Abstract
Genome sequences of two Synechococcus ecotypes inhabiting the Octopus Spring microbial mat in Yellowstone National Park revealed the presence of all genes required for nitrogenase biosynthesis. We demonstrate that nif genes of the Synechococcus ecotypes are expressed in situ in a region of the mat that varies in temperature from 53.5°C to 63.4°C (average 60°C); transcripts are only detected at the end of the day when the mat becomes anoxic. Nitrogenase activity in mat samples was also detected in the evening. Hitherto, N2 fixation in hot spring mats was attributed either to filamentous cyanobacteria (not present at >50°C in these mats) or to heterotrophic bacteria. To explore how energy-generating processes of the Synechococcus ecotypes track natural light and O2 conditions, we evaluated accumulation of transcripts encoding proteins involved in photosynthesis, respiration, and fermentation. Transcripts from photosynthesis (cpcF, cpcE, psaB, and psbB) and respiration (coxA and cydA) genes declined in the evening. In contrast, transcripts encoding enzymes that may participate in fermentation fell into two categories; some (ldh, pdhB, ald, and ackA) decreased in the evening, whereas others (pflB, pflA, adhE, and acs) increased at the end of the day and remained high into the night. Energy required for N2 fixation during the night may be derived from fermentation pathways that become prominent as the mat becomes anoxic. In a broader context, our data suggest that there are critical regulatory switches in situ that are linked to the diel cycle and that these switches alter many metabolic processes within the microbial mat.
Keywords: Synechococcus, diel regulation, fermentation, nitrogenase, Yellowstone National Park
Microbial mats are highly structured microbial communities that populate a variety of environments (1). Cyanobacteria are key players for the biogeochemistry of photosynthetic mats as they are the predominant primary producers in these systems, and many can reduce molecular nitrogen (N2) to ammonium. Cyanobacterial energy generation within microbial mats involves photosynthesis and aerobic respiration during the day and mainly fermentative metabolism during the night, when such mats become anoxic.
The microbial mat communities of alkaline siliceous hot springs in Yellowstone National Park have been studied in great detail (2–4). The mats are strictly prokaryotic in composition and represent stable communities containing photoautotrophic, photoheterotrophic, chemoautotrophic, and heterotrophic organisms. At temperatures between 50°C and 72°C, the 1-mm thick upper green layer of the mat contains unicellular cyanobacteria (Synechococcus spp.) (2, 4) embedded in a matrix of exopolymers and filamentous Chloroflexus/Roseiflexus-type phototrophic bacteria (5, 6). Thermophilic Synechococcus are an ecologically diverse group of organisms distributed along horizontal thermal and vertical light and O2 gradients in the mats (3, 4, 7, 8). Recently, genomes of two thermophilic Synechococcus isolates (designated Synechococcus OS-A and OS-B′), representing ecologically specialized populations in the microbial mat of Octopus Spring (9), were sequenced (A.-S.S., D.B., M.M.B., M.C.M., D.M.W., M.K., A.R.G., J. Heidelberg, and F. Cohan, unpublished data). Surprisingly, both genomes harbor a conserved nif gene cluster, suggesting that these organisms can fix N2. Hitherto, filamentous cyanobacteria growing at <50°C were the only thermophilic cyanobacteria implicated in N2 fixation in geothermal systems (10, 11). Few diazotrophic bacteria are thermophilic, and the highest temperature at which nitrogenase activity has been observed (64°C) was in the thermophilic, anaerobic archaeon Methanococcus thermolithotrophicus (12).
The arrangements of nif genes on the Synechococcus OS-A and OS-B′ chromosomes are essentially identical (the accession nos. for OS-A and OS-B′ genes are CP000239 and CP000240, respectively), encoding the catalytic nitrogenase subunits and the subunits involved in nitrogenase maturation/stability and in synthesis of the FeMo-cofactor (13–18). Nitrogenase is extremely O2 sensitive (19), and the problem of nitrogenase O2 sensitivity is acute in cyanobacteria as they perform oxygenic photosynthesis. To solve this problem, many filamentous cyanobacteria develop heterocysts, differentiated cells that exclude O2 (20, 21), have reduced linear photosynthetic electron transport (22), and have increased respiratory activity (23); these features of heterocysts help establish an environment favorable for N2 fixation (24). Nonheterocystous diazotrophic cyanobacteria can temporally (25) and spatially (26) separate photosynthesis from N2 fixation. For example, photosynthetic activity may be high during the day when cells harvest light energy for CO2 fixation, whereas N2 fixation occurs in the evening when there is little photosynthetic activity and respiration consumes most intracellular O2. Photosynthetic, respiratory, and nitrogenase activities in some cyanobacteria may also be under circadian control (27, 28).
