Abstract
Methods for transducing the cellular activities of mammalian cells into measurable electronic signals are important in many biotechnical applications, including biosensors, cell arrays, and other cell-based devices. This manuscript describes an approach for functionally integrating cellular activities and electrical processes in an underlying substrate. The cells are engineered with a cell-surface chimeric receptor that presents the nonmammalian enzyme cutinase. Action of this cell-surface cutinase on enzyme substrate self-assembled monolayers switches a nonelectroactive hydroxyphenyl ester to an electroactive hydroquinone, providing an electrical activity that can be identified with cyclic voltammetry. In this way, cell-surface enzymatic activity is transduced into electronic signals. The development of strategies to directly interface the activities of cells with materials will be important to enabling a broad class of hybrid microsystems that combine living and nonliving components.
Keywords: biomaterial, extracellular matrix, signal transduction
The development of strategies for controlling the interface between a cell and a material is important in a wide range of settings, including the enablement of cell-based sensing technologies, the growth of tissue-engineered products for medicine, and the preparation of substrates for studies of cell adhesion. For example, cell-based sensors are now important for screening compounds in drug discovery programs (1, 2) and are being developed as diagnostic tools to identify biowarfare agents in environmental samples (3). In both cases, cells are integrated into a microsystem to provide sensing functions that cannot be achieved with the common molecular strategies. The development of schemes for integrating the functions of the cell and the material particularly for the electronic materials that are used in microfabrication will further advance the goal of using living cells as components in hybrid microsystems. In this paper we report a strategy in which both cells and the electrode surfaces are engineered to specifically and directly interact with each other by way of a receptor–ligand interaction to produce electronic signals. This example is a significant alternative to current indirect methods for transducing cellular activities into electronic signals based on the electrical activity of excitable cells (4–6) or the use of bioluminescent (7, 8) or fluorescent (9–11) reporter proteins in that the technique we describe here constitutes a direct communication pathway from cell to electrode. Thus, the need for bulky optical instrumentation is eliminated, and the number of potentially noise-enhancing signal transduction steps is minimized. Here we demonstrate an approach that combines a biological modification of the cell surface and a chemical modification of an electrode surface to install a unique molecular transduction pathway that can convert a specific cellular activity into an electrical signal.
Our strategy relies on engineering the surface of a cell with a chimeric protein that can switch an electrode-tethered molecule from a nonelectroactive to an electroactive state. This switching results in measurable changes in the redox characteristics of the electrode, generating electrical signals when the cell-surface enzyme is placed in close proximity to the electrode (Fig. 1). To demonstrate this concept, we engineer the cell to present the enzyme cutinase, which we have previously used in its soluble form to switch functionalities on electrode surfaces from nonelectroactive to electroactive states (12). Cutinase is a fungal esterase that efficiently catalyzes the hydrolysis of acyl groups from appropriately designed surface-bound substrates. We use self-assembled monolayers presenting 4-hydroxyphenyl valerate as the substrate for the enzyme. Cutinase hydrolyzes the ester to give a hydroquinone, which can then be reversibly oxidized to give the corresponding benzoquinone. This redox cycle can be monitored by cyclic voltammetry, giving a direct electrical detection of the cutinase-derived product on the monolayer (Fig. 2b).
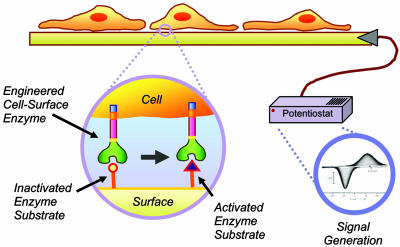
Fig. 1.
Schematic for transducing cellular activity into an electrical output. Adherent cells presenting a nonnative enzyme interact specifically with synthetic ligands on engineered surfaces. Enzymatic action switches a functionality on the electrode surface from a redox-inactive to a redox-active state, and this switching can be measured with cyclic voltammetry.
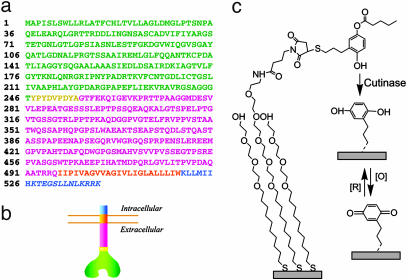
Fig. 2.
