaction, we examined cells cobombarded with P0CALP1–YN and YC–ASK1. Although 33 of the 41 transformed cells examined did not exhibit detectable YFP fluorescence, a faint YFP signal could be discerned in the nuclei of eight transformed cells (Fig. 2B). This result resembles the situation for cells bombarded with P0CA–YN and YC, where weak nuclear fluorescence was observed in only 9 of the 50 transformed cells examined (Fig. 2C). We conclude that the weak YFP signal sometimes obtained with P0CALP1 is nonspecific and that P0 interacts via its F-box motif with ASK1 in plant cells.
F-Box Motif Is Required for P0-Mediated Viral Pathogenicity.
We next asked whether the P0–SKP1 interaction is important for virus pathogenicity. As previously observed for BWYV (19), a CABYV mutant carrying a 14-nt deletion in the P0 ORF at a position upstream of the F-box domain (Δ14; Fig. 1A) was hypovirulent, accumulating ≈10 times less progeny viral RNA than plants infected with WT CABYV (data not shown). Similar low levels of progeny viral RNA accumulation were observed for a CABYV mutant carrying the LP2 mutation (data not shown).
The effect of the F-box mutation also was studied when P0 was expressed from an heterologous virus. In N. benthamiana infected with a Potato virus X (PVX)–P0BW chimera (see Fig. 6A, which is published as supporting information on the PNAS web site), accumulation of progeny viral RNA (Fig. 6B) in upper leaves was accompanied by severe necrosis of vascular tissue and death of the upper part of the plant (Fig. 6C). This result is in contrast to the mild leaf mosaic symptoms produced by PVX infection (Fig. 6C) but resembles the symptoms induced by other silencing suppressor proteins when expressed in the PVX background (20–22). When P0BWLP1 was substituted for P0BW in the chimera, the systemically infected leaves displayed only a few necrotic flecks (in addition to the mosaic symptoms typical of a PVX infection), and the plants survived (Fig. 6C). Collectively, these experiments implicate the association between P0 and plant SKP1 orthologs in the mechanism by which P0 enhances virus pathogenicity.
Depletion of SKP1 Ortholog(s) in N. benthamiana by Virus-Induced Gene Silencing (VIGS) Induces Resistance to BWYV.
If the P0–SKP1 interaction is important for virus pathogenicity, we reasoned that plants which produce less SKP1 should display heightened resistance to polerovirus infection. Arabidopsis lines carrying null mutations in both ASK1 and ASK2 are nonviable (23). Therefore, VIGS (24, 25) was used to lower SKP1 accumulation levels in the BWYV host N. benthamiana. The cDNA of an N. benthamiana SKP1 ortholog was cloned, and the encoded protein (NbSKP1) was shown to interact with P0BW and P0CA in the yeast two-hybrid system (data not shown). For the VIGS experiment, the sequence encoding all but the four N-terminal amino acids of NbSKP1 was inserted into the PVX genome. A long rather than a short NbSKP1 cDNA fragment was used to trigger VIGS in these experiments because we regard it as probable that, as in Arabidopsis, the SKP1-like molecules of N. benthamiana are encoded by a multigene family, and expression of a long cDNA fragment should increase the probability that transcripts of other members of the family that could otherwise complement NbSKP1 function will be targeted for silencing as well.
