Abstract
The regulation of initiation of DNA replication is crucial to ensure that the genome is replicated only once per cell cycle. In the Gram-positive bacterium Bacillus subtilis, the function of the YabA protein in initiation control was assigned based on its interaction with the DnaA initiator and the DnaN sliding clamp in the yeast two-hybrid and on the overinitiation phenotype observed in a yabA null strain. However, YabA is unrelated to known regulators of initiation and interacts with several additional proteins that could also be involved directly or not in initiation control. Here, we investigated the specific role of YabA interactions with DnaA and DnaN in initiation control by identifying single amino acid changes in YabA that disrupted solely the interaction with DnaA or DnaN. These disruptive mutations delineated specific interacting surfaces involving a Zn2+-cluster structure in YabA. In B. subtilis, these YabA interaction mutations abolished both initiation control and the formation of YabA foci at the replication factory. Upon coexpression of deficient YabA mutants, mixed oligomers formed foci at the replisome and restored initiation control, indicating that YabA acts within a heterocomplex with DnaA and DnaN. In agreement, purified YabA oligomerized and formed complexes with DnaA and DnaN. These findings underscore the functional association of YabA with the replication machinery, indicating that YabA regulates initiation through coupling with the elongation of replication.
Keywords: DnaA, DnaN, Gram-positive bacteria, initiation control
Chromosome replication is tightly regulated to ensure that initiation at the chromosomal origin takes place only once per cell cycle. In eubacteria, the widely conserved initiator protein DnaA acts at the chromosomal replication origin by opening the DNA duplex to allow the loading of the replication machinery (1). Bacteria evolved different mechanisms for initiation control that mostly regulate the activity and the availability of the initiator protein DnaA in the cell. In Escherichia coli, considered as a paradigm for initiation studies in eubacteria, the regulatory inactivation of DnaA (RIDA) acts after initiation to promote the switch from the active ATP-DnaA to the inactive ADP-DnaA form. RIDA is mediated by the Hda protein and requires the sliding-clamp DnaN (2, –5). A second level of regulation is mediated by the sequestration of the oriC region by the SeqA protein, which binds with a high affinity to hemimethylated GATC sequences within oriC, thus temporarily restraining reintitiation (6–8). The third level of initiation control involves the titration of the DnaA protein onto DnaA boxes at the datA locus (9, 10). Indeed, datA acts to promote the correct timing of initiation relative to the cell mass (11).
The mechanisms of initiation control characterized in E. coli are mostly conserved in the group of the enterobacteriaceae, and other bacterial species have developed different mechanisms. In Caulobacter crescentus, the DnaA-dependent initiation of replication is regulated through the temporal and spatial localization of the master regulatory protein CtrA, which prevents initiation in swarmer cells by binding to conserved sites within the origin, and coordinates DNA replication with the cell cycle progression (12, 13). In Bacillus subtilis, a model of Gram-positive bacteria including many human pathogens, the initiation is mediated by DnaA and involves an oriC sequence related to that of E. coli (14, 15). However, the regulation of initiation in B. subtilis is not well understood and appears to be very different from that of E. coli. First, no homologues of the Hda, SeqA, and Dam proteins and no locus similar to datA have been identified in B. subtilis and related bacteria. Second, the early stages of initiation of chromosomal replication require the physical interaction between DnaB and DnaD, two components of the B. subtilis primosome (16). It has been proposed that initiation could be controlled through the timely interaction of DnaB and DnaD at the cell membrane (17). Furthermore, DnaB and DnaD could exert their role in initiation control through a remodeling of chromosomal DNA at oriC (18). B. subtilis cells lacking yabA exhibit overinitiation and replication asynchrony, suggesting that YabA acts as a negative regulator of initiation (19, 20). YabA is conserved in low-GC Gram-positive bacteria and was found to interact in a yeast two-hybrid screen with several partners including DnaA and DnaN but also TlpA, a chemotactic transducer, and AcuB, an enzyme involved in acetoin utilization. In this context, the deletion of YabA, which abolished all of the interactions, does not suffice to establish the direct functional relationship between the interaction with DnaA and DnaN and initiation control. In E. coli, the interactions with DnaA and DnaN are required for Hda-mediated RIDA (21). The DnaA-ATP hydrolysis in RIDA requires critical arginine residues that could cooperate in a catalytic center created by the interaction between the homologous AAA+ domains of Hda and DnaA (21). However, in contrast to Hda, YabA shares no structural homology with DnaA, does not belong to the AAA+ family of ATPases, and does not contain any DnaN-binding consensus motif. These observations point to a different mechanism for YabA-mediated initiation control.
Here, we undertook a biochemical and genetic characterization of the role of the YabA interactions with DnaA and DnaN. We isolated single point mutations in YabA that were found to impair the specific interactions of YabA with either DnaA or DnaN. These disruptive mutations revealed important structural features of YabA and delineated specific interacting surfaces. Some of the mutations were transferred into B. subtilis, and their effects on initiation control and YabA subcellular localization were determined. We conclude from this analysis that YabA negatively regulates initiation in vivo by forming a ternary DnaA–YabA–DnaN complex that is associated with the replication factory during most of the cell cycle. In addition, the functional dissection of YabA illustrates a way to assign a role to a given interaction in a multipartner protein. Such an approach could be of general applicability, because protein networks reveal that most proteins have several interacting partners, suggesting that they could act in different protein assemblies to fulfil multiple functions in the cell.
Results
YabA, DnaA, and DnaN Form a Tripartite Complex in Vitro.