Engineered enzyme and surface chemistry schematic. (a and b) Amino acid sequence (a) and schematic (b) of the cell-surface cutinase construct showing the extracellular cutinase region (green), HA tag (yellow), fractalkine stalk (pink), and β1 integrin tail (orange transmembrane portion and blue intracellular tail, mutated portion in italics). (c) Surface chemistry scheme beginning with 4-hydroxy-(3-mercaptopropyl)phenyl valerate coupled to maleimide-presenting monolayers. Cutinase-mediated removal of the valerate-protecting group renders a hydroquinone that can be switched to and from a benzophenone with an oxidizing or reducing potential, respectively. The presence of the switchable redox pair can be measured with cyclic voltammetry.
We reasoned that cells producing cutinase on their surfaces would likewise have the ability to interact with protected hydroquinone moieties that are presented from an underlying substrate to generate electronic responses. Moreover, because cutinase is a fungal enzyme, its display on mammalian cells represents a unique and specific pathway for interacting with materials. We illustrate a proof of this concept by detailing the design and stable expression of cell surface cutinase in mammalian cells, the design and synthesis of enzyme substrate electrode surfaces, and the electrochemical signal evolution that arises when cutinase-expressing cells are cultured in contact with these surfaces.
Results
To express cutinase on the surfaces of mammalian cells, we constructed a vector coding for a multidomain chimeric protein in which cutinase is projected away from the cell membrane on the end of a rigid stalk domain. Intracellularly, this protein consists of a short, nonfunctional, 20-aa anchor derived from the β1 integrin (Fig. 2 a and b). To construct this protein, we PCR-amplified Fusarium solani pisi cutinase from the vector pCut22b(-Z) (13) and subcloned it into a previously developed construct that displays the enzyme carbonic anhydrase on cell surfaces (see Materials and Methods) (14). In this prior construct, carbonic anhydrase is projected from the cell surface on the end of a rigid 26-nm-long stalk derived from human fractalkine. Fractalkine is an extracellular membrane-bound chemokine whose chemokine domain is similarly projected from the cell by the stalk domain. Cutinase subcloning into this construct produced the protein shown in Fig. 2b: cutinase linked via its C terminus to the fractalkine stalk, which is in turn anchored in the cell membrane by the C-terminal tail of β1 integrin. We also we inserted an antibody target, the hemagglutinin (HA) nonapeptide epitope tag, between the cutinase and stalk domains, and we mutated the β1 integrin tail to prevent dominant-negative effects from the chimeric integrin tail acting within focal adhesions (see Fig. 2a and Materials and Methods).
Chinese hamster ovary (CHO) cells express this cutinase construct after transfection with Trojene transfection agent (Avanti Polar Lipids). Immunostaining with mouse anti-HA and anti-mouse-FITC showed that cutinase was expressed and localized to the extracellular side of the membrane, because nonpermeabilized cells exhibited the same level of expression as cells permeabilized with 1% Triton X-100. Nontransfected cells showed negligible antibody staining. Furthermore, stably transfected cells appeared to attach, spread, and proliferate at rates indistinguishable from untransfected cells, indicating that the construct’s cytoplasmic domain is not likely to interfere significantly with the cell’s native attachment machinery (Fig. 3a and b; see also the supporting information, which is published on the PNAS web site). Transiently transfected cells were propagated in selection medium (see Materials and Methods), and, after two rounds of fluorescence-assisted cell sorting, a stably transfected cell line was produced (85% stably transfected, as estimated by immunostaining) (Fig. 3a). We refer to this line as CHO-CUT cells.
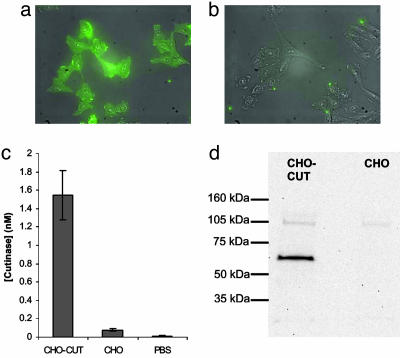
Fig. 3.