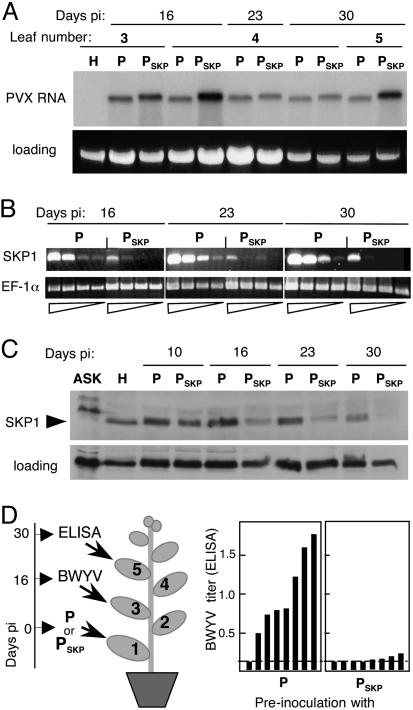
The PVX–NbSKP1 chimera transcript (PSKP) was rub-inoculated to a lower leaf (leaf 1, Fig. 3D) of young plants. As a control, plants also were inoculated with transcript of empty PVX vector (P). Symptoms of virus infection appeared on upper leaves of the inoculated plants by 8 days postinoculation (pi), and similar amounts of progeny viral RNA could be detected by Northern blot of total RNA extracted from the symptomatic leaves (Fig. 3A). Semiquantitative RT-PCR revealed that NbSKP1 mRNA accumulation in the PSKP-infected plants was reduced at 16–30 days pi to ≈5% of the levels observed in the P-infected plants (Fig. 3B). Western blot analysis using a SKP1-specific antiserum confirmed that NbSKP1 protein levels in the PSKP-infected plant were diminished (Fig. 3C). Starting at ≈15 days pi, the PSKP-inoculated plants developed a phenotype that is presumably a consequence of SKP1 depletion, including crinkling and epinasty of upper leaves and slight corkscrewing of the stem.
Fig. 3.
Depletion of SKP1 in N. benthamiana by VIGS provokes resistance to BWYV. (A) Northern blot analysis of PVX (P) and PVX-NbSKP1 (PSKP) progeny RNA in upper leaves of N. benthamiana at different times pi using a 32P-labeled RNA probe complementary to the 3′-terminal PVX RNA sequence. Leaf positions are indicated in D. Lane H is RNA from a healthy plant. (B) Inhibition of NbSKP1 transcript accumulation by VIGS. Reverse transcription followed by PCR with primers specific for the 3′ noncoding region of NbSKP1 mRNA was carried out on total RNA from leaf 4 (16 and 23 days pi) or 5 (30 days pi) of P- or PSKP-infected plants. In the dilution series, the amounts of cDNA template used for PCR were 1/10, 1/50, and 1/250 of that used in each left-hand lane. Products of RT-PCR amplification of a portion of the Elongation Factor 1α (EF-1α) sequence from the same RNA samples are shown as a control. (C) Inhibition of SKP1 accumulation by VIGS. NbSKP1 levels in leaf 3 were monitored at different times after inoculation with P or PSKP by Western blot using an SKP1-specific antiserum. The left-hand lane (ASK) was loaded with a protein extract from A. thaliana and lane H with protein from healthy N. benthamiana. (D) BWYV titer in leaf 5 of plants that had been preinoculated with PVX or PVX–NbSKP1. The plants were aphid-inoculated with BWYV 16 days after infection with PVX or PVX–NbSKP1 and tested by ELISA for BWYV at 30 days. (Right) Bars represent the ELISA A405 value for each plant. The ELISA background is indicated by the horizontal dashed line. The cartoon in Left shows the relative positions of the leaves subjected to the different treatments.
To test the effect of the lower NbSKP1 levels on BWYV infection, the leaves in position 3 of P- and PSKP-infected plants were inoculated at 16 days pi with aphids viruliferous for BWYV, and the leaves at position 5 were tested for virus infection by ELISA 2 weeks later (30 days pi). Viruliferous aphids rather than agro-infection with an infectious cDNA clone were used to deliver the BWYV inoculum because an F-box protein–SKP1 interaction has been implicated in uncoating of T-DNA in the plant cell nucleus before its integration into the host genome (26). High levels of BWYV were present in seven of the eight plants preinoculated with PVX (P), but virus levels were near background in the plants preinoculated with PSKP (Fig. 3D). We conclude that not only P0, but also its interaction partner SKP1, is required for efficient BWYV infection.
Mutation of the P0 F-Box Motif Inhibits Suppression of Gene Silencing.