During the day, Synechococcus spp. in the Octopus Spring mat synthesize and excrete photorespiratory glycolate, which can feed heterotrophic and photoheterotrophic bacteria (29). Most of the photosynthate synthesized during the day is stored as glycogen, which can be converted to acetate during the night (30). Acetate that accumulates in the mat overnight can be assimilated by heterotrophic and photoheterotrophic bacteria as the sun rises (31, 32). This observation is consistent with the finding that many cyanobacteria can ferment polyglucose under anoxic conditions (33). A pronounced switch from daytime hyperoxic conditions in the mat to anoxia as evening approaches suggests that marked changes in cellular metabolism track the diel cycle and light conditions of the environment.
Here we explore in situ conditions that lead to the accumulation of nitrogenase transcripts and activity in Synechococcus populations inhabiting the alkaline hot springs of Yellowstone National Park. We also examine in situ expression of genes encoding proteins associated with key energy-generating processes in cyanobacteria, including photosynthesis, respiration, and fermentation. The results demonstrate that the levels of transcripts from many of these genes, as well as nitrogenase activity, are controlled over the diel cycle, tracking both oxic and light conditions.
Results
Nitrogenase Gene Cluster.
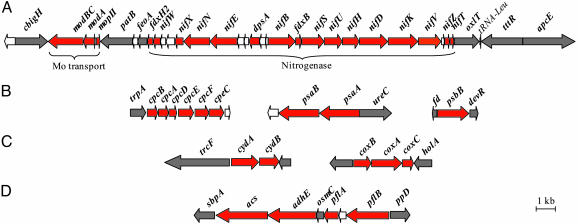
Genes encoding proteins for N2 fixation are clustered on the genomes of Synechococcus OS-A and OS-B′ (Fig. 1A). Genes required for nitrogenase biogenesis are on a 23-kbp region (from modBC to nifT), which is presented as part of a larger 32.5-kbp genomic region (Fig. 1A; from cbigH to apcE) that exhibits identical gene order in Synechococcus OS-A and OS-B′. Deduced amino acid sequences of the nitrogenase polypeptides of Synechococcus OS-A and OS-B′ are between 98% and 100% identical (Table 2, which is published as supporting information on the PNAS web site), which is very high relative to overall identity (88%) among putative orthologs in the two Synechococcus ecotypes. The best identities of the Synechococcus OS-A and OS-B′ Nif polypeptides are mostly with putative orthologs present in Anabaena, Nostoc, and Fischerella strains, ranging from just >50–86% (Table 2).
Fig. 1.
Clusters of genes involved in nitrogenase synthesis and energy metabolism in Synechococcus OS-B′. Red arrows, genes involved in N2 fixation (A), photosynthesis (B), respiration (C), and fermentation (D); gray arrows, genes not associated with these functions; white arrows, genes encoding hypothetical proteins. The functions of many of the different gene products are described in Table 1.
Nitrogenase Transcripts.
To examine in situ expression of nif genes, we used primer pairs [for RT-PCR and quantitative PCR (qPCR)] that would specifically detect nifH, nifD, and nifK transcripts (Table 1). Other primer pairs were designed to detect transcripts encoding photosynthesis, respiration, and fermentation proteins; the genes for some of these transcripts are present in small gene clusters (Fig. 1 B–D). Many of the photosynthesis, respiratory, and fermentation genes are not as similar between Synechococcus OS-B′ and OS-A ecotypes as are the nif genes (Table 1; e.g., ald and cpcF). However, primer pairs that were designed specifically for Synechococcus OS-B′ (except for cpcF primers) are likely to anneal to transcripts from other related Synechococcus ecotypes (3) within the mat.
Table 1.