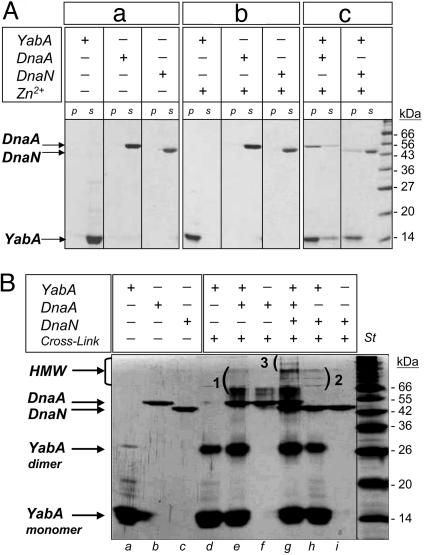
Three-hybrid studies indicated that YabA can form a ternary complex with DnaA and DnaN in yeast (19). To determine whether such a complex could form in vitro, the YabA, DnaA, and DnaN proteins from B. subtilis were purified to homogeneity (Fig. 1Aa). YabA self-assembled into a tetramer in solution, as determined by a combination of gel filtration and sedimentation on gradient sucrose approaches (data not shown). Interactions among YabA, DnaN, and/or DnaA were not detectable by gel filtration, suggesting that they are unstable or transient under the conditions tested. These interactions were then assayed by taking advantage of the property that, upon addition of a high concentration of Zn2+, the soluble YabA protein precipitated (Fig. 1Ab). The addition of Zn2+ to DnaA, DnaN, or DnaI alone did not yield any precipitate under these conditions. DnaI is a DnaA-homologous protein of the AAA+ family (22), and did not interact with YabA in the yeast two-hybrid (data not shown). Thus, DnaI was used as a control in the Zn2+-dependent precipitation assay. Addition of Zn2+ to protein mixtures containing YabA and either DnaA or DnaN induced the coprecipitation of most of the DnaA protein and part of the DnaN protein (Fig. 1Ac). Interestingly, after coincubation with DnaA, part of the YabA protein remained soluble, suggesting that the presence of DnaA somehow prevents the Zn2+-mediated precipitation of YabA. No precipitation of DnaI was observed in the presence of YabA (data not shown), indicating that Zn2+-dependent precipitation relies on a specific physical interaction. Together, these results show that YabA physically interacts with DnaA and DnaN in vitro, in agreement with the two-hybrid data.
Fig. 1.
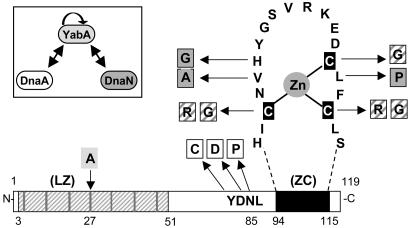
Interaction among YabA, DnaA, and DnaN in vitro. (A) Zinc precipitation assay. Purified YabA (14 kDa), DnaA (51 kDa), and DnaN (42 kDa) proteins (a). YabA precipitation upon addition of Zn2+ (b) was assayed in the presence of DnaA or DnaN (c). Soluble fractions and precipitates were analyzed by Coomassie-stained 12.5% SDS/PAGE. (B) Cross-linking experiment among YabA, DnaA, and DnaN. Glutaraldehyde-cross-linked products were analyzed by silver-stained 12.5% SDS/PAGE. Without (lanes a–c) and with (lanes d–i) glutaraldehyde. Incubation of YabA with DnaA (lane e) or/and DnaN (lane h) gave rise to new high-molecular-weight (HMW) species (1, 2, and 3, respectively). Note that DnaN, which is a dimer in solution, was not cross-linked under the conditions used.
The formation of a DnaA–YabA–DnaN ternary complex was assayed after cross-linking with glutaraldehyde (Fig. 1B). High-molecular-weight complexes were detected when YabA was incubated with either DnaA or DnaN, indicating that YabA interacted with DnaA and DnaN, in keeping with the Zn2+-precipitation assays. Of note, although YabA is tetrameric in solution, only dimers were detected after cross-linking under our experimental condition, suggesting that the YabA monomers are not symmetrical within the tetramer. A slight increase in the amount of cross-linked YabA dimer was observed in the presence of DnaA, but not of DnaN, suggesting that the interaction with DnaA could stabilize the YabA dimer. When YabA was incubated with both DnaA and DnaN, specific high-molecular-weight complexes were detected that were not present when YabA was incubated with the two proteins separately (Fig. 1B3). These results suggest that YabA forms a ternary complex with DnaN and DnaA in vitro, in agreement with the three-hybrid data (19).
Mutations Disrupting Physical Interaction with DnaA and DnaN Reveal Structural Features of YabA.