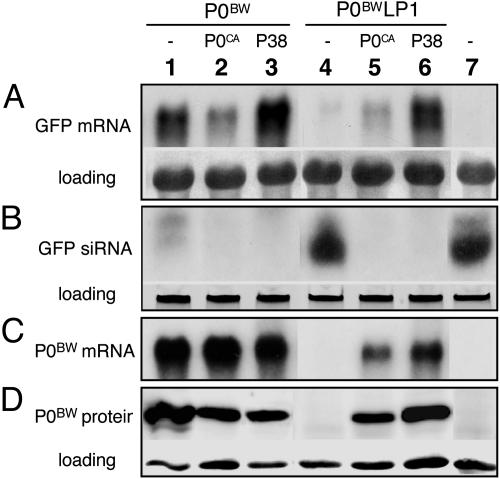
The above observations support a model in which interaction between P0 and one or more plant SKP1 orthologs is required for efficient polerovirus infection, but they fall short of establishing a direct link between the P0–SKP1 interaction and the silencing suppressor activity of P0. To address this question, we compared the ability of P0BW and P0BWLP1 to suppress RNA silencing induced by ectopic expression of a foreign transcript in an agro-infiltration assay (27). The assay employs N. benthamiana line 16c, which expresses GFP from a transgenic locus. Infiltration of a leaf with Agrobacteria harboring a pBin–GFP binary construct results in initial high levels of expression of GFP transcript in the infiltrated zone, which subsequently triggers PTGS-mediated degradation of the transcript. By 5 days postinfiltration, most of the GFP transcript expressed from the transgene and the agro-infiltrated pBin-GFP in the patch was degraded (Fig. 4A, lane 7), and GFP transcript-specific siRNAs appeared (Fig. 4B, lane 7). When Agrobacteria harboring a plasmid-expressing P0BW were coinfiltrated into a leaf along with the pBin–GFP-containing Agrobacteria, high levels of GFP transcript (Fig. 4A, lane 1) and low levels of GFP-specific siRNA were observed in the patches (Fig. 4B, lane 1). Note that the P0BW transcript is also abundant (Fig. 4C, lane 1) even though, like the GFP transcript, it is ectopically expressed and is expected to be a trigger for and a target of PTGS. The silencing suppressor protein encoded by the transcript, however, would “protect” it from degradation.
Fig. 4.
The LP1 mutation inhibits the silencing suppressor activity of P0BW but does not destabilize the protein. Leaves of N. benthamiana line 16c were infiltrated with a mixture of Agrobacteria strains containing pBin–GFP plus either empty vector (pBin61, lane 7) or pBin61 expressing the silencing suppressor protein(s) indicated at the top (lanes 1–6). Total RNA was extracted 5 days later from the agro-infiltrated patches and analyzed for the presence of GFP transcript (A), GFP transcript-derived siRNAs (B), and P0BW transcript (C) by Northern blot using specific 32P-labeled probes. Total protein was extracted from the same patches and analyzed for P0BW protein by Western blot using a P0BW-specific polyclonal antiserum (D). Loading controls are shown below each blot.
When P0BWLP1 was substituted for WT P0BW in the agro-infiltration assay, GFP transcript levels (Fig. 4A, lane 4) were almost as low as in patches infiltrated with Agrobacteria containing the empty vector pBin61 (Fig. 4A, lane 7). Accumulation of P0BWLP1 transcript was also low (Fig. 4C, lane 4), as expected if the LP1 mutation abolishes P0s silencing suppressor activity. Similar results were obtained when the silencing suppressor activities of P0CA and P0CALP1 were compared (data not shown).