Primers used for RT-PCR and qPCR analysis of Synechococcus OS-B′ transcript levels
| Gene | Function | Forward primer | Identity, % | Reverse primer | Identity, % |
|---|---|---|---|---|---|
| nifH | nitrogenase iron protein | GGAAGAAAACGGAGCCTAC | 94.7 | CGCCAGAGTAGGCGTATTTC | 100 |
| nifD | nitrogenase molybdenum-iron protein α chain | GCAGTGGCCAAGAGAGTTTC | 100 | CAATCAGGGCCACATCATAAG | 100 |
| nifK | nitrogenase molybdenum-iron protein β chain | CTTCTGCAGTGCAAGCAAAC | 100 | CATGGACAAAGGGCAGAGTTC | 100 |
| psbB | PSII chlorophyll binding protein | CTGCCCAAGTCTCTGCTCTTC | 90 | CAGGATAACCGGGAAGGTCTC | 85 |
| psaB | PSI reaction center protein | CACGTAGGCTGGGACAACTTC | 95.2 | GTCCGTCAGCCACAGAGAC | 100 |
| cpcE | phycocyanobilin lyase α subunit | CTGGGAAGCCCTTATCGAG | 84 | GTCGTAGGCTGGAGTTGGAG | 75 |
| cpcF | phycocyanobilin lyase β subunit | CTCAACTGGCAGCAGAATTG | 85 | CGTCAATATGGCTCAGCAAG | 55 |
| coxA | cytochrome c oxidase subunit I | GAACTTCCTGCTGAGCCTTG | 80 | GTAAAGGCACCGTTGTACCG | 90 |
| cydA | cytochrome d quinol oxidase subunit I | CAACCCCTTCATGGTCAACAG | 80 | CGCTCAAATGGCCGATATAC | 90 |
| pflB | pyruvate formate lyase | GGCGTTTTTGACGCCTATAC | 85 | GGTCTCTTCGTCCATCGTGTC | 94.1 |
| adhE | CoA-linked acetaldehyde dehydrogenase-alcohol dehydrogenase | CAATGCCCTGATGATTACCC | 100 | GTTTCCGCTTGAGTTCTTCG | 80 |
| acs | acetyl-CoA synthase | GAAAAAGTGCAGCAAACCCTTG | 68 | GGTGAGGGTGTGGGAGTAGAG | 100 |
| pflA | pyruvate formate lyase activating enzyme | GCACACGGCTTTGGATACTTC | 90 | GGAACCAGCACAAAACGAATC | 100 |
| ldh | l-lactate dehydrogenase | CCTTTTGGTGGTGAGCAATC | 70 | CCACCTCGCTATCTCCATG | 94.7 |
| pdhB | pyruvate dehydrogenase β subunit | GGAGAAGATGTCGGCCATTA | 85 | CATTGTTGGCAATTTGGTTG | 95 |
| ald | aldehyde dehydrogenase | CAGATCTGATCGAGGCTCGTG | 80 | GCGAAAGAGGGAGGTGTAGC | 85 |
| ackA | acetate kinase | GAGCTTAGATCGGCTGGATG | 100 | CAGTCCTCTTGGGCTGAGATC | 85 |
The transcripts analyzed encode proteins associated with nitrogenase, photosynthesis, respiration, and fermentation. The gene name, protein name/function, primer sequence, and identities (%) of primer sequences to orthologous sequences of Synechococcus OS-A are presented. The accession nos. for all OS-A and OS-B′ genes are CP000239 and CP000240, respectively.
Samples were collected from the Octopus Spring mat at six different time points and photon irradiances on Nov. 5, 2004 (Fig. 2A). Total RNA from each sample was used to evaluate nifH, nifD, and nifK transcript levels by RT-PCR (Fig. 2B Upper). The RNA preparation exhibited no detectable contamination with genomic DNA, as assessed by performing amplification reactions in the absence of reverse transcriptase (data not shown). Almost no transcript from nifH, nifD, or nifK was detected at time point 1 (T1) and T2, when ambient light levels were high. Transcripts for all three nif genes were detected at T3, T4, T5, and T6; at T3, the irradiance had declined to ≈50 μmol·m−2·s−1, whereas, at T4–T6, it approached 0. Quantitative analyses of transcript levels by qPCR (Fig. 2B Lower) showed that nif transcripts were barely detected at T1 and T2, increased sharply at T3 (≈40- to ≈120-fold the T1 value), and then declined as the evening progressed (to between 5- and 25-fold the T1 value). These findings suggest that the nif genes are only expressed when light levels are low and the mat is near anoxic.
Fig. 2.
Accumulation of nif transcripts in Octopus Spring mat samples during an afternoon to nighttime transition on Nov. 5, 2004. (A) Light intensity at the sampling times T1 (13:40), T2 (15:24), T3 (16:35), T4 (17:36), T5 (19:00), and T6 (21:00). (B) RT-PCR (Upper) and qPCR (Lower) analyses of nitrogenase transcript levels. Samples were from the top 2 mm in a region of the mat where the temperature ranged from 53.5°C to 63.4°C. Error bars on graphs indicate the mean ± SD.
Nitrogenase Activity.
To determine whether nitrogenase activity also increased in the mat toward evening, we performed in situ nitrogenase assays by using the top 2 mm of core samples from Octopus Spring (June 2005) and simultaneously monitored O2 profiles and nif transcript levels (Fig. 3). From 16:00 to 18:00 h, when incident irradiance was mostly above 900 μmol·m−2·s−1, nitrogenase activity was not detected and high rates of cyanobacterial photosynthesis caused hyperoxic conditions in the upper part of the mat. As photon irradiance declined, O2 concentration in the mat decreased dramatically. Irradiance at time 20:05 h was only ≈31 μmol·m−2·s−1, whereas O2 penetration was <0.1 mm, and high nitrogenase activity (relative to daytime levels) was detected in mat samples. Sampling at later times showed that the mat remained anoxic during the night, and nitrogenase activity was sustained. Transcripts for the nitrogenase subunits were only detected as the light levels declined and the mat became anoxic, consistent with the results presented in Fig. 2; almost no nif transcripts were detected at 18:00 h, whereas high transcript levels were observed at 20:30 and 23:00 h (Fig. 3 Bottom). Nitrogenase activity was only detected at the time when nitrogenase transcripts began to accumulate. Transcripts encoding photosynthetic proteins such as PsaB showed an opposite trend relative to nif transcripts; they were high during the day and low in the evening (Fig. 3 Bottom). Nitrogenase activity and cyanobacterial nif gene expression were also detected in mat samples collected from 60–61°C sites in Mushroom Spring (a nearby spring with similar mat communities as in Octopus Spring) during the evening (A.-S.S., D.B., D.M.W., E.B., J.W.P., M.K., A.R.G., and S. Jensen, unpublished data).