Characterization of transfected cells. (a and b) Stably transfected CHO-CUT cells (a) were highly fluorescent upon staining with mouse anti-HA tag and anti-mouse IgG-FITC in comparison with untransfected CHO cells (b). Shown are overlays of fluorescence and phase-contrast images. CHO-CUT cells also attached and spread in the same manner as untransfected CHO cells. (c) Cutinase activity of cells suspended in PBS at 100,000 cells per ml was much greater for CHO-CUT cells than for CHO cells. (d) Western blotting showed the expression of the full-length cutinase construct.
We verified the expression of the cutinase construct (56.4-kDa expected molecular mass) with Western blotting (Fig. 3d). CHO-CUT lysates exhibited one band at 62 kDa and a broader, weaker band at 115 kDa. These bands were not present in untransfected CHO cells. The higher-molecular-mass band likely represents a differentially glycosylated version of the cutinase construct because only a single band at 62 kDa was observed after lysates were digested with the deglycosylating enzyme PNGase F. Again, this band was not present in lysates prepared from nontransfected CHO cells (Fig. 3d). This high level of glycosylation of the cutinase construct is consistent with the known O-glycosylation of the fractalkine stalk (15), which may also be the reason that the observed molecular mass of 62 kDa is slightly higher than the predicted value of 56.4 kDa, because a residual amount of glycosylation may remain after PNGase F treatment.
To verify that the expressed construct contained active cutinase, we used an activity assay based on the enzymatic processing of the soluble dye precursor p-nitrophenyl butyrate (16). We found that suspended CHO-CUT cells possessed a substantially higher cutinase activity than untransfected CHO cells (Fig. 3c), corresponding to a cutinase activity of 9 × 106 active cutinase molecules per cell. Untransfected CHO cells did exhibit a small amount of activity compared with an acellular control, possibly as a result of the presence of other cellular esterases or small amounts of soluble esterases from the serum-containing culture medium.
We prepared cutinase-active substrates for cell adhesion by immobilizing 4-hydroxy-(3-mercaptopropyl)phenyl valerate to a maleimide-terminated self-assembled monolayer (SAM) (Fig. 2c) (17). Tri(ethylene glycol) groups also present on the monolayer are intended to reduce nonspecific adsorption of protein and increase the likelihood that the valerate groups remain accessible to the cell-surface cutinase. Monolayers included this tri(ethylene glycol) moiety in densities ranging from 70% to 95%, the balance being maleimide-thiol-conjugated 4-hydroxyphenyl valerate. MALDI-TOF MS showed that the immobilized 4-hydroxyphenyl valerate was a good substrate for soluble cutinase, because the valerate-protecting groups were completely removed from surfaces presenting 30% 4-hydroxyphenyl valerate after 10 min of incubation in a solution containing 100 nM soluble cutinase ([M+Na]+: unreacted, 1,142.6 expected and 1,143.6 observed; reacted, 1,058.5 expected and 1,059.0 observed) (Fig. 4a). Sodium adducts of disulfides were the major products observed with MALDI (12, 18, 19). Furthermore, cyclic voltammetry showed the appearance of redox peaks for the hydroquinone when monolayers presenting 30% 4-hydroxyphenyl valerate were incubated with cutinase (100 nM cutinase, 10 min at room temperature; see the supporting information) (12). These experiments reiterate that surfaces presenting 4-hydroxyphenyl valerate are effective cutinase substrates and that an added maleimide linkage between the electroactive species and the electrode surface does not diminish the ability to measure the reduction and oxidation of the benzoquinone–hydroquinone pair with cyclic voltammetry.