The foregoing observations are consistent with the hypothesis that a functional F-box motif is required for P0 silencing suppressor activity. The possibility remains open, however, that the LP1 mutant is for some reason less stable than the WT protein in planta, even though a functional F-box is usually a destabilizing protein motif (11). Thus, an unlikely but possible alternative scenario would be that failure of P0BWLP1 to suppress silencing is because of low accumulation levels rather than loss of silencing suppressor activity. To test this hypothesis, we added to the agro-infiltration assay mix a binary vector expressing a second RNA silencing suppressor, P38 of Turnip crinkle virus (TCV) (28). Addition of pBin–P38 stabilized the GFP and the P0BWLP1 transcripts (Fig. 4 A and C, lane 6) so that they accumulated to ≈65% of the level observed in patches agro-infected with WT pBin–P0BW plus pBin–P38 (Fig. 4 A and C, lane 3). Importantly, Western blot analysis of protein in the patches revealed that P0BWLP1 accumulated in amounts comparable with those observed for P0BW (Fig. 4D, compare lanes 3 and 6). Similar results were obtained when P0CA was used as the secondary silencing suppressor. Although the degree of protection afforded by P0CA was lower than with P38, the P0BW and P0BWLP1 proteins accumulated to similar levels (compare Fig. 4D, lanes 2 and 5). Together, these experiments establish that the inability of P0BWLP1 to suppress RNA silencing is a consequence of loss of silencing suppressor activity rather than instability of the mutant protein, and we can therefore conclude that the F-box motif is required for the silencing suppressor activity of P0.
Discussion
Many animal viruses exploit the cell’s ubiquitination/proteolysis machinery to inhibit host responses or more generally alter the cellular environment to favor infection (29). These viruses typically act at the ubiquitination step, either by expressing a novel E3 ligase with appropriate properties from the virus genome or by altering the specificity of a host E3 ligase. Examples of the latter strategy are the Vif protein of HIV-1 (HIV-1) and the Adenovirus proteins E4orf6 and E1B55K, which direct their target proteins (APOBEC3G and p53, respectively) to a Cul5-containing SCF-like complex (30, 31). None of these viral proteins, however, is a conventional F-box protein, and, indeed, only one viral protein other than P0 has been demonstrated to interact with a SKP1 ortholog. This protein is the Faba bean necrotic yellows virus protein CLINK, which may deregulate the host cell cycle in favor of viral DNA replication by targeting a pRB-like protein (32).
The most straightforward interpretation of our findings is that polerovirus silencing protein P0 acts as an F-box protein that targets an essential component of the host small RNA-dependent RNA degradation virus defense pathway. We cannot rule out the possibility that the P0–SKP1-ortholog complex recognizes and inactivates its target protein by simple sequestration, but, given the ability of the P0–ASK complex to assemble with AtCUL1, we regard it as more likely that P0 incorporates its substrate protein into an SCF complex for ubiquitination. Addition of ubiquitin chains to a target protein by the SCF generally destines it for degradation by the proteasome, although other scenarios that do not involve target degradation cannot be eliminated (33). It is furthermore possible that the P0–SKP1 complex could act indirectly, perhaps as an antagonist of a cellular F-box protein, which normally degrades a negative regulator of the silencing pathway. Discrimination among these various possibilities should be facilitated once the ultimate target or targets of P0 in the silencing pathway is identified. It also will be interesting to determine whether other viral silencing suppressor proteins employ an E3 ubiquitin ligase (although not necessarily an SCF E3 ligase) and ubiquitin-mediated processes to target proteins of the PTGS pathway in the cytosol.
F-box proteins generally interact with their targets via a C-terminal domain that is often, but not always, a known protein-interaction motif such as a leucine-rich repeat (LRR) or a kelch domain (11). No such motif is present in P0, but P0CA and P0BW both contain a C-terminal-proximal sequence (K/R)IYGEDGX3FWR (related sequences are present in other polerovirus P0s), which could represent a previously undescribed type of substrate interaction domain. Our future studies will address the problem of determining which component of the host silencing machinery is the target of the putative SCFP0 E3 ubiquitin ligase.
Materials and Methods
Gene Constructs, Virus Infection, and Agro-Infection.