Fig. 3.
In situ nitrogenase activity, photon irradiance, oxygen penetration, and transcript abundance measured in the Octopus Spring microbial mat on June 23, 2005. (Top) Incident photon irradiance (small black circles) and nitrogenase activity (large gray circles) as a function of time of day. Vertical bars represent error, whereas horizontal bars indicate incubation period. (Middle) Depth distribution of oxygen concentration in the microbial mat at 934, 29.3, and 0.0 μmol ·m−2·s−1, respectively. (Bottom) qPCR examination of nifHDK and psaB transcript levels at times indicated. Error bars are as in Fig. 2.
Transcripts for Proteins Involved in Energy Metabolism.
Nitrogenase requires 16 ATP per N2 reduced. The ATP used for N2 fixation in mats could be from photosynthesis, which would still be occurring as light levels decline and the consumption of O2 by respiration becomes more rapid than O2 production. Fermentation of carbohydrates that accumulate as a consequence of photosynthetic CO2 fixation in the day may also generate energy during the evening.
We used specific primer pairs (Table 1) to evaluate in situ transcript levels from genes encoding proteins important for photosynthesis, respiration, and fermentation during the November 2004 experiment (Fig. 4A–D). Based on RT-PCR, transcripts for photosynthetic proteins showed an opposite trend relative to nif transcripts (compare Fig. 4A with Fig. 2B); they were high during the day and low in the evening. For genes associated with photosynthesis (cpcF and cpcE, phycocyanin lyase; psaB, subunit of PS I reaction center; and psbB, PS II antenna chlorophyll binding protein), transcripts were high at T1 and T2, decreased somewhat by T3, and then became barely detectable by T4. However, the psbB transcript was still detectable in mat samples collected in the evening (Fig. 4A, T4–T6).
Fig. 4.
Transcript levels in Octopus Spring mat samples collected on Nov. 5, 2004 at sampling times T1–T6, as in Fig. 2. RT-PCR was performed to examine accumulation of transcripts from genes involved in photosynthesis (cpcF, cpcE, psaB, and psbB) (A), respiration (coxA and cydA) (B), and fermentation (pflB, pflA, adhE and acs transcript increase during the night, whereas ldh, pdhB, ald and ackA transcripts decrease during the night) (C and D). (E) Quantification of transcript levels analyzed by RT-PCR (A–D) by using qPCR. Samples were from the top 2 mm in a region of the mat where the temperature ranged from 53.5°C to 63.4°C. Error bars are as in Fig. 2.
Transcripts for proteins involved in respiration (coxA, for subunit I of cytochrome aa3-type cytochrome c oxidase and cydA, for subunit I of the cytochrome quinol oxidase) appeared to remain nearly constant (coxA) or to decline slightly in the evening (cydA), based on RT-PCR (Fig. 4B). In contrast, two distinct patterns of gene expression were associated with putative fermentation genes (1, 33) (see Table 1). Transcripts encoding pyruvate formate lyase (pflB), pyruvate formate lyase activating enzyme (pflA), CoA-linked alcohol/acetaldehyde dehydrogenase (adhE), and acetyl-CoA synthase (acs) increased during the evening when the mat was anoxic (Fig. 4C); genes encoding these transcripts are colocalized on the genomes (Fig. 1D), and all or some may be cotranscribed. Transcripts for lactate dehydrogenase (ldh), pyruvate dehydrogenase (pdhB), and to some extent aldehyde dehydrogenase (ald) and acetate kinase (ackA) (genes for these proteins are not clustered on the genome) declined during the night (Fig. 4 C and D).