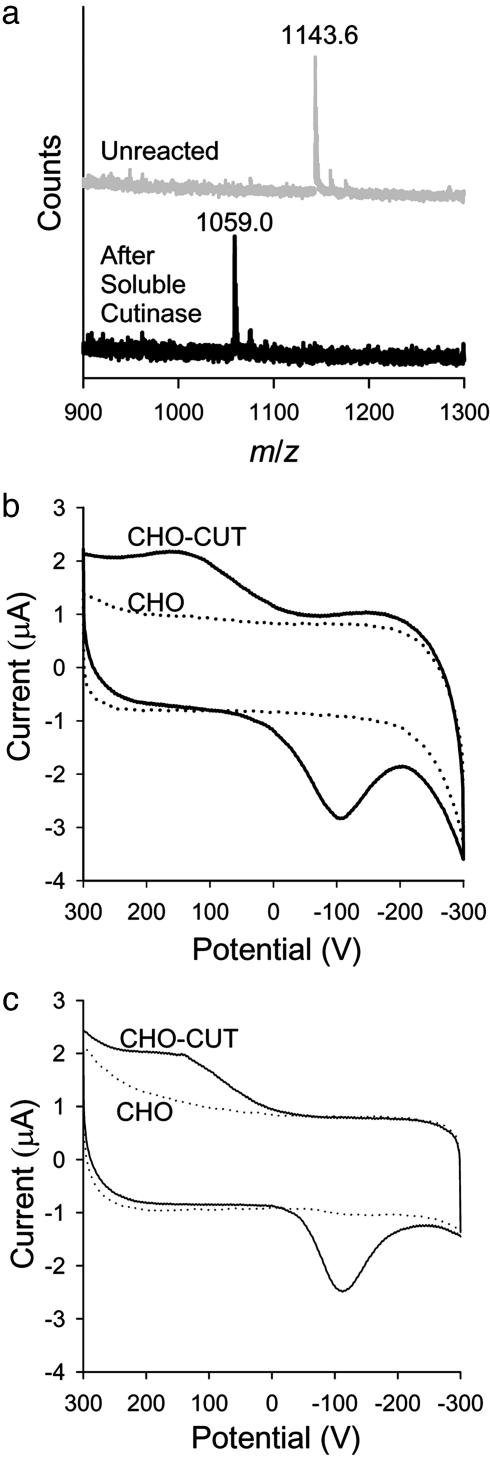
Fig. 4.
Surface and electrochemical characterization. (a) MALDI-TOF MS showing that addition of 100 nM soluble cutinase to 30% 4-hydroxyphenyl valerate monolayers completely removes the valerate protecting group. (b and c) Cyclic votammogram showing the appearance of redox peaks for CHO-CUT cells but not CHO cells after 4 h of cell culture on 30% 4-hydroxyphenyl valerate monolayers (b) and after 40 min, when these monolayers had been preadsorbed with fibronectin before cell seeding (c).
We next cultured transfected cells on the monolayers to characterize the cell-derived signaling on the monolayer. We formed culture wells on monolayers by attaching a silicone cylinder with vacuum grease such that the monolayer acted as the working electrode, a Ag∕AgCl electrode served as the reference electrode, and a platinum wire acted as the counter electrode. Serum-supplemented cell growth medium was used as the electrolyte. We found that when cells were seeded onto 30% 4-hydroxyphenyl valerate monolayers, they attached and spread within the first few hours of culture. Concomitant with cell attachment and spreading, we observed the appearance of redox peaks in the cyclic voltammagrams of CHO-CUT cell systems but not for untransfected CHO cells (Fig. 4b). Oxidation peaks were located at −105 V, and reduction peaks were located at 165 V. These redox peaks correspond to the peaks generated for hydroquinone monolayers and for 4-hydroxyphenyl valerate monolayers incubated with soluble cutinase (see the supporting information). Based on comparison with hydroquinone monolayers (supporting information), the conversion of the acylhydroquinone to the electroactive reporter group is low (≈15%). We did not determine whether this low yield was due to obstruction of acylhydroquinone groups by adsorbed proteins, by a restricted availability of the cell-surface cutinase enzyme due to clustering, or due to a chemical reaction that consumes the quinone reporter group. The observation that untransfected CHO cells did not generate a measurable redox signal, however, indicates a cutinase-specific signaling mechanism for CHO-CUT cells. This conclusion can be further justified in light of fluorescence imaging showing that CHO cells and CHO-CUT cells appeared to attach and spread to similar degrees (Fig. 3 a and b; see also the supporting information).