Plasmid constructions are described in detail in Supporting Materials and Methods, which is published as supporting information on the PNAS web site. Production and inoculation of infectious chimera transcripts of PVX were as described in ref. 10. A. thaliana were infected with CABYV by agro-inoculation (34). N. benthamiana were infected with BWYV by using 30 viruliferous Myzus persicae per leaf and a 4-day inoculation access period (35). The aphids were confined to a single leaf by using a clip-on cage. BWYV levels were assayed by double antibody-sandwich ELISA on leaf tissue extracts with a BWYV-specific antiserum (Loewe Biochemica, Sauerlach, Germany). Western blot analysis of NbSKP1 levels used an antiserum raised against a peptide corresponding to the N-terminal sequence of ASK1 (36). The loading control was obtained with an anti-Cdc2 (PSTAIRE) polyclonal antibody (Santa Cruz Biotechnology). Detection of NbSKP1 transcript by semiquantitative PCR is described in Supporting Materials and Methods.
Agro-infiltration of leaves of N. benthamiana line 16c (34) and Northern and Western blot analysis were performed as described in refs. 10 and 37 with TRIzol (Invitrogen) used for siRNA extraction. Radioactivity in bands was quantified with a Phosphorimager Bas1000 (Fujix, Kyoto, Japan).
Two-Hybrid Assay.
Two-hybrid screening of an A. thaliana cDNA library (Clontech) was carried out in AH109 as described by the Clontech Matchmaker Protocol with a first round of selection on dropout plates lacking histidine, tryptophan, and leucine (−HWL) followed by more stringent selection on plates lacking adenine as well as the three above-mentioned amino acids (−AHWL). BD–P0 fusion proteins were detected by Western blot using a monoclonal antibody (Roche) directed against a myc epitope encoded by the spacer sequence between the BD and P0 coding regions. In bridging assays, ASK1 and ASK2 were expressed from pVT–U102 (38).
Pull-Down Experiments.
GST–ASK1, GST–ASK2, and GST in E. coli extracts were immobilized on glutathione-Sepharose beads, which then were incubated with [35S]methionine-labeled P0CA. A detailed description is provided in Supporting Materials and Methods.
Bimolecular Fluorescence Complementation.
The P0CA and P0CALP1 coding sequences were isolated as BamHI–EcoRI restriction fragments and cloned into pENTR3C (Invitrogen). The resulting entry vectors were used to introduce the P0 sequences into the split YFP destination vectors by Gateway Scientific (St. Louis) technology to obtain P0–YN, P0–YC, and P0LP1–YN constructs. 35S–CPRF2–CFP, YN–ASK1, YC–ASK1, and YC–EID1 are described elsewhere (17). The different constructs were transformed into 3-day-old, dark-grown mustard seedlings by particle bombardment (17). Images were recorded 5 h after bombardment with an Axioskop II fluorescence microscope (Zeiss) by using CFP- and YFP-specific filters and 9-ms and 2-s exposure times, respectively.
Supplementary Material
Acknowledgments
We thank David Baulcombe (The Sainsbury Laboratory, Norwich, U.K.) and Olivier Voinnet (Institut de Biologie Moléculaire des Plantes du Centre National de la Recherche Scientifique) for transgenic plants and constructs; Ahmed Ghannam (Institut de Biologie Moléculaire des Plantes du Centre National de la Recherche Scientifique) for materials and advice concerning semiquantitative PCR; and Patrice Dunoyer, Christophe Himber, Marie-Claire Criqui, Thomas Potuschak, and Alexis Thomann for suggestions. M.P. was supported by Société Française d’Exportation des Ressources Éducatives, and K.M. was funded by European Union Grant HPRN-CT-2002-00333. Additional support was provided by Centre National de la Recherche Scientifique.
