Abstract
Cyclin D1 is a multifaceted regulator of both transcription and cell-cycle progression that exists in two distinct isoforms, cyclin D1a and D1b. In the prostate, cyclin D1a acts through discrete mechanisms to negatively regulate androgen receptor (AR) activity and thus limit androgen-dependent proliferation. Accordingly, cyclin D1a is rarely overexpressed in prostatic adenocarcinoma and holds little prognostic value in this tumor type. However, a common polymorphism (A870) known to facilitate production of cyclin D1b is associated with increased prostate cancer risk. Here we show that cyclin D1b is expressed at high frequency in prostate cancer and is up-regulated in neoplastic disease. Furthermore, our data demonstrate that, although cyclin D1b retains AR association, it is selectively compromised for AR regulation. The altered ability of cyclin D1b to regulate the AR was observed by using both in vitro and in vivo assays and was associated with compromised regulation of AR-dependent proliferation. Consistent with previous reports, expression of cyclin D1a inhibited cell-cycle progression in AR-dependent prostate cancer cells. Strikingly, cyclin D1b significantly stimulated proliferation in this cell type. AR-negative prostate cancer cells were nonresponsive to cyclin D1 (a or b) expression, indicating that defects in AR corepressor function yield a growth advantage specifically in AR-dependent cells. In summary, these studies indicate that the altered AR regulatory capacity of cyclin D1b contributes to its association with increased prostate cancer risk and provide evidence of cyclin D1b-mediated transcriptional regulation.
Keywords: corepressor, G870A, polymorphism, cell cycle, thyroid hormone receptor β
Prostate cancer (PCa) is the most frequently diagnosed malignancy among men in the U.S., and treatment of disseminated PCa is based on their requirement for androgen (1). Current therapies aim to reduce endogenous androgen production and/or inhibit the activity of the androgen receptor (AR) (2) and are initially effective. However, incurable, recurrent tumors result from aberrant AR reactivation (3). Thus, understanding the mechanisms by which AR activity is controlled is pivotal for improving PCa management.
AR is a member of the nuclear receptor superfamily and regulates genes critical to PCa initiation, progression, and treatment (4). AR activation is initiated by ligand (androgen) binding, which stimulates interaction of the receptor N and C termini and stabilizes active receptor conformation (5). AR activation triggers nuclear translocation, wherein AR binds specific DNA sequences termed androgen responsive elements (AREs) residing in genes involved in growth and survival of this tumor type (6, 7). Target gene activation is enhanced by recruitment of coactivators that promote receptor stabilization, facilitate DNA binding, or render DNA more accessible to the transcriptional machinery (8). This action is opposed by a select group of corepressor proteins that foster a state of inactive transcription (8).
Previously, we and others identified cyclin D1a as a critical AR corepressor (9–11). Cyclin D1a is well characterized as a cell-cycle regulator, interacting with cyclin-dependent kinase (CDK)4/6 to form an active kinase complex that promotes S-phase entry (12). Accordingly, cyclin D1a is overexpressed in many tumor types, and aberrant cyclin D1a–CDK4 activity leads to inappropriate proliferative signaling (13). However, cyclin D1a is rarely overexpressed in PCa and has no independent prognostic value in this tumor type (14, 15). Mouse models of PCa demonstrate that cyclin D1a expression is reduced as a function of tumor progression (16), and cyclin D1a has been reported to be sequestered in the cytoplasm in human disease (16). Together, these data suggest that cyclin D1a function may negatively impact PCa growth and progression. Emerging evidence suggests that this role of cyclin D1 is likely attributed to cell-cycle-independent functions.
Cyclin D1a expression levels increase in response to androgen stimulation in PCa cells, and subsequent CDK4 activation is essential for ensuing cell-cycle progression (17). However, accumulated cyclin D1a binds directly to and inhibits AR, thus limiting further cellular proliferation (9–11). This function of cyclin D1a is independent of CDK association and is consistent with an emerging role for cyclin D1a in transcriptional regulation (17–19). Cyclin D1a association and regulation with a growing number of transcription factors (e.g., thyroid hormone receptor β, estrogen receptor α, signal transducer and activator of transcription 3, and Sp1) is suspected to contribute to its function in tumorigenesis (18). Association between endogenous AR and cyclin D1a occurs in cells derived from PCa and liver, and this interaction significantly represses ligand-dependent AR activity at equimolar ratios of AR to cyclin D1a (11, 20, 21). Cyclin D1a transcriptional regulation of the AR is manifested through at least two discrete mechanisms. First, we and others have shown that cyclin D1a binds histone deacetylase 3 (HDAC3) to mediate transcriptional repression (22, 23). Second, cyclin D1a directly binds the AR N terminus and prevents formation of the active receptor conformation (21). These two cyclin D1a activities are effective in repressing AR transcriptional potential on a wide spectrum of target genes (20). Recently, the cyclin D1a repressor domain (RD) was mapped to a region (amino acids 142–253) that is both required and sufficient for AR inhibition and repression of androgen-dependent proliferation in PCa cells (22). Combined, these data suggest that androgen-induced elevation of cyclin D1a serves initially to stimulate cellular proliferation; but upon CDK saturation, cyclin D1a uses discrete mechanisms to attenuate subsequent AR activity. Supporting this negative feedback hypothesis, AR activity is lowest at the G1/S transition of the cell cycle, where cyclin D1a levels peak (24). Despite the importance of cyclin D1a in the regulation of AR activity, no mutants of cyclin D1a compromised for repression have been identified in PCa.
A polymorphism (G/A870) of the cyclin D1 locus has recently been associated with increased cancer risk for a wide variety of tumors, including PCa (25–27). Individuals harboring the A870 allele demonstrate an increased propensity to produce an alternatively spliced cyclin D1 mRNA (transcript b). Translation of transcript b results in the production of a variant of the cyclin D1a protein, termed cyclin D1b, with a divergent C terminus (25). Although individuals harboring the “A/A” genotype have an increased propensity to produce transcript b, the levels of expression can be markedly variant and transcript a can still be detected (25, 28). Given the discordance between genotype and cyclin D1b production, assessment of genotype alone may not provide an accurate assessment of risk. Recent investigations have demonstrated that cyclin D1b harbors distinct functions from cyclin D1a with regard to cell-cycle control and cellular transformation (29, 30). However, the influence of cyclin D1b on transcriptional regulation had yet to be assessed.