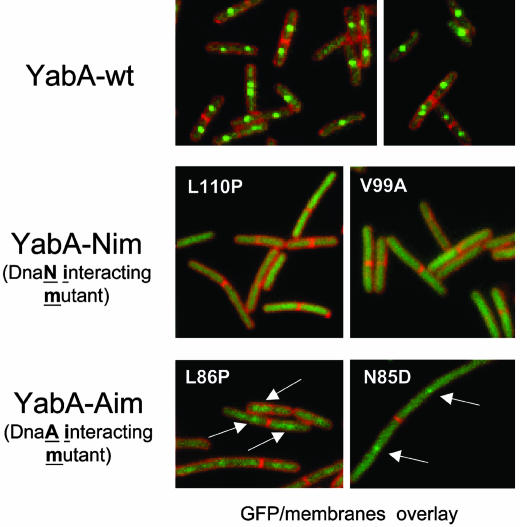
YabA is a small protein of 119 amino acids, with an unusual structural organization (Fig. 2A) composed of a canonical leucine zipper (LZ) at its N terminus (residues 1–51) and a potential zinc cluster (ZC) at its C terminus (residues 80–119). The LZ motifs are known to mediate specific protein dimerization (23). In a YabA dimer, each monomer could contribute three correctly positioned cysteine residues to form a six-cysteine cluster that could coordinate two Zn2+ ions (24). To investigate the relationship between the predicted structural features of YabA and its capacity to interact with its partners and to study the role of the interactions with DnaA and DnaN in initiation control, we designed a strategy termed “functional dissection” that consists in isolating amino acid changes in YabA that disrupt specifically selected interactions. The functional dissection of YabA involved both the random and site-directed mutagenesis of the yabA coding sequence, as described in Materials and Methods. Haploid yeast colonies expressing mutated YabA proteins fused to the GAL4 DNA-binding domain (baits) were arrayed on plates. The YabA mutants were then tested for their capacity to interact with YabA, DnaA, and DnaN expressed as fusion with the GAL4 activation domain (prey) in a yeast two-hybrid mating assay (see Materials and Methods). Thirty-five YabA mutant baits (3.3%) that did not interact with DnaA and/or DnaN but retained YabA self-interaction were isolated. These mutants were then subjected to a second interaction assay with the TlpA and AcuB preys (see Fig. 7, which is published as supporting information on the PNAS web site), so that they were not affected in their ability to interact with the other YabA partners identified in the network (19). The yabA alleles from the mutant baits that had retained the capacity to interact with the YabA, TlpA, and AcuB were entirely sequenced. Mutations generating single amino acid changes in YabA were identified (see Table 1, which is published as supporting information on the PNAS web site). Among these mutants, three single substitutions Y83C, N85D, and L86P were found to impair interaction with DnaA, and two single mutations V99A and L110P affected specifically the interaction with DnaN (Fig. 2 and Table 1). Additionally, the single changes C97R and C112R abolished interactions with both DnaA and DnaN. Site-directed mutagenesis was also performed in the YabA bait to change the heptadic L13, L27, and L41 residues to alanine in the LZ motif, the conserved residues C97, C109, and C112 to glycine in the potential ZC domain, and the poorly conserved residue H100 to glycine (Fig. 2). The effects of these mutations on interactions were determined as described above and are listed in Table 1.
Fig. 2.
Mapping of DnaA- and DnaN-interacting mutants. Schematic representation of YabA with the predicted LZ and the potential ZC domains. The positions of the leucine residues in LZ are represented by gray bars. The Cs in the black boxes indicate the key cysteine residues in ZC. The letters correspond to amino acids in the one-letter code. Amino acid substitutions in YabA affecting the interaction with DnaA (white), DnaN (gray), both DnaA and DnaN (white and gray diagonals), and self-interaction (light gray) are indicated. A representation of the YabA-interacting partners used in this study is shown. Arrows represent interactions detected by the yeast two-hybrid assay and are oriented from bait to prey.
Altogether, this mutational analysis identified key residues in YabA required for interaction and likely delineated interacting surfaces. The DnaA interaction mutations, hereafter named YabA-Aim, are clustered adjacent to the ZC motif (Fig. 2). The DnaN interaction mutations, hereafter named YabA-Nim, are located in the ZC-motif (Fig. 2). Interestingly, mutations affecting the conserved cysteine residues abolished both the interaction with DnaA and DnaN, suggesting that the ZC-like structure is important for these two interactions. Among mutations affecting the LZ domain, the L41A mutation did not have any detectable effect, whereas the L13A substitution prevented interaction with TlpA and AcuB without affecting interaction with DnaA and DnaN. In contrast, the L27A mutation drastically affected YabA self-interaction together with all of the other interactions (Table 1 and Fig. 7), suggesting that this residue is critical for the formation of the coiled-coil LZ structure. Also, in the screening of random YabA mutants, the loss of self-interaction was always associated with the loss of all the other interactions. These findings suggest that the formation of the LZ, which mediates YabA oligomerization, is a prerequisite for protein interactions.
Impaired Subcellular Localization of YabA Interaction Mutants in B. subtilis.
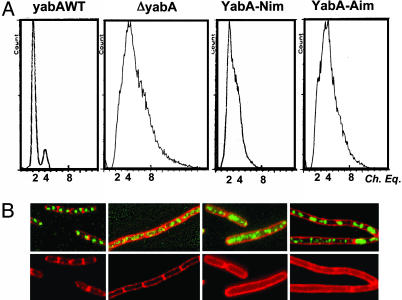
The next step in the functional dissection of YabA was the transfer of interaction mutations in B. subtilis to determine their effects on YabA function. First, we investigated the subcellular localization of YabA by fusing the GFP to the N terminus of YabA. The gfp–yabA construct was used to replace the chromosomal yabA and was also inserted ectopically at the amyE locus in a ΔyabA strain. The fusion to GFP appeared to stabilize YabA, because higher amounts of GFP-YabA relative to untagged YabA were detected by Western blot analysis (see Fig. 8, which is published as supporting information on the PNAS web site). The GFP-YabA protein restored initiation control (see below), indicating that it was fully functional. The localization patterns of GFP-YabA expressed from the chromosomal and the ectopic loci were identical upon fluorescence microscopy examination, and the ectopic gfp–yabA construct was used for further localization studies and mutation transfer. GFP-YabA formed one and two foci per cell, with single foci located at midcell (Fig. 3; and see Fig. 8A). The YabA localization profile did not match that of Spo0J, which marks the oriC region (25, 26), but exhibited a replisome-like distribution (Fig. 8C). Furthermore, under our experimental conditions, the YabA foci always colocalized with DnaX, an essential component of the replication machinery (Fig. 8B). Two YabA-Nim mutants (V99A and L110P) and two YabA-Aim mutants (N85D and L86P) were fused to the GFP to determine their subcellular localizations (Fig. 3). After their ectopic expression in a ΔyabA strain, the YabA-Nim proteins were dispersed throughout the cytoplasm under all the growth conditions tested. The YabA-Aim proteins were also dispersed, although weak fluorescent patches could be observed over the diffuse background. Because the YabA-Nim and YabA-Aim mutations did not modify the amount of the GFP-YabA protein in the cell (see Fig. 9C, which is published as supporting information on the PNAS web site), these results suggest that the localization of YabA at midcell may require a dual interaction with DnaA and DnaN.