Although monolayers that present oligo(ethylene glycol) groups are among the best inert surfaces (that is, they prevent nonspecific protein adsorption and cell attachment), the introduction of ligands at densities >5% can compromise this inert character. Our use of monolayers that presented the 4-hydroxyphenyl valerate substrate at densities up to 30% would therefore be expected to give rise to some degree of protein adsorption. Although an adsorbed layer of proteins would facilitate cell attachment, it could potentially obstruct electrical signaling between the cells and the monolayer. To address this issue, we first preadsorbed fibronectin (50 μg·ml−1 for 1 h at 37°) to a monolayer presenting 4-hydroxyphenyl valerate at a density of 30%. Instead of occluding the substrate from the cell-bound cutinase, however, we found that redox peaks were generated much more quickly than without fibronectin preadsorption. Acceleration in cell attachment and spreading was also observed for fibronectin-coated monolayers, and it is most likely that this increase in attachment speed is responsible for the more rapid signal generation. Notably, signal intensities that were observed after 4 h from non-fibronectin-coated monolayers were produced in only 40 min for fibronectin-coated monolayers (Fig. 4c). Untransfected CHO cells showed a very slight but measurable electrochemical response with fibronectin preadsorption.
Discussion
These results show that by engineering both the cell surface and the electrode surface, a specific pathway can be engineered to transduce cell activity directly into a measurable electrochemical signal. Interestingly, protein adsorption does not completely block access of the enzyme substrate and signal generation, although it may mask a fraction of the reporter groups. In either case, it is clear that the cell-surface cutinase can at least partly displace the adsorbed layer of protein and access the substrates on the monolayers. It is possible that this access is facilitated by the fractalkine stalk, which projects cutinase 26 nm away from the cell surface and might be able to project the enzyme more readily through an adsorbed protein layer than a construct lacking such a stalk (15). Alternatively, the signal enhancement we observed with fibronectin preadsorption may also be due to the increased local concentration of cutinase at the surface produced by rapid adhesion of the cell. Also, we cannot rule out that a subset of the cutinase is released from the cell surface and acts on the surface; however, this seems unlikely given the strong attachment-dependent nature of signal evolution and the fact that fluorescence staining shows a large amount of cell-associated cutinase (Fig. 3a). Without the requirement of monolayers containing at least 90% oligoethylene glycol moieties, there is the capacity in the monolayer to include a higher density of 4-hydroxyphenyl valerate or biospecificity-enhancing ligands such as adhesive peptides. In addition, without the need for a nonfouling surface, surface chemistries other than SAMs would be amenable to this technique. In this way, routes for increasing the sensitivity, specificity, and applicability of the system are relatively unhindered.
When introducing novel receptors into cells, it is important to establish that the receptor does not interfere with normal cellular processes. The construct ultimately used in our studies for generating electrical signals was identified through a series of optimizations, a brief discussion of which is revealing. Our initial expectation in designing the extracellular cutinase construct was that using the native β1 integrin tail fragment would localize this construct to focal adhesions, the protein assemblies that provide for integrin-mediated cell attachment, and therefore position the cutinase domain in close contact with its substrates on electrode surfaces. Although CHO cells expressed the unmutated integrin-containing construct and localized the cutinase to the extracellular side of the membrane, immunostaining showed that the cells were rounded and poorly attached to their culture substrates. To test whether the cutinase activity or the integrin domain was responsible for causing this cell rounding and poor adhesion, we produced two mutant constructs. The first construct, produced by deleting Gly-146 through Thr-207, lacked cutinase activity. The second construct, the β1 integrin mutant ultimately used in our electrochemistry experiments reported above (Fig. 2a), was produced by inserting a frame shift after Lys-527. When cells were transfected with the cutinase mutant construct, they again became rounded and poorly attached, as they had with the unmutated construct. Cells transfected with the integrin mutant, however, showed wild-type spreading behavior and high levels of surface-bound cutinase expression, which led us to conclude that the β1 integrin tail, originally intended to localize the protein to focal adhesions, instead disrupted cell attachment, and it reiterates the necessity of designing novel engineered proteins such that their expression does not affect the cell in undesirable ways.