Given the importance of cyclin D1 in modulating AR function, we analyzed the prevalence and consequence of cyclin D1b in PCa. We observed a striking representation of the A allele and cyclin D1B production in both PCa cells and tumors. To delineate the mechanisms by which cyclin D1b may influence PCa risk, the ability of this variant to regulate AR function was investigated. Using multiple techniques, we demonstrate that although cyclin D1b retains AR binding function it is selectively compromised for AR regulation. The biological consequence of this defect is that cyclin D1b stimulates androgen-dependent proliferation, in contrast to the inhibition of cell-cycle progression mediated by cyclin D1a. AR-negative PCa cells were refractory to regulation by both cyclin D1 isoforms, demonstrating specificity of cyclin D1 action. Together, these data suggest that cyclin D1b contributes to PCa risk through compromised AR regulation and indicate that cyclin D1b expression may yield a significant growth advantage for AR-positive PCa cells. Moreover, these data are the first to demonstrate that cyclin D1b harbors transcriptional regulatory functions distinct from cyclin D1a.
Results
G/A870 Polymorphism and Cyclin D1b Expression.
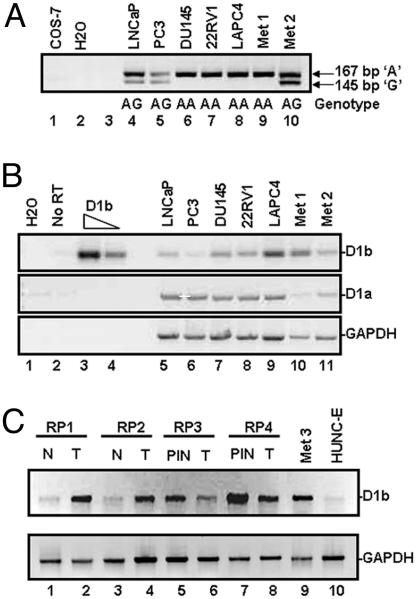
The A870 polymorphism of cyclin D1 is associated with increased PCa risk (26, 27). However, the prevalence of this genotype in established PCa models and association with cyclin D1b production had yet to be assessed. Initially, genotyping was performed by means of RFLP analysis (strategy shown in Fig. 7A, which is published as supporting information on the PNAS web site) by using genomic DNA isolated from PCa cells (LNCaP, PC3, DU145, LAPC4, and 22RV1) and two different PCa lymph node metastases (Met-1 and Met-2). For a negative control, COS-7 cells were used, because this cyclin D1 locus does not amplify with human-specific primers. As shown in Fig. 1A, Met-1 and three of the PCa cell lines (DU145, 22RV1, and LAPC4) were homozygous for the A allele (Fig. 1A, lanes 6–9). A heterozygous genotype was observed in Met-2, LNCaP, and PC3 cells (Fig. 1A, lanes 4, 5, and 10). These data demonstrate that the A allele is predominant in PCa cell lines and tumor samples. However, the A870 polymorphism does not necessitate transcript b production. Specifically, it has been documented that individuals harboring the AA genotype still produce cyclin D1a (25) and that the G allele can generate cyclin D1b (25). Thus, determination of transcript b expression is essential, because this parameter has significantly increased the predictive value as compared with analysis of the genotype alone for some tumor types (28). To date, no study has assessed the relative expression of cyclin D1b in PCa. To monitor cyclin D1b expression, RT-PCR analysis of PCa cell lines and tumors was performed by using primers specific for transcript a or b (strategy shown in Fig. 7B). PCR amplification of pRC-cyclin-D1b plasmid is shown as a positive control (Fig. 1B, lanes 3 and 4), and no PCR product was observed in the absence of reverse transcriptase (Fig. 1B, lane 2). The four AA homozygous PCa samples (DU145, LAPC4, 22RV1, and Met-1) exhibited increased expression of cyclin D1b transcript (Fig. 1B, lanes 7–10) in relation to their heterozygous counterparts (LNCaP, PC3, and Met-2) (Fig. 1B, lanes 5, 6, and 11). However, expression of both cyclin D1a and D1b transcript was seen in all cell lines, thus verifying that the cyclin D1 genotype is not predictive of the relative transcript ratio. GAPDH was used as an internal control for all PCRs (Fig. 1B Bottom). Further investigation into cyclin D1b expression was performed by using tissue obtained from four different radical prostatectomies and matching tissue from the same patient [classified to be normal or prostatic intraepithelial neoplasia (PIN), as shown]. In addition, a third-lymph-node metastisis (Met-3) and an immortalized prostatic epithelial cell line, HUNC-E, were also analyzed for cyclin D1b expression. All tumor samples showed cyclin D1b expression, consistent with previous results (Fig. 1C). Interestingly, matched normal tissue and HUNC-E cells showed lower expression of cyclin D1b, but PIN (precancerous lesions) demonstrated increased expression. Taken together, these findings show a predominance of both the A allele and cyclin D1b transcript in PCa models and confirm that the AA genotype does not predict exclusive production of cyclin D1b.
Fig. 1.
The AA genotype and transcript b are prevalent in PCa cells. (A) DNA was harvested and analyzed by using the strategy outlined in Fig. 7A. Data are representative of three independent experiments, and determined genotypes are shown. (B) RT-PCR detection of cyclin D1 isoform mRNA in PCa cell lines and tumor samples. A duplicate sample of 22Rv1 RNA, incubated in the absence of reverse transcriptase, was included as a control (lane 2). For each PCR, generated cDNA or cyclin D1b-encoding plasmid (lanes 3 and 4) was used in combination with the specific primers. (C) mRNA was isolated from tumor and normal or PIN lesions from the same patient (lanes 1–8), a prostatic metastatic tumor sample (lane 9), and HUNC-E cells (lane 10). RT-PCR analysis for D1b and GAPDH expression was performed as in B.
Cyclin D1b Maintains Efficient AR Interaction.