We also used qPCR to quantify transcript abundance. As observed by RT-PCR (Fig. 4), transcripts for photosynthesis genes (psbB, psaB, cpcE, and cpcF) were high during the day relative to the evening (Fig. 4E Top Left); by T4, the transcripts for these genes were barely detectable. Transcripts from genes encoding proteins involved in respiration (coxA and cydA) also declined during the evening (Fig. 4E Top Right). It was not clear from the RT-PCR results (Fig. 4B) that the transcript encoding CoxA decreased as light levels declined, but the qPCR results showed an ≈3-fold decline in transcript abundance at all times after 16:35 h (T3); because of amplification saturation, the RT-PCR does not report true abundance differences. In contrast, and in accord with RT-PCR results, transcripts for the linked set of fermentation genes (pflB, pflA, adhE, and acs; Fig. 4E Bottom Left and Middle Left) increased in the evening, whereas transcripts for the others (ldh, ald, pdhB, and ackA; Fig. 4E Middle Right) declined. For the former set of fermentation-associated transcripts, once the transcript levels increased, they remained high into the night.
Discussion
The combined use of genomic information, gene expression analysis, and ecophysiological methods has provided insights into the ecology of Synechococcus-dominated hot spring mats. We demonstrated that the Synechococcus nif genes are expressed in situ as the mat becomes anoxic under declining photon irradiance in the late afternoon and that N2 fixation increases in parallel with increasing transcript levels. N2 fixation detected in earlier studies was attributed to heterocystous cyanobacteria, which only live in the lower temperature regions of the mat, or to heterotrophic bacteria (10, 12, 34). Although we cannot exclude the participation of heterotrophic bacteria in N2 fixation at 53.5°C–63.4°C in this mat, our results demonstrate that expression of Synechococcus nif genes occurs in concert with considerable levels of nitrogenase activity. These results strongly suggest that the unicellular Synechococcus ecotypes fix N2 at elevated temperatures within the hot spring mats. Irradiance levels of <50 μmol·m−2·s−1 corresponded with expression of nif genes in the mat. At higher photon irradiances, when cyanobacterial O2 production exceeds O2 consumption by the various microbes in the top few millimeters of the mat, O2 accumulates within the mat and inhibits nitrogenase activity (and possibly the expression of nif genes).
As seen in intertidal (35) and hypersaline cyanobacterial mats (36), the nitrogenase genes of Synechococcus are expressed during the night, as in the Octopus Spring mat. Together with the activity of other N2-fixing bacteria, fixation by cyanobacteria may alleviate N-limitation in such mats. However, the absence of O2 in the mats at these times limits energy production to that generated by fermentative metabolism. Fermentation does not produce nearly as much chemical bond energy as respiration (fermentation produces 2–4 ATPs per glucose metabolized), so the rate at which ATP is produced during the night would likely limit cyanobacterial N2 fixation.
It is unclear how the nitrogenase genes are regulated in thermophilic cyanobacteria. In diazotrophic bacteria except for cyanobacteria, nitrogenase is often regulated by the NifLA proteins, regulatory elements that sense and respond to changing environmental O2 levels (37). We have not detected genes encoding NifLA-like proteins on the Synechococcus OS-A and OS-B′ genomes, but the genomes do contain genes encoding proteins that regulate nitrogen-associated processes, including the glnB (for PII) and ntcA genes (18). The PII polypeptide can function as a global regulator of nitrogen utilization, whereas cyanobacterial NtcA appears to activate genes encoding proteins involved in the transport and assimilation of various nitrogen compounds (38). NtcA is homologous to transcriptional regulators belonging to the cAMP receptor protein family and directly binds to the promoter regions at specific sites with the consensus nucleotide sequence GTAN8TAC. We localized this sequence upstream of several genes in the genomes of Synechococcus OS-A and OS-B′; for example, the sequence was localized upstream of genes for nitrite reductase (nirA), the PII protein, the ammonium transporter (amt), and NtcA itself. However, there is no clear consensus sequence upstream of the nif genes, so it is uncertain what regulatory protein modulates nif gene expression.
Transcripts for proteins involved in photosynthesis and respiration were abundant during the day when O2 levels in the mat are high, but decreased, in many cases dramatically, as the mat became anoxic in the evening. Maintaining some transcripts (and proteins) for fully active photosynthetic and respiratory systems throughout the night could be advantageous for the cyanobacteria. At daybreak, an intact photosynthetic and respiratory capacity would allow for the production of moderate levels of ATP and rapid consumption of photosynthetically produced O2, thereby sustaining N2 fixation activity. In preliminary experiments, a strong burst of nitrogenase activity was observed during sunrise in mat samples from Mushroom Spring (A.-S.S., D.B., D.M.W., E.B., J.W.P., M.K., A.R.G., and S. Jensen, unpublished data). A similar burst of N2 fixation in temperate microbial mats, occurring soon after sunrise, has been reported in refs. 1 and 39.