Fig. 3.
DnaN- and DnaA-interacting mutants are impaired for localization. Overlay of fluorescence signals from GFP-YabA (green) and FM5–95 membrane dye (red) in living cells expressing YabA mutants that have lost their ability to interact with DnaN (YabA-Nim) or with DnaA (YabA-Aim). The nature of the mutation in YabA is indicated in white on the pictures. Arrows point to the fluorescent patches and weak foci observed in strains expressing the GFP YabA-Aim proteins.
Impaired Overinitiation and Replication Asynchrony in YabA Interaction Mutants in B. subtilis.
The effect of the YabA interaction mutations on the regulation of initiation in B. subtilis cells was assessed by flow cytometry. The YabA-Aim and YabA-Nim mutations were transferred into a yfp–yabA construct that replaced the wild-type yabA locus in strain CRK6000 (see Table 2, which is published as supporting information on the PNAS web site). This strain background was used for flow cytometry analyses because it did not form cell chains under the experimental conditions used. Exponentially growing cultures were treated with chloramphenicol to inhibit initiation and to allow completion of ongoing rounds of replication (27). This runout replication assay measures the number of chromosome equivalents per cell that corresponds to the number of origins present at the time of drug treatment. The yfp–yabA strain exhibited a profile similar to that of the wild type (yabA+, data not shown) with cells harboring two or four origins (Fig. 4A), indicating that replication was synchronous and that the yfp–yabA fusion was functional. In contrast, the ΔyabA strain exhibited a broaden distribution of origins, with some cells containing >8 origins (Fig. 4A). These results are in keeping with another study (20). Thus, overinitiation and asynchronous replication take place in the ΔyabA cells. We observed a similar phenotype in the yabA-Nim and the yabA-Aim mutants, although the yabA-Nim had a less pronounced defect (Fig. 4A), indicating that yabA interaction mutants were deficient for initiation control. This finding was confirmed by fluorescence microscopy by using the Spo0J-GFP fusion to visualize the origin region. The yabA-Aim and yabA-Nim mutations were introduced in the chromosomal yabA locus of a 168 strain expressing the Spo0J-GFP fusion (Table 2) by using a method that leaves no scar other than the desired mutation in the chromosome (28). Two or four Spo0J regularly positioned foci were observed in the wild-type cells, whereas in the ΔyabA cells, a higher number of foci were present throughout the cells (Fig. 4B), showing overinitiation in the mutant strain, as described in ref. 19. This result correlates well with the flow-cytometry data and indicates that YabA negatively regulates the initiation of replication. In the yabA-Nim and the yabA-Aim mutant strains, a higher number of Spo0J foci (≫4) were observed in most of the cells, which were also elongated, particularly for the yabA-Aim mutant (Fig. 4B). This cell filamentation was observed in the early exponential phase and disappeared at later stages of growth. These observations indicate that overinitiation and asynchronous replication are taking place in the yabA-Nim and yabA-Aim mutants, in agreement with the flow-cytometry analysis. Altogether, these results suggest that the initiation control mediated by YabA requires the capacity to interact with DnaA and DnaN.
Fig. 4.
Asynchronous and overinitiation in interacting mutant strains. Two distinct assays were used: (A) Flow-cytometry analysis of DNA contents in yabA mutant cells. Comparison of histograms of DNA content in the CRK6000 background for wild-type yfp–yabA+, mutant ΔyabA yfp–yabA–Aim (N85D), and ΔyabA yfp–yabA–Nim (L110P) strains. (B) Visualization of origin regions. Spo0J-GFP foci were used to monitor in the 168 background the localization pattern of the origin region in the yabA+, ΔyabA, yabA-Aim, and yabA-Nim strains. Overlay of Spo0J-GFP foci (green) and FM5–95 membrane staining (red).
Functional Complementation of the YabA-Aim and YabA-Nim Mutant Proteins Restores Midcell Localization.
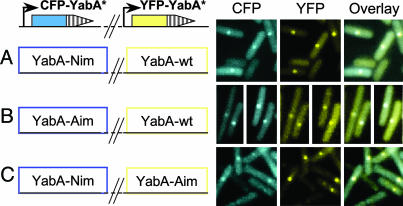
We have shown that YabA mutations disrupting interactions with DnaA and DnaN in the yeast two-hybrid system drastically affected YabA localization and initiation control in B. subtilis. Because YabA oligomerizes, we reasoned that the YabA-Aim and YabA-Nim mutant proteins could associate to form a mixed complex carrying sites for interaction with DnaA and DnaN. If such YabA mixed complexes are able to interact with DnaA and DnaN in the cell, they should localize at midcell. To test this hypothesis, a YabA-Aim (N85D), a YabA-Nim (V99A), and the wild-type YabA proteins were fused to CFP and to yellow fluorescent proteins (YFPs). The cfp–yabA and yfp–yabA derivatives were introduced at the ectopic amyE locus and at the chromosomal yabA locus, respectively, to generate strains that coexpress various combinations of YabA proteins in the same cell.