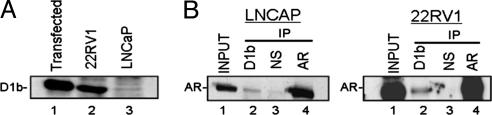
Given the importance of cyclin D1a in the regulation of androgen-dependent proliferation, we investigated whether cyclin D1b maintains prostate-specific attributes. As expected, cyclin D1b exhibited nuclear localization and did not influence ligand-induced AR nuclear translocation (Fig. 8A, which is published as supporting information on the PNAS web site). Analysis of two AR-positive PCa lines with divergent genotypes (A/G:LNCaP, and AA:22Rv1) confirmed endogenous cyclin D1b expression (Fig. 2A). Coimmunoprecipitation experiments revealed an association between endogenous AR and cyclin D1b in both cell types (Fig. 2B), and the AR-cyclin D1b association was recapitulated by using both ectopically expressed proteins (Fig. 8B) and in vitro binding assays (Fig. 8C). These findings revealed that cyclin D1 exon 5 is not required for cyclin D1b binding, consistent with maintenance of the RD (22) in cyclin D1b, and identified cyclin D1b as a putative AR regulatory protein.
Fig. 2.
Cyclin D1b associates with the AR in PCa cells. (A) Immunoblot of cyclin D1b in transfected C33A cells or endogenous expression in LNCaP and 22Rv1 cells. (B) Lysates from LNCaP and 22RV1 cells were immunoprecipitated with antibodies specific for AR, cyclin D1b, or a nonspecific control. The precipitated complex was immunoblotted for AR expression.
Cyclin D1b Exhibits Dual Mechanisms of AR Repression.
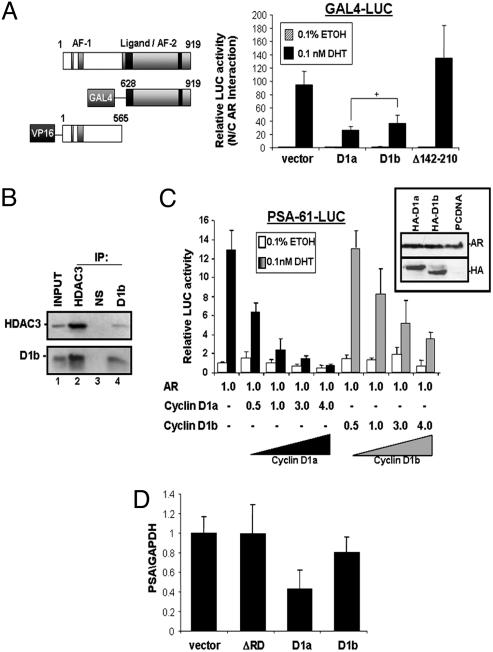
Because cyclin D1b associated directly with the AR (Figs. 2B, 8B, and 8C), mechanistic similarity to cyclin D1a was investigated by assessing the ability to recruit histone deacetylases and abrogate AR N/C-terminal interaction (10, 21). The ability of cyclin D1b to regulate AR N/C-terminal interactions was assessed by using a well characterized mammalian two-hybrid system (Fig. 3A) (31). As previously described, cells were transfected with GAL4-AR628–919, VP-16-AR1–565, Gal4-luciferase reporter, β-gal, and expression plasmid encoding cyclin D1a, D1b, cyclin D1Δ142–210, or vector (21). There was no significant difference between the ability of cyclin D1a and D1b to block the AR N/C interaction (Fig. 3A), indicating that cyclin D1b harbors some AR regulatory ability. A subdeletion (Δ142–210) of the RD that does not regulate N/C interactions has no effect on this system. To ensure that cyclin D1 had no independent effect on the VP16 moiety, experiments were repeated with a constitutively active Gal4-VP16 fusion. As shown in Fig. 9, which is published as supporting information on the PNAS web site, ligand treatment and expression of cyclin D1a or D1b had no effect on VP16 activity, demonstrating specificity of cyclin D1 action. To assess cyclin D1b association with HDAC3, cells were transfected as indicated and lysates were immunoprecipitated with antibodies specific for HDAC3, cyclin D1b, or control serum. A specific interaction was observed between cyclin D1b and HDAC3 (Fig. 3B, lanes 2 and 4), and in vitro studies confirmed these results (Fig. 8D). Together, these data verify that cyclin D1b functions to recruit HDAC3 and inhibit AR N/C interactions. Thus, both transcriptional regulatory functions of cyclin D1a are preserved in the cyclin D1b variant.
Fig. 3.
Cyclin D1b is compromised for AR corepressor activity. (A) Impact on AR N/C-terminal interactions was assessed in CV1 cells transfected with Gal4-LUC, β-gal, the depicted constructs encoding the mammalian-2-hybrid system (Top), and cyclin D1a, cyclin D1b, or vector. After transfection, cells were treated for 24 h with either vehicle or 5th-dihydrotestosterone (DHT) as indicated. After stimulation, cells were analyzed for relative luciferase activity. Results shown represent the average induction with standard deviations (+, P > 0.05). (B) COS7 cells transfected with HDAC3 and cyclin D1b were analyzed as in Fig. 2. (C) CV1 cells were transfected as indicated and analyzed in A. Relative DNA used is shown. (Inset) Relative expression of AR- and HA-tagged proteins transfected at a 1:3 ratio. (D) LNCaP cells were transfected with pBABE-Puro and pCDNA, cyclin D1a, cyclin D1b, or cyclin D1-ΔRD. mRNA from puromycin resistant cells was harvested and subjected to quantitative real-time RT-PCR. Data represent the average of at least three independent points performed in triplicate with standard deviations.
Cyclin D1b Exhibits Compromised AR Corepressor Activity on the Prostate-Specific Antigen (PSA) Promoter.