To examine the possibility that Synechococcus might perform fermentation in the dark, we studied expression of a number of genes implicated in cyanobacterial fermentation pathways (1, 33). It is interesting that genes possibly associated with fermentation processes during the day differ from those active in the evening. The pflB, pflA, adhE, and acs genes are clustered on the genome (Fig. 1D), and the transcripts from these genes increase in parallel during the evening (as noted in Fig. 1D, some genes may be part of a single transcriptional unit). The PflB and PflA proteins are involved in the fermentation pathway that generates acetyl-CoA and formate from pyruvate whereas AdhE converts acetyl-CoA into ethanol. The Synechococcus ecotypes examined in this study thus appear to have the potential to synthesize formate and ethanol during the night. Transcripts from the ldh, pdhB, and, to some extent, adh and ackA are high in the day and decline in the evening. Most of these transcripts appear to be present (especially the ald transcript) over the entire diel cycle, although the levels are significantly higher in the day. These transcript profiles suggest that the Synechococcus ecotypes may have a potential to form lactate and possibly acetate during the day and also during the transition period when the cyanobacteria switch from “light” to “dark” metabolism. These putative fermentation proteins may also be involved in metabolic pathways other than fermentation.
It is interesting to consider these results in the context of past work on the formation and fate of fermentation products in alkaline hot spring cyanobacterial mats and the nature of the interactions of organisms within mat communities. Acetate was measured as the major product of fermentation that accumulated when mat samples were shifted from light, oxic conditions to dark, anoxic conditions. Smaller amounts of propionate and even smaller amounts of butyrate and valerate were detected, but more reduced fermentation products (e.g., ethanol and lactate) did not accumulate (31). The production of acetate (and CO2) occurred within hours after mat samples were transferred to the dark; acetate production was linked to glycogen fermentation by Synechococcus (30). Under conditions used for these cited experiments, the production and consumption of fermentation products by all members of the mat community would occur simultaneously. For instance, the production of CO2 associated with fermentation of glycogen by Synechococcus may require the presence of a second formate-using organism, because so far we have found no evidence that the Synechococcus genomes contain the gene encoding formate hydrogenlyase, which converts formate to H2 and CO2. The generation of acetyl-CoA in fermenting Synechococcus cells via pyruvate formate lyase, rather than by the pyruvate dehydrogenase reaction (which generates reduced pyridine nucleotide), would help protect the NAD+ pool. This finding highlights the issue that changes in transcript levels in one organism may not be reflected by accumulation of a specific “predicted” end product in the environment because of metabolite exchange processes that occur within the mat consortium. Furthermore, the failure to observe lactate accumulation may be a consequence of the utilization of this metabolite by other organisms in the mat community (31). The production of reduced fermentation products like lactate and ethanol can be decreased by syntrophic interactions between fermentative and H2-consuming organisms (e.g., acetogenic and sulfate-reducing bacteria and methanogenic archaea) in these mats (31). Interspecies H2 transfer allows fermentative microorganisms to reoxidize NADH by reducing protons to H2, alleviating the need for regenerating NAD+ by reduction of high-energy intermediates; more rapid regeneration of NAD+ would allow for elevated rates of fermentation and ATP production, which in turn could stimulate N2 fixation. However, extensive searches of the Synechococcus genomes have not identified genes encoding enzymes necessary for interspecies H2 transfer (e.g., NADH:ferredoxin oxidoreductase and hydrogenase).
Our findings highlight one of the most interesting and novel aspects of microbial mats: The individuals in this consortium of organisms have metabolic capabilities that are strongly integrated into “community metabolism” (30–32). This integration in turn suggests strong evolutionary constraints because of obligate interactions among the physiologically integrated bacterial populations. To further resolve the intriguing networks of interaction and to understand how they shape and define microbial communities and the evolution of ecotypes, we are taking an interdisciplinary approach in which molecular and genomic analyses are interfaced with ecological and evolutionary questions and methodologies.
Methods
Sample Collection.
Samples were collected on Nov. 5, 2004 and June 23, 2005 from the southernmost effluent channel of Octopus Spring. The temperature of Octopus Spring exhibited a temporal cycle of ≈5 min, with a 53.5–63.4°C temperature range at the sampling site, and an average temperature of 60°C over the entire sampling period. The mat was ≈1 cm thick, with Synechococcus populating the top 1-mm layer (other cyanobacteria were not observed by microscopic analysis). Cores (1 cm in diameter) were collected, and the top 2 mm of each was excised, frozen in liquid N2, and stored for nucleic acid extraction. Samples were collected at six time points during the diel cycle in November 2004 (see Fig. 2) and at four time points in June 2005 (see Fig. 3). Microprofiles of O2 concentration were measured with a Clark-type O2 microsensor operated by an automated profiling system (Unisense, Aarhus, Denmark) (40, 41), and the downwelling photon irradiance at each sampling time was measured by using a LI-1400 datalogger (Li-Cor, Lincoln, NE).
RNA Extraction.