When expressed alone in the cell, the YabA-Aim and YabA-Nim proteins tagged with cyan fluorescent protein (CFP) or YFP were mostly dispersed in the cell, whereas the CFP- and YFP-tagged wild-type YabA protein formed bright foci at midcell (data not shown), as observed with the GFP fusions (Fig. 3). The coexpression of YFP-YabA promoted the relocalization in midcell foci of most of the dispersed CFP-YabA-Nim and CFP-YabA-Aim proteins (Fig. 5), indicating that the YabA-Nim and YabA-Aim mutant proteins are able to form mixed complexes with the wild-type YabA in vivo. Remarkably, the coexpression of the two dispersed CFP-YabA-Nim and YFP-YabA-Aim promoted the formation of foci localized at midcell containing both CFP and YFP fluorescence (Fig. 5). The foci were not a consequence of protein overproduction in the cell, because the coexpression of a YFP- and CFP-tagged YabA-Aim yielded a diffuse localization profile but did not promote the formation of foci (data not shown). These results indicate that the YabA-Nim and YabA-Aim proteins form mixed complexes that localize at the cell center. This restored localization is associated with the capacity of the mixed YabA complexes to interact with both DnaA and DnaN and suggests that YabA localization in wild-type cells requires interaction with both DnaA and DnaN.
Fig. 5.
Restoration of YabA-Nim localization byYabA-Aim. YabA-Nim and YabA-Aim proteins fused to CFP (blue) were coexpressed with a wild-type YabA fused to YFP (yellow) (A and B, respectively). YabA-Nim and YabA-Aim proteins fused to CFP (blue) or YFP (yellow), respectively, were also coexpressed in the same cells (C). Because the expression of the CFP-fusions was inducible by xylose, the strains were grown on plates containing a low amount (0–0.1%) of xylose to avoid quenching of the YFP signal by the CFP derivative. Using cells with YFP- or CFP-YabA foci, we verified that there was no detectable level of fluorescence crossover of YFP into the CFP channel and vice versa (data not shown).
Functional Complementation of the YabA-Aim and YabA-Nim Mutant Proteins Restores Initiation Control.
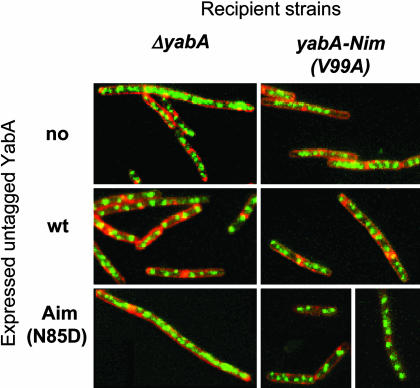
To determine whether the YabA-Aim/YabA-Nim mixed complexes also restored initiation control, the origin regions were visualized with Spo0J-GFP in cells coexpressing various combinations of YabA proteins. The YabA and YabA-Aim (N85D) proteins were placed under the control of an isopropyl β-d-thiogalactoside (IPTG)-inducible promoter and expressed from replicative plasmids (see Table 3, which is published as supporting information on the PNAS web site.) that were introduced into the ΔyabA and the yabA-Nim (V99A) strains. In absence of YabA (no IPTG or in the presence of an empty plasmid), both strains exhibited the expected overinitiation phenotype (Fig. 6). Analysis of the GFP-Spo0J foci distribution in both strains revealed that 38–50% of the cells contained multiple foci (≫8) indicating that aberrant overinitiation occurred (see Table 4, which is published as supporting information on the PNAS web site). Upon induction of the wild-type YabA protein (40 μM IPTG, Fig. 8D), ≈65% of the cells in both ΔyabA and yabA-Nim strains exhibited a normal distribution with two and four foci, and only 17% of the cells exhibited aberrant overinitiation (Fig. 6 and Table 4). This proportion of cells displaying normal initiation control corresponded to the maximum level of complementation that could be reached under the experimental conditions used. These results indicate that the yabA-Nim mutant is recessive. Remarkably, the induction of the YabA-Aim protein complemented the initiation defects of the yabA-Nim strain (60% of cells with two and four foci, 16% with multiple foci, Table 4) almost as efficiently as the wild-type YabA protein, whereas YabA-Aim protein enhanced overinitiation in the ΔyabA strain. The induction of the YabA-Nim protein in the yabA-Nim and the ΔyabA strains produced similar distributions of Spo0J foci, indicating that the YabA-Nim overexpression did not restore initiation control (Table 4). Thus, the coexpression of the deficient YabA-Nim and YabA-Aim mutant proteins restores initiation control in most of the cells. This functional complementation was also observed by monitoring the nucleoid relative DNA content in the cells (see Fig. 10, which is published as supporting information on the PNAS web site), likely mediated by the formation of mixed complexes of YabA mutant proteins that are able to interact with both DnaA and DnaN and suggests that, in wild-type cells, the regulation of initiation mediated by YabA requires its interaction with both DnaA and DnaN.
Fig. 6.