Mounting evidence suggests that the CDK-independent functions of cyclin D1a in transcriptional regulation play important roles in cancer cells (18, 19). However, the ability of cyclin D1b to exert these functions had yet to be determined. Our data demonstrate that cyclin D1b harbors activities that modulate transcription. To examine the functional consequence of these activities in PCa cells, the impact on AR activity was assessed. Initially, reporter assays were performed by using the well characterized PSA-61-LUC reporter, which harbors both an androgen-responsive proximal promoter and a distal enhancer (32). As described, AR-negative cells were cotransfected with a PSA reporter plasmid, AR, and increasing concentrations of either HA-D1a or HA-D1b expression plasmid. Cyclin D1a was a potent, dose-dependent inhibitor of AR action (Fig. 3C) (21), consistent with previous reports. Specificity of this action was demonstrated in that a cyclin D1 mutant incapable of binding AR (cyclin D1-ΔRD) had no impact on PSA promoter activity (ref. 22 and data not shown). However, cyclin D1b was significantly compromised for repression of this AR target. As shown, AR activity was elevated between 2- and 5-fold in cyclin D1b as compared with cyclin D1a-expressing cells (Fig. 3C). The difference in AR attenuation was not due to decreased expression of cyclin D1b (Fig. 3C Inset). These results show that cyclin D1b exhibits compromised ability to regulate AR function at the PSA locus. Further investigation into the effects of cyclin D1a and D1b expression on the endogenous PSA locus was performed by using quantitative real-time PCR. As shown in Fig. 3D, LNCaP cells transfected with cyclin D1-ΔRD or vector control showed no alteration of PSA expression, whereas cyclin D1a expectedly reduced PSA mRNA levels. By contrast, cyclin D1b was significantly compromised for regulating PSA expression, thus validating the studies performed on ectopic promoters.
Cyclin D1b Regulates Transcription in a Manner Distinct from Cyclin D1a.
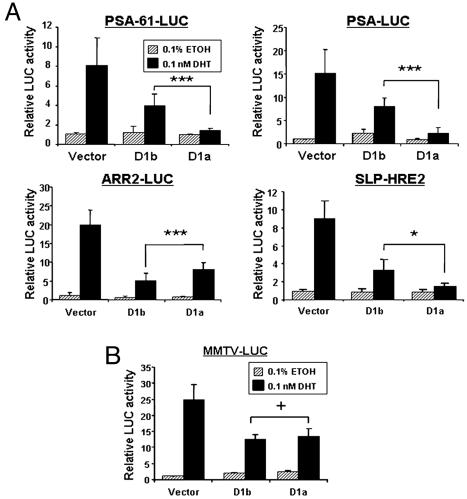
Because cyclin D1b demonstrated weakened repressor capability on the PSA-61-LUC reporter, the specificity of this effect was assessed. Experimental conditions were first validated by using the PSA-61-LUC reporter and untagged cyclin D1 constructs (Fig. 4A), wherein cyclin D1b was inefficient as a repressor (similar to Fig. 3 C and D). Identical conditions were used to assess cyclin D1b impact on the PSA-LUC reporter, which lacks the distal enhancer region and associated AREs (33). Again, cyclin D1b failed to repress AR activity to the capacity of cyclin D1a (Fig. 4A). To analyze a larger spectrum of validated AR targets, cyclin D1b-mediated regulation of probasin (ARR2) and sex-limited protein (SLP) expression was assessed (33, 34). As shown, SLP-HRE2-LUC inhibition was compromised in a manner comparable with the PSA promoter, resulting in a 2.2-fold increase in AR activity as compared with cyclin D1a (Fig. 4A). Thus, cyclin D1b was compromised for AR regulation on multiple targets. By contrast, cyclin D1b showed a statistically significant increase (≈16%) in repressor activity on the probasin (ARR2-LUC) reporter when compared with cyclin D1a (Fig. 4A). These data indicate that cyclin D1b harbors altered transcriptional activities as compared with cyclin D1a and also demonstrate that the poor repressor functions observed with PSA and SLP regulation cannot be attributed to nonspecific effects. Last, no difference in AR corepression was observed on the MMTV-LUC reporter, which contains three hormone responsive elements recognized by both the AR and other nuclear receptors (e.g., GR) (Fig. 4B) (35). For MMTV regulation, the data clearly show that cyclin D1a and cyclin D1b activities are indistinguishable. The cyclin D1-ΔRD construct had no effect on SLP or MMTV activity, as expected and consistent with our previous observations (ref. 22 and data not shown). Interestingly, cyclin D1b demonstrated an increased ability to repress thyroid hormone receptor β (Fig. 10, which is published as supporting information on the PNAS web site). Together, these data demonstrate that cyclin D1b can regulate transcription and that its ability to modulate AR activity is altered as compared with cyclin D1a.
Fig. 4.
Cyclin D1b displays promoter-specific regulation of the AR. (A) Reporter assays were performed as in Fig. 3C by using ARE-containing promoters (∗, P < 0.05; ∗∗∗, P < 0.001). (B) CV1 cells were transfected as in A by using the MMTV-LUC reporter (+, P > 0.05).
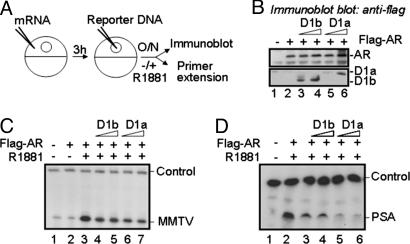
To validate this hypothesis, a Xenopus oocyte system was used. Previous work has established Xenopus laevis oocytes as an excellent model system for studying transcriptional regulation by the AR (36, 37). In this system, reporter DNA introduced into the nuclei of Xenopus oocytes is assembled into chromatin through a replication-coupled (ssDNA template) or replication-independent (dsDNA) pathway in a template-dependent manner (38). We exploited this system to examine the relative effect of cyclin D1a and D1b on AR activation as depicted (Fig. 5A). For protein expression, the indicated mRNAs and subsequent reporter constructs (MMTV-LTR-CAT or PSA-61-LUC) were injected into the oocyte cytoplasm. After incubation with R1881 or vehicle, oocytes were collected. The expression and effect of cyclin D1a and D1b on AR-dependent transcription was revealed by immunoblotting (Fig. 5B) and primer extension (Fig. 5 C and D), respectively. As shown, AR activity on the MMTV-CAT reporter was enhanced ≈14-fold with the addition of R1881 and inhibited by the addition of either cyclin (Fig. 5C). However, cyclin D1a was 35% more effective as a repressor than cyclin D1b on this promoter. Similar results were observed on PSA-61-LUC, wherein cyclin D1a was 63% more effective than D1b (Fig. 5D). Thus, cyclin D1b repressor activity was compromised on both AR targets in vitro but again demonstrated a more severe defect on the PSA-61-LUC. Combined, these data confirm that cyclin D1b regulates transcription in a manner distinct from cyclin D1a.