Abbreviations
- PTGS
posttranscriptional gene silencing
- SKP1
S-phase kinase-related protein 1
- ASK
Arabidopsis SKP1-like
- YFP
yellow fluorescent protein
- CFP
cyan fluorescent protein
- BD
binding domain
- CABYV
Cucurbit aphid-borne yellows virus
- BWYV
Beet western yellows virus
- PVX
Potato virus X
- AD
activation domain
- CUL1
Cullin1
- pi
postinoculation
- YN
N-terminal YFP
- YC
C-terminal YFP
- VIGS
virus-induced gene silencing
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Baulcombe D. Nature. 2004;431:356–363. doi: 10.1038/nature02874. [DOI] [PubMed] [Google Scholar]
- 2.Zamore P. D., Haley B. Science. 2005;309:1519–1524. doi: 10.1126/science.1111444. [DOI] [PubMed] [Google Scholar]
- 3.Silhavy D., Burgyan J. Trends Plant Sci. 2004;9:76–83. doi: 10.1016/j.tplants.2003.12.010. [DOI] [PubMed] [Google Scholar]
- 4.Roth B. M., Pruss G. J., Vance V. B. Virus Res. 2004;102:97–108. doi: 10.1016/j.virusres.2004.01.020. [DOI] [PubMed] [Google Scholar]
- 5.Voinnet O. Nat. Rev. Genet. 2005;6:206–220. doi: 10.1038/nrg1555. [DOI] [PubMed] [Google Scholar]
- 6.Silhavy D., Molnar A., Lucioli A., Szittya G., Hornyik C., Tavazza M., Burgyan J. EMBO J. 2002;21:3070–3080. doi: 10.1093/emboj/cdf312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vargason J. M., Szittya G., Burgyan J., Tanaka Hall T. M. Cell. 2003;115:799–811. doi: 10.1016/s0092-8674(03)00984-x. [DOI] [PubMed] [Google Scholar]
- 8.Ye K., Malinina L., Patel D. J. Nature. 2003;426:874–878. doi: 10.1038/nature02213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mayo M. A., Ziegler-Graff V. Adv. Virus Res. 1996;46:413–460. doi: 10.1016/s0065-3527(08)60077-9. [DOI] [PubMed] [Google Scholar]
- 10.Pfeffer S., Dunoyer P., Heim F., Richards K. E., Jonard G., Ziegler-Graff V. J. Virol. 2002;76:6815–6824. doi: 10.1128/JVI.76.13.6815-6824.2002. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 11.Cardozo T., Pagano M. Nat. Rev. Mol. Cell Biol. 2004 2004;5:739–751. doi: 10.1038/nrm1471. [DOI] [PubMed] [Google Scholar]
- 12.Petroski M. D., Deshaies R. J. Nat. Rev. Mol. Cell Biol. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- 13.Moon J., Parry G., Estelle M. Plant Cell. 2004;16:3181–3195. doi: 10.1105/tpc.104.161220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Farras R., Ferrando A., Jasik J., Kleinow T., Okresz L., Tiburcio A., Salchert K., del Pozo C., Schell J., Koncz C. EMBO J. 2001;20:2742–2756. doi: 10.1093/emboj/20.11.2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Risseeuw E. P., Daskalchuk T. E., Banks T. W., Liu E., Cotelesage J., Hellmann H. Estelle, M., Somers D. E., Crosby W. L. Plant J. 2003;34:753–767. doi: 10.1046/j.1365-313x.2003.01768.x. [DOI] [PubMed] [Google Scholar]
- 16.Zhao D., Ni W., Feng B., Han T., Petrasek M. G., Ma H. Plant Physiol. 2003;133:203–217. doi: 10.1104/pp.103.024703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stolpe T Süsslin, C., Marrocco K., Nick P., Kretsch T., Kircher S. Protoplasma. 2005;226:137–146. doi: 10.1007/s00709-005-0122-6. [DOI] [PubMed] [Google Scholar]
- 18.Hu C. D., Chinenov Y., Kerppola T. D. Cell. 2002;9:789–798. doi: 10.1016/s1097-2765(02)00496-3. [DOI] [PubMed] [Google Scholar]