The cyclic voltammetry experiments shown here demonstrate that electrochemical signals are transduced between the engineered cells and the monolayer, but this work does not address all of the quantitative and mechanistic aspects of this system. For example, cyclic voltammograms for the monolayer-tethered hydroquinone show that this redox couple is not strictly reversible. Although this redox couple has been used extensively in designing functional monolayers (12, 20–22), the factors that give rise to the deviation of the couple from the expected form are not clear. Furthermore, the kinetics for a membrane-confined enzyme acting on molecules immobilized on a surface to which the cell is attached are certain to display features that are distinct from the corresponding kinetics with a soluble enzyme. Future work is necessary to address these issues.
This technology represents an important step in establishing molecular communication channels between cells and materials. This biospecific enzyme–substrate system now enables the direct electronic monitoring of a specific cellular activity, the production of the cutinase construct. In the example presented here, we demonstrated that the presence of constitutively expressed cutinase on cell surfaces could be detected electronically through the enzymatic processing of self-assembled monolayer substrates. This system could be easily extended by cloning the cutinase construct out of the vector for constitutive expression and into a vector for which expression could be induced in response to certain factors, such as biowarfare agents or molecules of pharmacological interest. In this way, cutinase expression would then occur in response to a specific applied stimulus, and the presence of the analyte could be directly measured electronically, without the necessity of the optical systems required by other techniques, such as fluorescence or chemiluminescence. Such a system could form the basis of a specific and compact biosensor.
Materials and Methods
Plasmid Construction.
Plasmids were maintained and propagated in E. coli (TOP10F′, Novagen). Restriction enzymes were purchased from New England Biolabs unless otherwise noted. PCR mutagenesis was performed to amplify cutinase from pCut22b(-Z) (13) and remove the internal KpnI site. The following primer pairs were used: 5′-CGCTGCCCAGCCGGCTCTAGACATGGGCCTGCC-3′∕5′-GCTAGGACCGAGGGTCCCCAAGTTGCCCGTC-3′ and 5′GACGGGCAACTTGGGGACCCTCGGTCCTAGC-3′∕5′-CGGAGCTCGAATTCGGTACCTCCTCCAGC-3′. The two fragments produced were then amplified with the two outside primers, and the resulting bridged fragment was digested with KpnI and XbaI. This fragment was then ligated with T4 DNA ligase into the construct reported by Kato and Mrksich (14). Before subcloning, the internal NgoMIV site in the previously reported construct was changed to an XbaI site using the QuikChange site-directed mutagenesis kit (Stratagene) and the following primers: 5′-CATCTGACTGTCCTGCTGGCCGGTCTAGAGTCACACTGG-3′ and 5′-CCAGTGTGACTCTAGACCGGCCAGCAGGACAGTCAGATG-3′. For immunostaining, a 33-bp oligonucleotide fragment corresponding to the HA tag (amino acid sequence YPYDVPDYA) was inserted at the KpnI site, and the entire leader-cutinase–HA tag–stalk–integrin construct was cloned into the pCI-neo vector (Promega) with NheI and NotI. The cutinase-inactive mutant was produced by digestion with BsgI and religation. The β1-integrin-inactive mutant was produced by the introduction of two nucleotides (AA) after His-526 by quick-change site-directed mutagenesis and the following primers: GCTGATATGGAAGCTTTTAATGATAATTCATAAGACAGAAGGGAGTTTGC and GCAAACTCCCTTCTGTCTTATGAATTATCATTAAAAGCTTCCATATCAGC. This frameshift resulted in the generation of an early stop codon 38-bp downstream from the mutation. This construct with the truncated integrin sequence was used in all electrochemistry experiments. The construct consisted of a 1,617-bp gene coding for a 56.4-kDa protein.
Transfection and Characterization of CHO-CUT Cells.