Fig. 5.
Cyclin D1b demonstrates promoter-specific repression of AR activity in vitro. (A) Xenopus oocytes were injected and treated as shown here and as described in Material and Methods. (B) Injected oocytes containing increasing amounts of flag-cyclin D1a, flag-cyclin D1b, or vector were lysed and analyzed for AR or cyclin expression by anti-flag immunoblot. (C and D) Oocytes were injected and treated as depicted in A with either MMTV-CAT or PSA-61-LUC. Primer extension to detect control and experimental mRNAs was conducted as described.
Cyclin D1b Fails to Repress Androgen-Dependent Growth.
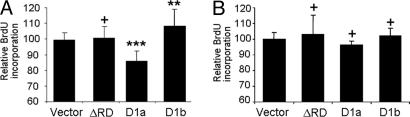
Our data indicate that, although cyclin D1b retains some AR modulatory function, its corepressor activity is significantly altered. We have previously shown that elevated cyclin D1a inhibits endogenous AR activity and AR-dependent proliferation (10, 22). Therefore, we investigated the effect of cyclin D1b on PCa proliferation. For these studies, AR-positive/AR-dependent LNCaP cells were transfected with the indicated expression plasmids and allowed to recover before the addition of BrdUrd for 18 h. Transfected (GFP-positive) cells were stained and scored for BrdUrd incorporation. Consistent with previous studies, cyclin D1a expression inhibited proliferation (14.2% as compared with control) in androgen-dependent PCa cells whereas the ΔRD mutant had no effect (Fig. 6A). Surprisingly, cyclin D1b failed to repress proliferation and actually stimulated cell-cycle progression in these cells (8.2%) (Fig. 6A). Growth regulation by both cyclin D1a and D1b was also monitored in the AR-negative PCa cells (PC3), wherein no significant difference in proliferation was noted upon ectopic expression of either cyclin (Fig. 6B). These data demonstrate the specificity of cyclin D1b action and confirm that cyclin D1a and D1b have differential effects on androgen-dependent PCa growth. Combined, these data indicate that cyclin D1b expression may render a specific growth advantage in AR-positive PCa cells.
Fig. 6.
Regulation of androgen-dependent growth is compromised by cyclin D1 polymorphism. (A) LNCaP cells were transfected with H2B-GFP alone, H2B-GFP and ΔRD, GFP-cyclin D1a, or GFP-cyclin D1b. After labeling and fixation, cells were stained for BrdUrd incorporation. For each condition, >150 cells were counted on each of at least six coverslips. Bars represent relative percent BrdUrd incorporation, and error bars represent the standard deviation. (B) PC3 cells were transfected and analyzed as in A for BrdUrd incorporation (∗∗, P < 0.01; ∗∗∗, P < 0.001 compared with vector; +, P > 0.05 compared with vector).
Discussion
Herein we define a mechanism by which a cyclin D1 variant, favored by the A870 polymorphism, may increase PCa risk. We demonstrate that the A allele is present at high frequency in all tested PCa cell lines and tumor tissues and that transcript b was expressed in all tumor and PIN tissues tested (Fig. 1). Moreover, although cyclin D1b was found in association with AR in PCa cells (Fig. 2), AR modulation was markedly distinct from that observed with cyclin D1a. First, cyclin D1b was selectively compromised for regulation of multiple AR target genes (Figs. 3–5). Second, the biological consequence of this disparity was revealed in that, whereas cyclin D1a significantly attenuated androgen-dependent proliferation in PCa cells, cyclin D1b promoted cell-cycle progression (Fig. 6A). No effect on proliferation was observed in AR-negative PCa cells (Fig. 6B), indicating that regulation of androgen-dependent growth is mediated through the AR. Combined, these data are the first to identify a transcriptional regulatory function of cyclin D1b and demonstrate that cyclin D1b harbors activities distinct from cyclin D1a. Moreover, these studies suggest that cyclin D1b expression may yield a significant growth advantage in AR-positive PCa cells, thus revealing one potential mechanism by which the A870 allele may alter PCa risk.
Cyclin D1b Selectively Modulates AR Activity.
A major finding of this study is that cyclin D1b is produced in both neoplastic disease of the prostate and PCa cells and that cyclin D1b selectively regulates AR function. Numerous AR comodulators have previously been reported to demonstrate promoter specificity, although the underlying mechanism behind such action is poorly understood. For example, the ARIP3 and PIAS1 coactivators potentiate AR activity on promoters containing minimal AREs, yet they fail to repress probasin expression (39). In addition, N/C-terminal AR interactions are required for activity of the full-length PSA and probasin promoters but not the MMTV or SLP promoters (40). The disparity in AR regulation by cyclin D1a versus D1b was observed both in mammalian cell assays and on chromatinized reporters in Xenopus oocytes, wherein the ratio of cyclin D1 repression (D1a/D1b) on the PSA-61-LUC promoter was twice as high as on the MMTV promoter (Fig. 5). ARE sequence may play a role in determining promoter specificity, because AR binding affinity is response-element-specific (41). As shown, cyclin D1a effectively represses transcription on all tested ectopic and endogenous AR target genes (Figs. 3–5) (20). In contrast, cyclin D1b displayed compromised ability to regulate the AR. It is unlikely that N/C-terminal interactions play a role in cyclin D1b promoter specificity, because the AR activation of both the SLP and MMTV promoters does not require this association (40). Because the AR inhibitory activity of cyclin D1a and site of HDAC3 recruitment have been previously mapped to RD (22), and cyclin D1b maintains this region, it is not surprising that HDAC3 binding to this isoform was intact. However, the relative recruitment and requirement of HDAC3 at each target promoter has yet to be addressed. Future studies will be needed to reveal the mechanisms by which cyclin D1b selectively regulates AR target gene expression.