Complementation of YabA-Nim initiation defect by YabA-Aim. Spo0J-GFP foci were observed in exponentially growing ΔyabA and yabA-Nim cells harboring a plasmid expressing the wild-type YabA or the YabA-Aim proteins from an IPTG-inducible Pspac promoter (as indicated at left). Optimal complementation was achieved with 40 μM IPTG, corresponding to the level of YabA protein visualized in Fig. 8D. An analysis of the distribution Spo0J foci in the cell population is presented in Table 4.
Discussion
Here, we investigated specifically the biological role of the interactions of YabA with DnaA and DnaN by isolating from a yeast two-hybrid assay single amino acid changes in YabA that disrupted only the interaction with either DnaA or DnaN. We found that the identified YabA-Nim and YabA-Aim disruptive mutations were clustered at or near residues important for the integrity of the predicted ZC structure. Together with the observation that mutations affecting the conserved cysteine residues disrupted the interactions with both DnaA and DnaN, these results suggest that the ZC structure is essential for interaction. In addition, a single mutation at the center of the N-terminal LZ structure resulted in the loss of self-interaction and of all of the interactions with the YabA partners. These results suggested that the LZ mediates the formation of YabA oligomers, in which a specific ZC structure is required for the interaction with DnaA and DnaN. This relationship between YabA structure and protein interaction established with the yeast two-hybrid was supported by the biochemical evidence. Indeed, the purified YabA protein is a tetramer in solution, likely formed by the asymmetric association of two YabA dimers. This hypothesis is in agreement with the observation that the YabA tetramer has an elongated shaped (data not shown). The propensity of YabA, but not of DnaA, DnaN, or DnaI, to precipitate in the presence of high concentration of Zn2+ is indicative of its ability to chelate zinc. Furthermore, the addition of Zn2+ to protein mixtures containing YabA triggered the precipitation of DnaA and DnaN but not that of the control AAA+ protein DnaI. Thus, the Zn2+-dependent coprecipitation assay revealed interaction of YabA with DnaA and DnaN. Furthermore, crosslinking of a mixture of the three proteins in solution (no added Zn2+) revealed specific high-molecular-weight complexes, suggesting that a ternary DnaA–YabA–DnaN complex forms in vitro, in agreement with yeast trihybrid data (19). Altogether, these results suggest that, in YabA oligomers, a specific Zn2+-containing structure is required for interaction with DnaA and DnaN, and the YabA-Aim and YabA-Nim mutations could alter the interaction surfaces on this structure.
The wild-type YabA protein formed one or two foci per cell that colocalized with the replication factory under our experimental conditions. This observation partly corroborates a recent study showing that YabA colocalizes with the replication factory at a late stage of growth (20). In contrast, the YabA-Aim and YabA-Nim mutant proteins did not form foci and were mostly dispersed in cytoplasm. Combination of runout replication/flow-cytometry assay and visualization of the origin regions bound by Spo0J-GFP revealed that both the yabA-Aim and yabA-Nim mutant strains exhibited overinitiation, suggesting that they were impaired in initiation control. Remarkably, complementation between DnaA- and DnaN-interaction mutants restored both YabA replisome-like localization and initiation control. Altogether, these data indicate that the YabA-Aim and YabA-Nim mutant proteins are properly folded in the B. subtilis cell, and each mutant has an interaction deficiency that can be complemented by the other within a functional heterocomplex. These findings are consistent with the formation of DnaA–YabA–DnaN complexes in vitro. The initiation control and the YabA subcellular localization were affected simultaneously by all of the YabA mutations tested and restored simultaneously by cross-complementation, suggesting YabA down-regulates initiation as part of a multimeric complex with DnaN and DnaA and that its association with the replication factory is essential for its function. The functional association of YabA with the replication factory reveals that the control of initiation is tightly coupled to the elongation step of DNA replication.
The regulation of DNA replication at the initiation step is crucial to prevent overreplication that would be deleterious for the genome integrity. In different organisms, such as E. coli, B. subtilis and C. crescentus, the control of DNA replication at the initiation step is mediated by different regulatory proteins. In E. coli, the well characterized DnaA-related Hda protein regulates initiation by promoting the hydrolysis of DnaA-ATP to produce the inactive DnaA-ADP (3, 29, 30). In vitro, this hydrolysis requires Hda interaction with DnaA and with DnaN loaded onto the DNA and involves a conserved arginine-containing motif in the AAA+ proteins (21, 31). Whether in B. subtilis YabA also promotes a RIDA-like switch is not known. However, because YabA is not related to the AAA+ family of proteins and considering the structural requirement for its activity, it is likely that YabA acts in initiation control by a different mechanism. Remarkably, the common feature between the unrelated YabA and Hda proteins is to regulate initiation by coupling it with elongation. Therefore, it appears that the coupling of initiation regulation with the elongation step of replication has been evolutionarily conserved in eubacteria.
The localization of YabA at the replisome factory and not at the origins reveals an interesting aspect of its mode of action. During the B. subtilis cell cycle, the origins duplicate close to the cell center, and sister oriC rapidly migrate apart from each other toward the cell poles (26, 32–34). In contrast, the replication factory is quasi-stationary at midcell (35). Thus, origins are located away from the replication factory and from YabA foci during most of the cell cycle, suggesting that YabA is likely not involved in preventing the untimely firing of distant origins, and other levels of initiation control must be present in B. subtilis, as hinted by the functional interplay between DnaB and DnaD at the cell membrane (16–18). A possibility is that the YabA oligomers could associate with DnaN at the replisome, and could trap the DnaA protein released from the origin immediately after initiation, thus preventing a refiring of the sister oriC. Further work is required to unravel this mechanism of initiation control present in a large group of bacteria.