CHO cells were purchased from American Type Culture Collection and maintained according to their instructions. Cells were transfected with Trojene cationic lipid transfection reagent (Avanti Polar Lipids) according to the manufacturer’s protocol for 24 h and propagated in CHO growth medium supplemented with 0.5 mg·ml−1 G-418 sulfate (catalog no. 10131, GIBCO). Cells were subsequently sorted by FACS on the 10th day after transfection with a fluorescent anti-HA antibody (catalog no. A488–101L, Covance), propagated for 15 days more, and sorted a second time with FACS on day 25 after transfection. Cell lines prepared in this way (CHO-CUT cells) exhibited >85% stable transfection, as measured by immunostaining and fluorescence microscopy.
Cutinase Activity and Western Blotting.
Equal numbers of CHO and CHO-CUT cells were pelleted, lysed in gel loading buffer (2% sodium dodecyl sulfate∕0.1% bromophenol blue∕0.1% 2-mercaptoethanol∕10% glycerol∕50 mM Tris, pH 6.8), electrophoresed on 4–20% Tris·HCl polyacrylamide gel, and transferred to nitrocellulose, which was probed with anti-HA primary and goat anti-mouse-horseradish peroxidase secondary antibodies. Membranes were analyzed by chemiluminescent detection (kit no. RPN2109, Amersham Pharmacia). PNGase F (New England Biolabs) was used according to the manufacturer’s instructions to deglycosylate proteins for Western blotting. Cutinase activity was measured by gently trypsinizing CHO and CHO-CUT cells, resuspending them at 100,000 cells per ml and performing a previously described colorimetric assay based on p-nitrophenyl butyrate (16).
Monolayer Synthesis.
4-hydroxy-(3-mercaptopropyl)phenyl valerate was synthesized in seven steps from commercially available reagents (see the supporting information). Maleimide-terminated and oligoethylene glycol-terminated alkanethiolates were synthesized as described in ref. 17. All intermediates gave satisfactory 1H NMR spectra. SAMs were prepared by overnight incubation in 1 mM total alkanethiolate in ethanol. Conjugation of 4-hydroxy-(3-mercaptopropyl)phenyl valerate was performed by adding a 2 mM solution in dimethylformamide to preformed SAMs terminated with malemide groups at a density of 5–30%. This reaction was carried out for 3 h, after which monolayers were rinsed several times with ethanol.
MS.
The monolayers were analyzed on a Voyager-DE Biospectroscopy mass spectrometer (Applied Biosystems) using α-cyanohydroxycinnamic acid (saturated solution in 1:1 water∕acetonitrile) as the matrix. The SAMs were rinsed with deionized water, dried under N2, spotted directly with 1 μl of matrix solution, and analyzed immediately.
Electrochemistry.
Cyclic voltammetry was performed with a Bioanalytical Systems (West Lafayette, IN) CV-50W potentiostat. All experiments used a custom-designed electrochemical cell, with the monolayer as the working electrode, a Pt wire as the counter electrode, and a Ag∕AgCl reference electrode. Cells were seeded at a density of 270,000 cells per 2.25 cm2 well, which rapidly formed confluent monolayers upon cell attachment. Cultures were tested directly, with CHO culture medium as the electrolyte. Scans were performed at a rate of 100 mV·s−1.
Supplementary Material
Acknowledgments
We thank Woon-Seok Yeo for assistance with synthesis, Joshua Maurer for helpful discussions, and Jacob Basak for synthesis of maleimide alkanethiolates. This work was supported by the National Institutes of Health and by an Army Multidisciplinary Research Program of the University Research Initiative grant. The National Science Foundation Materials Research Science and Engineering Center provided use of its facilities.