Cyclin D1b Demonstrates Altered Regulation of Androgen-Dependent Proliferation.
Previous studies report that cyclin D1a repression of AR activity modulates androgen-dependent PCa proliferation. As such, the presence of a negative feedback loop has been proposed wherein androgen-induced cyclin D1a expression serves to limit the rate of further cell-cycle progression (9, 10, 17, 22). This hypothesis is supported by a large body of evidence, including the observations that AR activity is inversely correlated with cyclin D1 expression (24), that cyclin D1 expression is lost as a function of prostate PCa progression in the transgenic adenocarcinoma of the mouse prostate (TRAMP) model, and that cyclin D1 has been reported to be sequestered to the cytoplasm in advanced prostate tumors (16). Here we show that cyclin D1b not only lacks the ability to attenuate proliferation in AR-positive PCa cells, but actually stimulates cell-cycle progression in this cell type. The data presented suggest that the altered transcriptional regulatory functions of cyclin D1b influence its affect on proliferation and that cyclin D1b expression may yield a specific growth advantage in AR-positive PCa cells. Our studies revealed that cyclin D1b was ineffective at repressing AR activity on selected AR targets, including one used clinically to monitor PCa progression (PSA). By contrast, cyclin D1b demonstrated enhanced corepressor activity on a target involved in a developmental process (SLP) and on a thyroid hormone receptor-specific target. Together, these data indicate that the specificity of cyclin D1b action may act disparately on specific classes of AR target genes. Unfortunately, relatively few direct AR target genes have been identified and validated, and the principle targets involved in mediating androgen-dependent proliferation remain elusive. Thus, the present study indicates that it will be essential to identify the specific cohort of AR target genes that are disparately regulated by both cyclin D1a and cyclin D1b.
In summary, our study highlights the importance of analyzing cyclin D1b expression and function in PCa. We show that the A870G polymorphism is present and that transcript b is produced in the majority of PCa cells examined. Our data establish cyclin D1b as a modifier of gene transcription and demonstrate that this function of cyclin D1b may have critical consequence for cancer cells. We show that cyclin D1b is selectively compromised for AR modulation and that this attribute of cyclin D1b may yield a specific growth advantage to AR-positive PCa cells. This study reveals a facet of cyclin D1b function and puts forth a potential mechanism through which cyclin D1b may alter PCa risk. Given the ability of cyclin D1b to increase risk or outcome in a number of distinct tumor types, these data also provide the impetus to determine the contribution of transcriptional control in the analysis of cancer-related cyclin D1b activity.
Materials and Methods
Cell Culture.
PCa cells were obtained and cultured as described (10, 20, 22). LAPC4 cells were obtained from C. Sawyers (University of California, Los Angeles) (42). For steroid-free conditions, phenol red free media was used with charcoal dextran-treated serum.
Tumor Sample Collection.
Three inguinal lymph nodes with metastatic prostate carcinoma and four radical prostatectomy tissues were obtained from University Hospital (Cincinnati). After pathological examination, fresh samples were divided into 1-cm3 pieces and immediately frozen.
Plasmids.
Most plasmid constructs used were previously described (20–22, 30, 31, 33, 36). pCDNA-HA D1b was amplified by PCR flanked by XbaI and XmaI restriction sites and inserted into the corresponding sites of MS2-Flag. pRC-D1a was amplified by PCR flanked by BamHI sites. The resulting PCR product was inserted into MS2-Flag. The integrity of all constructs was verified by sequencing. SLP-HRE2-LUC was a generous gift from F. Claussens (University of Leuven, Louvain, Belgium) and was described in ref. 34.
PCR-RFLP Analysis.
Genomic DNA was extracted from cell culture lines and tumor tissues by using the DNAeasy purification kit (Qiagen, Valencia, CA). Isolated DNA was quantified, and 300 ng of DNA was used in a PCR as described (43). Products were extensively digested with ScrFI and resolved by agarose gel electrophoresis.
RT-PCR.
RNA was isolated by using TRIzol from cultured cell lines or isolated tumor tissue. PCR amplification of cyclin D1b and GAPDH was performed as described (20, 43). Cyclin D1a amplification used the previously published D1b forward primer (43) and the reverse primer 5′-GCGGATCCTCAGATGTCCACGTCCCG for 30 cycles (94°C for 1 min, 51°C for 1 min, 72°C for 1 min). Resulting products were resolved by agarose gel electrophoresis. Real-time PCR was performed on cDNA prepared from LNCaP cells transfected with indicated plasmids and pBabe-puro by means of the Lipofecten method. Transfected cells were rapidly selected in puromycin followed by RNA extraction. Applied Biosystems gene expression assays for PSA and GAPDH were used for relative expression and quantitated via the ΔΔCt method as per manufacturer’s protocol using the Applied Biosystems 7500F PCR system.
Reporter Assays.
CV1 cells were seeded in steroid-free conditions and transfected as indicated with a total of 4 μg of DNA by using the calcium phosphate method (44). For each transfection, 0.5 μg of CMV-β-gal, 0.75 μg of reporter, and 0.5 μg of SG5-AR were added. Cyclin D1a, cyclin D1b, or pCDNA (empty vector) constructs were included as indicated. After transfection, cells recovered for 18 h before stimulation with either hormone (0.1 nM 5th-dihydrotestosterone or vehicle). Cells were harvested and monitored for luciferase and β-gal (internal control for transfection efficiency) activity as described (10). Data shown represent the average and standard deviation of a minimum of six independent data points. Appropriate P values were obtained by using ANOVA followed by Newman–Keuls multiple-comparison post tests.
Cell Proliferation Assays.
For BrdUrd assays, cells were seeded on coverslips and transfected with 2 μg of GFP-D1a, GFP-D1b, cyclin D1-ΔRD/H2B-GFP, or H2B-GFP alone by means of Lipofectin. BrdUrd staining was performed as described (22).
Protein–Protein Interaction Studies.