Mat samples that had been frozen and stored at −80°C were dispersed by vortexing them in 1 ml of 10 mM NaOAc (pH 4.5) and 0.5 g of glass beads (150–212 μm, Sigma-Aldrich) in a 2-ml screw cap microfuge tube. Cells were harvested by centrifugation (11,750 × g) for 1 min at 4°C, the cell pellet resuspended in 250 μl of 10 mM NaOAc (pH 4.5), 37.5 μl of 500 mM Na2-EDTA (pH 8.0), and RNA was extracted as described in ref. 42. Isolated RNA was treated twice with 7 units per reaction of RNase-free DNase (Qiagen, Valencia, CA) for 20 min, the reaction was stopped with 1 vol of phenol:chloroform (1:1), and the RNA in the aqueous layer was precipitated during a 20-min incubation at −20°C after the addition of 0.1 vol of 10 M LiCl and 2.5 vol of 100% ethanol. RNA concentration was determined by absorption at 260 nm and with the ribogreen RNA quantification kit (Molecular Probes).
RT-PCR.
Specific primers were designed to amplify regions (≈200 nucleotides) of Synechococcus OS-B′ nifH, nifK, nifD, psaB, psbB, cpcA, cpcF, cpcE, coxA, cydA, pflB, pflA, adhE, acs, ldh, pdhB, ald, and ackA genes. The accession no. for all OS-B′ genes is CP000240. The primer pairs used are given in Table 1. Primers were annealed to 100 ng of total RNA extracted from mat samples and extended for 45 min at 55°C by using 200 units of reverse transcriptase (RT) (Superscript III, Invitrogen). Two microliters of the RT reaction was used for the subsequent PCR; each PCR contained 50 units of TaqDNA polymerase (Qiagen, Valencia, CA) and 5% dimethyl sulfoxide. The PCR program included 1 cycle of 95°C for 1 min, 30 cycles of 94°C for 10 s, 55°C for 30 s, 72°C for 30 s, and a final incubation at 72°C for 10 min. Amplified products were analyzed by electrophoresis in a 1.2% agarose gel.
qPCR.
Levels of specific transcripts in total mRNA from the mat samples were quantified by RT-PCR by using the Engine Opticon system (Bio-Rad). Single-stranded cDNA, synthesized in a reverse transcriptase reaction by using DNase-treated RNA (as described above), served as the template for qPCR amplifications, which were performed by using the DyNAmo HS SYBR green qPCR kit (Finnzymes, Espoo, Finland). The specific amplification protocol was 1 cycle at 95°C for 10 min, 44 cycles at 94°C for 10 s, 56°C for 15 s, 72°C for 8 s, and a final incubation of 72°C for 10 min. We determined both the absolute (43) and relative (normalized to the T1 sample, collected at 13:30 h) levels of each specific RNA among all environmental samples.
Nitrogenase Activity.
Nitrogenase activity was assayed by the acetylene reduction technique (44). Core mat samples (1.13 cm2) were collected, and the top 2 mm was removed and placed in 10-ml serum vials that were sealed under a constant stream of argon. Two milliliters of argon-sparged Octopus Spring water was added, and the vials were incubated for 15 min in the hot spring at in situ temperature and photon irradiance. The nitrogenase assay was then initiated with the injection of 1 ml of O2-free acetylene, and vials were incubated in the spring for an additional 2 h before each reaction was stopped with 0.2 ml of formaldehyde. Acetylene injections to initiate the assays were done at 16:00, 18:05, 21:05, and 23:00 h (15 min after each preincubation) when the irradiances were 919, 31, 0, and 0 μmol·m−2·s−1, respectively. Each assay was performed in triplicate. The nitrogenase activity reported was the total activity over the 2-h incubation; samples used for measuring transcript abundance were taken at the end of each incubation period, except for one sample, which was taken at 20:30 h instead of 20:05 h. Ethylene production in samples was measured by gas chromatography (Shimadzu GC-8A with a flame ionization detector).
Supplementary Material
Acknowledgments
We thank Fred Cohan (Wesleyan University) and John Heidelberg (Institute of Genomic Research) for the work they have done on genome analysis; Natalia Khuri for discussions concerning gene identification and for building a web site, which has been instrumental in our analysis of the genes present on the Synechococcus OS-A and OS-B′ genomes; Eric Becraft and Chris Klatt for help in the field; and the U.S. National Park Service and personnel from Yellowstone National Park for their permission to conduct this work and their helpful assistance. A.R.G. and D.B. thank Lucas Stal, Paul Ludden, and Luis Rubio for helpful and exciting discussions. The research was funded by the Frontiers in Integrative Biology Program for National Science Foundation Grant EF-0328698. M.K. acknowledges additional support from the Danish Natural Science Research Council, and J.W.P. acknowledges support from National Institutes of Health National Institute of General Medical Sciences Grant GM069938 and the National Aeronautics and Space Administration Thermal Biology Institute.