- 19.Ziegler-Graff V., Brault V., Mutterer J. D., Simonis M. T., Herrbach E., Guilley H., Richards K. E., Jonard G. Mol. Plant–Microbe Interact. 1996;9:501–510. [Google Scholar]
- 20.Pruss G., Ge X., Shi X. M., Carrington J. C., Bowman Vance V. Plant Cell. 1997;9:859–868. doi: 10.1105/tpc.9.6.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brigneti G., Voinnet. O Li, W. X., Ji L. H., Ding S. W., Baulcombe D. C. EMBO J. 1988;17:6739–6746. doi: 10.1093/emboj/17.22.6739. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22.Yelina N. E., Savenkov E. I., Solovyev A. G., Morozov S. Y., Valkonen J. P. J. Virol. 2002;76:12981–12991. doi: 10.1128/JVI.76.24.12981-12991.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu F., Ni W., Griffith M. E., Huang Z., Chang C., Peng W., Ma H., Xie D. Plant Cell. 2004;16:5–20. doi: 10.1105/tpc.017772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baulcombe D. C. Curr. Opin. Plant Biol. 1999;2:109–113. doi: 10.1016/S1369-5266(99)80022-3. [DOI] [PubMed] [Google Scholar]
- 25.Burch-Smith T. M., Anderson J. C., Martin G. B., Dinesh-Kumar S. P. Plant J. 2004;39:734–746. doi: 10.1111/j.1365-313X.2004.02158.x. [DOI] [PubMed] [Google Scholar]
- 26.Tzfira T., Vaidya M., Citovsky V. Nature. 2004;431:87–92. doi: 10.1038/nature02857. [DOI] [PubMed] [Google Scholar]
- 27.Voinnet O., Lederer C., Baulcombe D. C. Cell. 2000;103:157–167. doi: 10.1016/s0092-8674(00)00095-7. [DOI] [PubMed] [Google Scholar]
- 28.Qu F., Ren T., Morris T. J. J. Virol. 2003;77:511–522. doi: 10.1128/JVI.77.1.511-522.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Banks L., Pim D., Thomas M. Trends Biochem. Sci. 2003;28:452–459. doi: 10.1016/S0968-0004(03)00141-5. [DOI] [PubMed] [Google Scholar]
- 30.Yu X., Yu Y., Liu B., Luo K., Kong W., Mao P., Yu X. F. Science. 2003;302:1056–1060. doi: 10.1126/science.1089591. [DOI] [PubMed] [Google Scholar]
- 31.Querido E., Blanchette P., Yan Q., Kamura T., Morrison M., Boivin D., Kaelin W. G., Conaway R. C., Conaway J. W., Branton P. E. Genes Dev. 2001;15:3104–3117. doi: 10.1101/gad.926401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aronson M. N., Meyer A. D., Gyorgyey J., Katul L., Vetten H. J., Gronenborn B., Timchenko T. J. Virol. 2000;74:2967–2972. doi: 10.1128/jvi.74.7.2967-2972.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Welchman R. L., Gordon C., Mayer R. J. Nat. Rev. Mol. Cell Biol. 2005;6:599–609. doi: 10.1038/nrm1700. [DOI] [PubMed] [Google Scholar]
- 34.Voinnet O., Vain P., Angell S., Baulcombe D. C. Cell. 1998;95:177–187. doi: 10.1016/s0092-8674(00)81749-3. [DOI] [PubMed] [Google Scholar]
- 35.Bruyère A., Brault V., Ziegler-Graff V., Simonis M. T., Van den Heuvel J. F., Richards K., Guilley H., Jonard G., Herrbach E. Virology. 1997;230:323–334. doi: 10.1006/viro.1997.8476. [DOI] [PubMed] [Google Scholar]
- 36.Potuschak T., Lechner E., Parmentier Y., Yanagisawa S., Grava S., Koncz C., Genschik P. Cell. 2003;115:679–689. doi: 10.1016/s0092-8674(03)00968-1. [DOI] [PubMed] [Google Scholar]
- 37.Dunoyer P., Lecellier C. H., Parizotto E. A., Himber C., Voinnet O. Plant Cell. 2004;16:1235–1250. doi: 10.1105/tpc.020719. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 38.Vernet T., Dignard D., Thomas T. Y. Gene. 1987;52:225–233. doi: 10.1016/0378-1119(87)90049-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.