Immunoprecipitation assays were performed as described by using antibodies specific for AR (22), HDAC3 (H-99, Santa Cruz Biotechnology), cyclin D1a (AB-3, NeoMarkers), cyclin D1b (described in Fig. 11, which is published as supporting information on the PNAS web site), and preimmune serum as the nonspecific control. Immunoblotting of precipitated complexes was performed with identical antibodies, with the exception of HDAC3 (B12, Santa Cruz Biotechnology). In vitro binding assays were performed as described (10).
Transcriptional Analyses in Xenopus Oocytes.
cDNAs for AR, cyclin D1a, and/or D1b were injected into Xenopus oocytes for expression as described (36). mRNA was generated by using linearized DNA templates and an SP6 Message Machine kit. The preparation of stage VI Xenopus oocytes and microinjection were performed essentially as described (36). For transcriptional analysis, individual or mixture of mRNAs were injected into the cytoplasm of the oocytes for protein synthesis (500 ng/μl for AR and 250 ng/μl and 500 ng/μl for D1a and D1b, 18.4 nl per oocyte), and followed 2–3 h later by nuclear injection of the MMTV-CAT or PSA-Luc reporter (50 ng/μl, 18.4 nl per oocyte). Injected oocytes were incubated at 18°C overnight in modified Barth’s solution in the presence or absence of 50 nM R1881. Transcriptional analysis by primer extension was performed essentially as described (37) by using a CAT-specific or luciferase-specific primer for detection of transcripts from the MMTV-CAT or PSA-LUC reporter. As an internal control, primer extension of the endogenous histone H4 mRNA was performed as described (37).
Supplementary Material
Acknowledgments
We thank the K.E.K. and E.S.K. laboratories for critical reading of the manuscript. This work was supported by National Institutes of Health (NIH) Grants CA 099996 and CA 093404 (to K.E.K.), the Center for Environmental Genetics (NIH/National Institute on Environmental Health Sciences Grant P-30-ES06096), NIH Grant CA106471 (to E.S.K.), NIH Grant DK065264 (to J.W.), the Albert J. Ryan Foundation, and NIH Training Grant T32 ES07250-16.
Glossary
Abbreviations:
- AR
androgen receptor
- PCa
prostate cancer
- ARE
androgen responsive element
- SLP
sex-limited protein
- PSA
prostate-specific antigen
- HDAC3
histone deacetylase 3
- CDK
cyclin-dependent kinase
- RD
repressor domain
- PIN
prostatic intraepithelial neoplasia.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Denmeade S. R., Isaacs J. T. Nat. Rev. Cancer. 2002;2:389–396. doi: 10.1038/nrc801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Salesi N., Carlini P., Ruggeri E. M., Ferretti G., Bria E., Cognetti F. J. Exp. Clin. Cancer Res. 2005;24:175–180. [PubMed] [Google Scholar]
- 3.Feldman B. J., Feldman D. Nat. Rev. Cancer. 2001;1:34–45. doi: 10.1038/35094009. [DOI] [PubMed] [Google Scholar]
- 4.Trapman J., Brinkmann A. O. Pathol. Res. Pract. 1996;192:752–760. doi: 10.1016/S0344-0338(96)80097-5. [DOI] [PubMed] [Google Scholar]
- 5.He B., Kemppainen J. A., Wilson E. M. J. Biol. Chem. 2000;275:22986–22994. doi: 10.1074/jbc.M002807200. [DOI] [PubMed] [Google Scholar]
- 6.Pratt W. B., Toft D. O. Endocr. Rev. 1997;18:306–360. doi: 10.1210/edrv.18.3.0303. [DOI] [PubMed] [Google Scholar]
- 7.Kallio P. J., Palvimo J. J., Mehto M., Janne O. A. J. Biol. Chem. 1994;269:11514–11522. [PubMed] [Google Scholar]
- 8.Heinlein C. A., Chang C. Endocr. Rev. 2002;23:175–200. doi: 10.1210/edrv.23.2.0460. [DOI] [PubMed] [Google Scholar]
- 9.Knudsen K. E., Cavenee W. K., Arden K. C. Cancer Res. 1999;59:2297–2301. [PubMed] [Google Scholar]
- 10.Petre C. E., Wetherill Y. B., Danielsen M., Knudsen K. E. J. Biol. Chem. 2002;277:2207–2215. doi: 10.1074/jbc.M106399200. [DOI] [PubMed] [Google Scholar]
- 11.Reutens A. T., Fu M., Wang C., Albanese C., McPhaul M. J., Sun Z., Balk S. P., Janne O. A., Palvimo J. J., Pestell R. G. Mol. Endocrinol. 2001;15:797–811. doi: 10.1210/mend.15.5.0641. [DOI] [PubMed] [Google Scholar]
- 12.Mittnacht S. Curr. Opin. Genet. Dev. 1998;8:21–27. doi: 10.1016/s0959-437x(98)80057-9. [DOI] [PubMed] [Google Scholar]
- 13.Motokura T., Arnold A. Curr. Opin. Genet. Dev. 1993;3:5–10. doi: 10.1016/s0959-437x(05)80334-x. [DOI] [PubMed] [Google Scholar]
- 14.Han E. K., Lim J. T., Arber N., Rubin M. A., Xing W. Q., Weinstein I. B. Prostate. 1998;35:95–101. doi: 10.1002/(sici)1097-0045(19980501)35:2<95::aid-pros2>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 15.Aaltomaa S., Eskelinen M., Lipponen P. Prostate. 1999;38:175–182. doi: 10.1002/(sici)1097-0045(19990215)38:3<175::aid-pros1>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 16.Maddison L. A., Huss W. J., Barrios R. M., Greenberg N. M. Prostate. 2004;58:335–344. doi: 10.1002/pros.10341. [DOI] [PubMed] [Google Scholar]