Materials and Methods
Bacterial Strains and Plasmids.
The B. subtilis strains used in this study are listed in Table 2, and the plasmids constructs are listed in Table 3.
Functional Dissection of yabA.
Site-directed mutagenesis of yabA was performed by PCR amplification and joining by using oligonucleotides carrying the desired mutations. Random mutagenesis was achieved by PCR amplification under mutagenic conditions. pGBDU-yabA mutant baits were constructed by gap-repair recombination in yeast. About 1,000 independent transformants were organized in 96-well format on synthetic complete media lacking uracyl (SC-U) plates to form a library of yabA mutant baits, and the library was mated with PJ69–4α strains containing the preys pGAD-yabA, pGAD-dnaN, and pGAD-dnaASID (amino acids 71–331). Additional information is provided in Supporting Materials and Methods, which is published as supporting information on the PNAS web site.
Construction of yabA Point Mutations and GFP Fusions.
Point mutations were transferred in the chromosomal yabA locus as described in Supporting Materials and Methods by using the one-step gene-replacement procedure described in ref. 28. The gfp–yabA and cfp–yabA fusion constructs were made as described in Supporting Materials and Methods. The yfp–yabA–Aim and yfp–yabA–Nim fusions at the chromosomal locus were constructed as described in ref. 20.
Fluorescence Microscopy.
For microscopic observations, cells (168 background) were grown in LB as described in Supporting Materials and Methods, mounted on agarose slides as described in ref. 26, and observed by using a Leica DMRA2 microscope. Images were acquired by using a Leica DC350F charge-coupled device camera.
Flow Cytometry.
Cells from the CRK6000 background were grown exponentially in minimal medium and treated with chloramphenicol (200 μg/ml) to inhibit new rounds of initiation (20). Incubation was continued for 5 h to complete ongoing chromosome replication. Cells were fixed with ethanol and analyzed by using a Bryte HS (Bio-Rad) flow cytometer as described in ref. 27.
Protein Purification and Zinc Precipitation Assay.
Purified proteins were obtained as described in Supporting Materials and Methods. YabA (25 μM), DnaA (2 μM), DnaN (2 μM), and DnaI (3 μM) were incubated alone or mixed in combination in a final volume of 50 μl in buffer (25 mM Tris, pH 8, 150 mM NaCl, and 0.5 mM DTT). After 2 hours of incubation on ice, ZnSO4 (100 μM final) was added, and the reaction mixtures were left at 4°C overnight. The samples were then centrifuged (13,000 × g at 4°C for 15 min), and pellets were resuspended in 50 μl of urea (2 M). The supernatants and resuspended pellets (15 μl) were analyzed by SDS/PAGE 12.5% and Coomassie blue staining.
Chemical Protein Cross-Linking.
Purified proteins were incubated mixed or separately on ice for 30 min in buffer D (22.5 μl). The final monomer concentrations were 8.5 μM YabA, 2.7 μM DnaA, and 2 μM DnaN. Cross-linking was initiated by the addition of glutaraldehyde (0.01% final concentration) for 20 min at 25°C. The reaction was terminated with glycine (0.2 M final concentration). The samples were analyzed by SDS/PAGE 12.5%, followed by a silver staining.
Supplementary Material
Acknowledgments
We thank E. d’Alençon for the construction of the pTYB-dnaN plasmid and T. Kanzaki for producing Western blots of YabA mutants. This work was supported, in part, by funding from the Ministry of Research and Technology of France.
Glossary
Abbreviations:
- CFP
cyan fluorescent protein
- IPTG
isopropyl β-d-thiogalactoside
- LZ
leucine zipper
- RIDA
regulatory inactivation of DnaA
- YFP
yellow fluorescent protein
- ZC
zinc cluster.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Kornberg A., Baker T. A. DNA Replication. 2nd Ed. New York: Freeman; 1992. [Google Scholar]
- 2.Mizushima T., Nishida S., Kurokawa K., Katayama T., Miki T., Sekimizu K. EMBO J. 1997;16:3724–3730. doi: 10.1093/emboj/16.12.3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nishida S., Fujimitsu K., Sekimizu K., Ohmura T., Ueda T., Katayama T. J. Biol. Chem. 2002;277:14986–14995. doi: 10.1074/jbc.M108303200. [DOI] [PubMed] [Google Scholar]
- 4.Katayama T., Sekimizu K. Biochimie. 1999;81:835–840. doi: 10.1016/s0300-9084(99)00213-8. [DOI] [PubMed] [Google Scholar]
- 5.Katayama T., Kubota T., Kurokawa K., Crooke E., Sekimizu K. Cell. 1998;94:61–71. doi: 10.1016/s0092-8674(00)81222-2. [DOI] [PubMed] [Google Scholar]
- 6.Taghbalout A., Landoulsi A., Kern R., Yamazoe M., Hiraga S., Holland B., Kohiyama M., Malki A. Genes Cells. 2000;5:873–884. doi: 10.1046/j.1365-2443.2000.00380.x. [DOI] [PubMed] [Google Scholar]