Glossary
Abbreviations:
- SAM
self-assembled monolayer
- CHO
Chinese hamster ovary
- HA
hemagglutinin
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Galietta L. V. J., Jayaraman S., Verkman A. S. Am. J. Physiol. 2001;281:C1734–C1742. doi: 10.1152/ajpcell.2001.281.5.C1734. [DOI] [PubMed] [Google Scholar]
- 2.Tucker C. L., Fields S. Nat. Biotechnol. 2001;19:1042–1046. doi: 10.1038/nbt1101-1042. [DOI] [PubMed] [Google Scholar]
- 3.Rider T. H., Petrovick M. S., Nargi F. E., Harper J. D., Schwoebel E. D., Mathews R. H., Blanchard D. J., Bortolin L. T., Young A. M., Chen J. Z., Hollis M. A. Science. 2003;301:213–215. doi: 10.1126/science.1084920. [DOI] [PubMed] [Google Scholar]
- 4.Voelker M., Fromherz P. Biophys. J. 2004;86:271A–271A. [Google Scholar]
- 5.Fromherz P., Offenhausser A., Vetter T., Weis J. Science. 1991;252:1290–1293. doi: 10.1126/science.1925540. [DOI] [PubMed] [Google Scholar]
- 6.Bonifazi P., Fromherz P. Adv. Mater. 2002;14:1190–1193. [Google Scholar]
- 7.Nivens D. E., McKnight T. E., Moser S. A., Osbourn S. J., Simpson M. L., Sayler G. S. J. Appl. Microbiol. 2004;96:33–46. doi: 10.1046/j.1365-2672.2003.02114.x. [DOI] [PubMed] [Google Scholar]
- 8.Simpson M. L., Sayler G. S., Patterson G., Nivens D. E., Bolton E. K., Rochelle J. M., Arnott J. C., Applegate B. M., Ripp S., Guillorn M. A. Sens. Actuators, B. 2001;72:134–140. doi: 10.1016/s0925-4005(00)00641-9. [DOI] [PubMed] [Google Scholar]
- 9.Dooley C. T., Dore T. M., Hanson G. T., Jackson W. C., Remington S. J., Tsien R. Y. J. Biol. Chem. 2004;279:22284–22293. doi: 10.1074/jbc.M312847200. [DOI] [PubMed] [Google Scholar]
- 10.Zhang J., Campbell R. E., Ting A. Y., Tsien R. Y. Nat. Rev. Mol. Cell Biol. 2002;3:906–918. doi: 10.1038/nrm976. [DOI] [PubMed] [Google Scholar]
- 11.Kuang Y., Biran I., Walt D. R. Anal. Chem. 2004;76:2902–2909. doi: 10.1021/ac0354589. [DOI] [PubMed] [Google Scholar]
- 12.Yeo W. S., Mrksich M. Angew. Chem., Int. Ed. 2003;42:3121–3124. doi: 10.1002/anie.200250862. [DOI] [PubMed] [Google Scholar]
- 13.Hodneland C. D., Lee Y. S., Min D. H., Mrksich M. Proc. Natl. Acad. Sci. USA. 2002;99:5048–5052. doi: 10.1073/pnas.072685299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kato M., Mrksich M. J. Am. Chem. Soc. 2004;126:6504–6505. doi: 10.1021/ja039058e. [DOI] [PubMed] [Google Scholar]
- 15.Fong A. M., Erickson H. P., Zachariah J. P., Poon S., Schamberg N. J., Imai T., Patel D. D. J. Biol. Chem. 2000;275:3781–3786. doi: 10.1074/jbc.275.6.3781. [DOI] [PubMed] [Google Scholar]
- 16.Kolattukudy P. E., Purdy R. E., Maiti I. B. Methods Enzymol. 1981;71:652–664. [Google Scholar]
- 17.Houseman B. T., Gawalt E. S., Mrksich M. Langmuir. 2003;19:1522–1531. [Google Scholar]
- 18.Su J., Mrksich M. Langmuir. 2003;19:4867–4870. [Google Scholar]
- 19.Su J., Mrksich M. Angew. Chem., Int. Ed. 2002;41:4715–4718. doi: 10.1002/anie.200290026. [DOI] [PubMed] [Google Scholar]
- 20.Yousaf M. N., Houseman B. T., Mrksich M. Proc. Natl. Acad. Sci. USA. 2001;98:5992–5996. doi: 10.1073/pnas.101112898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hickman J. J., Ofer D., Laibinis P. E., Whitesides G. M., Wrighton M. S. Science. 1991;252:688–691. doi: 10.1126/science.252.5006.688. [DOI] [PubMed] [Google Scholar]
- 22.Hong H. G., Park W., Yu E. J. Electroanal. Chem. 1999;476:177–181. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.