- 17.Knudsen K. E., Arden K. C., Cavenee W. K. J. Biol. Chem. 1998;273:20213–20222. doi: 10.1074/jbc.273.32.20213. [DOI] [PubMed] [Google Scholar]
- 18.Coqueret O. Gene. 2002;299:35–55. doi: 10.1016/s0378-1119(02)01055-7. [DOI] [PubMed] [Google Scholar]
- 19.Lamb J., Ramaswamy S., Ford H. L., Contreras B., Martinez R. V., Kittrell F. S., Zahnow C. A., Patterson N., Golub T. R., Ewen M. E. Cell. 2003;114:323–334. doi: 10.1016/s0092-8674(03)00570-1. [DOI] [PubMed] [Google Scholar]
- 20.Petre-Draviam C. E., Cook S. L., Burd C. J., Marshall T. W., Wetherill Y. B., Knudsen K. E. Cancer Res. 2003;63:4903–4913. [PubMed] [Google Scholar]
- 21.Burd C. J., Petre C. E., Moghadam H., Wilson E. M., Knudsen K. E. Mol. Endocrinol. 2005;19:607–620. doi: 10.1210/me.2004-0266. [DOI] [PubMed] [Google Scholar]
- 22.Petre-Draviam C. E., Williams E. B., Burd C. J., Gladden A., Moghadam H., Meller J., Diehl J. A., Knudsen K. E. Oncogene. 2005;24:431–444. doi: 10.1038/sj.onc.1208200. [DOI] [PubMed] [Google Scholar]
- 23.Lin H. M., Zhao L., Cheng S. Y. J. Biol. Chem. 2002;277:28733–28741. doi: 10.1074/jbc.M203380200. [DOI] [PubMed] [Google Scholar]
- 24.Martinez E. D., Danielsen M. J. Biol. Chem. 2002;277:29719–29729. doi: 10.1074/jbc.M112134200. [DOI] [PubMed] [Google Scholar]
- 25.Betticher D. C., Thatcher N., Altermatt H. J., Hoban P., Ryder W. D., Heighway J. Oncogene. 1995;11:1005–1011. [PubMed] [Google Scholar]
- 26.Koike H., Suzuki K., Satoh T., Ohtake N., Takei T., Nakata S., Yamanaka H. Anticancer Res. 2003;23:4947–4951. [PubMed] [Google Scholar]
- 27.Wang L., Habuchi T., Mitsumori K., Li Z., Kamoto T., Kinoshita H., Tsuchiya N., Sato K., Ohyama C., Nakamura A., et al. Int. J. Cancer. 2003;103:116–120. doi: 10.1002/ijc.10793. [DOI] [PubMed] [Google Scholar]
- 28.Bala S., Peltomaki P. Cancer Res. 2001;61:6042–6045. [PubMed] [Google Scholar]
- 29.Lu F., Gladden A. B., Diehl J. A. Cancer Res. 2003;63:7056–7061. [PubMed] [Google Scholar]
- 30.Solomon D. A., Wang Y., Fox S. R., Lambeck T. C., Giesting S., Lan Z., Senderowicz A. M., Conti C. J., Knudsen E. S. J. Biol. Chem. 2003;278:30339–30347. doi: 10.1074/jbc.M303969200. [DOI] [PubMed] [Google Scholar]
- 31.Wang Q., Lu J., Yong E. L. J. Biol. Chem. 2001;276:7493–7499. doi: 10.1074/jbc.M009916200. [DOI] [PubMed] [Google Scholar]
- 32.Cleutjens K. B., van der Korput H. A., van Eekelen C. C., van Rooij H. C., Faber P. W., Trapman J. Mol. Endocrinol. 1997;11:148–161. doi: 10.1210/mend.11.2.9883. [DOI] [PubMed] [Google Scholar]
- 33.Marshall T. W., Link K. A., Petre-Draviam C. E., Knudsen K. E. J. Biol. Chem. 2003;278:30605–30613. doi: 10.1074/jbc.M304582200. [DOI] [PubMed] [Google Scholar]
- 34.Verrijdt G., Schauwaers K., Haelens A., Rombauts W., Claessens F. J. Biol. Chem. 2002;277:35191–35201. doi: 10.1074/jbc.M205928200. [DOI] [PubMed] [Google Scholar]
- 35.No D., Yao T. P., Evans R. M. Proc. Natl. Acad. Sci. USA. 1996;93:3346–3351. doi: 10.1073/pnas.93.8.3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang Z. Q., Li J., Wong J. Mol. Endocrinol. 2002;16:924–937. doi: 10.1210/mend.16.5.0829. [DOI] [PubMed] [Google Scholar]
- 37.Huang Z. Q., Li J., Sachs L. M., Cole P. A., Wong J. EMBO J. 2003;22:2146–2155. doi: 10.1093/emboj/cdg219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Almouzni G., Wolffe A. P. Genes Dev. 1993;7:2033–2047. doi: 10.1101/gad.7.10.2033. [DOI] [PubMed] [Google Scholar]
- 39.Kotaja N., Aittomaki S., Silvennoinen O., Palvimo J. J., Janne O. A. Mol. Endocrinol. 2000;14:1986–2000. doi: 10.1210/mend.14.12.0569. [DOI] [PubMed] [Google Scholar]
- 40.He B., Lee L. W., Minges J. T., Wilson E. M. J. Biol. Chem. 2002;277:25631–25639. doi: 10.1074/jbc.M202809200. [DOI] [PubMed] [Google Scholar]
- 41.Claessens F., Verrijdt G., Schoenmakers E., Haelens A., Peeters B., Verhoeven G., Rombauts W. J. Steroid Biochem. Mol. Biol. 2001;76:23–30. doi: 10.1016/s0960-0760(00)00154-0. [DOI] [PubMed] [Google Scholar]
- 42.Klein K. A., Reiter R. E., Redula J., Moradi H., Zhu X. L., Brothman A. R., Lamb D. J., Marcelli M., Belldegrun A., Witte O. N., Sawyers C. L. Nat. Med. 1997;3:402–408. doi: 10.1038/nm0497-402. [DOI] [PubMed] [Google Scholar]
- 43.Sawa H., Ohshima T. A., Ukita H., Murakami H., Chiba Y., Kamada H., Hara M., Saito I. Oncogene. 1998;16:1701–1712. doi: 10.1038/sj.onc.1201691. [DOI] [PubMed] [Google Scholar]
- 44.Chen C., Okayama H. Mol. Cell. Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.