- 7.Lu M., Campbell J. L., Boye E., Kleckner N. Cell. 1994;77:413–426. doi: 10.1016/0092-8674(94)90156-2. [DOI] [PubMed] [Google Scholar]
- 8.von Freiesleben U., Rasmussen K. V., Schaechter M. Mol. Microbiol. 1994;14:763–772. doi: 10.1111/j.1365-2958.1994.tb01313.x. [DOI] [PubMed] [Google Scholar]
- 9.Kitagawa R., Ozaki T., Moriya S., Ogawa T. Genes Dev. 1998;12:3032–3043. doi: 10.1101/gad.12.19.3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ogawa T., Yamada Y., Kuroda T., Kishi T., Moriya S. Mol. Microbiol. 2002;44:1367–1375. doi: 10.1046/j.1365-2958.2002.02969.x. [DOI] [PubMed] [Google Scholar]
- 11.Morigen, Molina F., Skarstad K. J. Bacteriol. 2005;187:3913–3920. doi: 10.1128/JB.187.12.3913-3920.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gorbatyuk B., Marczynski G. T. Mol. Microbiol. 2005;55:1233–1245. doi: 10.1111/j.1365-2958.2004.04459.x. [DOI] [PubMed] [Google Scholar]
- 13.McGrath P. T., Viollier P., McAdams H. H. Curr. Opin. Microbiol. 2004;7:192–197. doi: 10.1016/j.mib.2004.02.012. [DOI] [PubMed] [Google Scholar]
- 14.Ogasawara N., Moriya S., von Meyenburg K., Hansen F. G., Yoshikawa H. EMBO J. 1985;4:3345–3350. doi: 10.1002/j.1460-2075.1985.tb04087.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moriya S., Ogasawara N., Yoshikawa H. Nucleic Acids Res. 1985;13:2251–2265. doi: 10.1093/nar/13.7.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bruand C., Velten M., McGovern S., Marsin S., Serena C., Ehrlich S. D., Polard P. Mol. Microbiol. 2005;55:1138–1150. doi: 10.1111/j.1365-2958.2004.04451.x. [DOI] [PubMed] [Google Scholar]
- 17.Rokop M. E., Auchtung J. M., Grossman A. D. Mol. Microbiol. 2004;52:1757–1767. doi: 10.1111/j.1365-2958.2004.04091.x. [DOI] [PubMed] [Google Scholar]
- 18.Zhang W., Carneiro M. J., Turner I. J., Allen S., Roberts C. J., Soultanas P. J. Mol. Biol. 2005;351:66–75. doi: 10.1016/j.jmb.2005.05.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Noirot-Gros M.-F., Dervyn E., Wu L. J., Mervelet P., Errington J., Ehrlich S. D., Noirot P. Proc. Natl. Acad. Sci. USA. 2002;99:8342–8347. doi: 10.1073/pnas.122040799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hayashi M., Ogura Y., Harry E. J., Ogasawara N., Moriya S. FEMS Microbiol. Lett. 2005;247:73–79. doi: 10.1016/j.femsle.2005.04.028. [DOI] [PubMed] [Google Scholar]
- 21.Su’etsugu M., Shimuta T. R., Ishida T., Kawakami H., Katayama T. J. Biol. Chem. 2005;280:6528–6536. doi: 10.1074/jbc.M412060200. [DOI] [PubMed] [Google Scholar]
- 22.Koonin E. V. Nucleic Acids Res. 1992;20:1997. doi: 10.1093/nar/20.8.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bornberg-Bauer E., Rivals E., Vingron M. Nucleic Acids Res. 1998;26:2740–2746. doi: 10.1093/nar/26.11.2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krishna S. S., Majumdar I., Grishin N. V. Nucleic Acids Res. 2003;31:532–550. doi: 10.1093/nar/gkg161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin D. C., Grossman A. D. Cell. 1998;92:675–685. doi: 10.1016/s0092-8674(00)81135-6. [DOI] [PubMed] [Google Scholar]
- 26.Glaser P., Sharpe M. E., Raether B., Perego M., Ohlsen K., Errington J. Genes Dev. 1997;11:1160–1168. doi: 10.1101/gad.11.9.1160. [DOI] [PubMed] [Google Scholar]
- 27.Ogura Y., Imai Y., Ogasawara N., Moriya S. J. Bacteriol. 2001;183:3833–3841. doi: 10.1128/JB.183.13.3833-3841.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fabret C., Ehrlich S. D., Noirot P. Mol. Microbiol. 2002;46:25–36. doi: 10.1046/j.1365-2958.2002.03140.x. [DOI] [PubMed] [Google Scholar]
- 29.Kato J., Katayama T. EMBO J. 2001;20:4253–4262. doi: 10.1093/emboj/20.15.4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Camara J. E., Skarstad K., Crooke E. J. Bacteriol. 2003;185:3244–3248. doi: 10.1128/JB.185.10.3244-3248.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Su’etsugu M., Takata M., Kubota T., Matsuda Y., Katayama T. Genes Cells. 2004;9:509–522. doi: 10.1111/j.1356-9597.2004.00741.x. [DOI] [PubMed] [Google Scholar]
- 32.Webb C. D., Teleman A., Gordon S., Straight A., Belmont A., Lin D. C., Grossman A. D., Wright A., Losick R. Cell. 1997;88:667–674. doi: 10.1016/s0092-8674(00)81909-1. [DOI] [PubMed] [Google Scholar]
- 33.Sharpe M. E., Errington J. Mol. Microbiol. 1998;28:981–990. doi: 10.1046/j.1365-2958.1998.00857.x. [DOI] [PubMed] [Google Scholar]
- 34.Webb C. D., Graumann P. L., Kahana J. A., Teleman A. A., Silver P. A., Losick R. Mol. Microbiol. 1998;28:883–892. doi: 10.1046/j.1365-2958.1998.00808.x. [DOI] [PubMed] [Google Scholar]
- 35.Lemon K. P., Grossman A. D. Science. 1998;282:1516–1519. doi: 10.1126/science.282.5393.1516. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.