Glossary
Abbreviations:
- qPCR
quantitative PCR
- Tn
time point n.
Footnotes
References
- 1.Stal L. J. In: The Ecology of Cyanobacteria. Whitton B. A., Potts M., editors. Dordrecht, The Netherlands: Kluwer; 2000. pp. 61–120. [Google Scholar]
- 2.Brock T. D. Thermophilic Microorganisms and Life at High Temperatures. New York: Springer; 1978. [Google Scholar]
- 3.Ward D. M., Ferris M. J., Nold S. C., Bateson M. M. Microbiol. Mol. Biol. Rev. 1998;62:1353–1370. doi: 10.1128/mmbr.62.4.1353-1370.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ward D. M., Castenholz R. W. In: The Ecology of Cyanobacteria. Whitton B. A., Potts M., editors. Dordrecht, The Netherlands: Kluwer; 2000. pp. 37–59. [Google Scholar]
- 5.Nübel U., Bateson M. M., Vandieken V., Wieland A., Kühl M., Ward D. M. Appl. Environ. Microbiol. 2002;68:4593–4603. doi: 10.1128/AEM.68.9.4593-4603.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Castenholz R. W., Pierson B. K. Ecology of Thermophilic Anoxygenic Phototrophs. Dordrecht, The Netherlands: Kluwer; 1995. [Google Scholar]
- 7.Ramsing N. B., Ferris M. J., Ward D. M. Appl. Environ. Microbiol. 2000;66:1038–1049. doi: 10.1128/aem.66.3.1038-1049.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferris M. J., Kühl M., Wieland A., Ward D. M. Appl. Environ. Microbiol. 2003;69:2893–2898. doi: 10.1128/AEM.69.5.2893-2898.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Allewalt J. P., Bateson M. M., Revsbech N. P., Slack K., Ward D. M. Appl. Environ. Microbiol. 2005;72:544–550. doi: 10.1128/AEM.72.1.544-550.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stewart W. D. P. Phycologia. 1970;9:261–268. [Google Scholar]
- 11.Wickstrom C. E. J. Phycol. 1980;16:436–443. [Google Scholar]
- 12.Belay N., Sparling R., Daniels L. Nature. 1984;312:286–288. doi: 10.1038/312286a0. [DOI] [PubMed] [Google Scholar]
- 13.Jacobson M. R., Cash V. L., Weiss M. C., Laird N. F., Newton W. E., Dean D. R. Mol. Gen. Genet. 1989;219:49–57. doi: 10.1007/BF00261156. [DOI] [PubMed] [Google Scholar]
- 14.Rangaraj P., Ludden P. W. J. Biol. Chem. 2002;277:40106–40111. doi: 10.1074/jbc.M204581200. [DOI] [PubMed] [Google Scholar]
- 15.Hu Y., Fay A. W., Dos Santos P. C., Naderi F., Ribbe M. W. J. Biol. Chem. 2004;279:54963–54971. doi: 10.1074/jbc.M408983200. [DOI] [PubMed] [Google Scholar]
- 16.Kim S., Burgess B. K. J. Biol. Chem. 1996;271:9764–9770. doi: 10.1074/jbc.271.16.9764. [DOI] [PubMed] [Google Scholar]
- 17.Johnson D. C., Dos Santos P. C., Dean D. R. Biochem. Soc. Trans. 2005;33:90–93. doi: 10.1042/BST0330090. [DOI] [PubMed] [Google Scholar]
- 18.Rubio L. M., Ludden P. W. J. Bacteriol. 2005;187:405–414. doi: 10.1128/JB.187.2.405-414.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fay P. Microbiol. Rev. 1992;56:340–373. doi: 10.1128/mr.56.2.340-373.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murry M. A., Wolk C. P. Arch. Microbiol. 1989;151:469–474. [Google Scholar]
- 21.Wolk C. P., Ernst A., Elhai J. In: The Molecular Biology of Cyanobacteria. Bryant D. A., editor. Boston: Kluwer; 1994. pp. 769–823. [Google Scholar]
- 22.Tel-Or E., Stewart W. D. P. Proc. R. Soc. London Ser. B; 1977. pp. 61–86. [Google Scholar]
- 23.Valladares A., Herrero A., Pils D., Schmetterer G., Flores E. Mol. Microbiol. 2003;47:1239–1249. doi: 10.1046/j.1365-2958.2003.03372.x. [DOI] [PubMed] [Google Scholar]
- 24.Golden J. W., Yoon H. S. Curr. Opin. Microbiol. 2003;6:557–563. doi: 10.1016/j.mib.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 25.Reddy K. J., Haskell J. B., Sherman D. M., Sherman L. A. J. Bacteriol. 1993;175:1284–1292. doi: 10.1128/jb.175.5.1284-1292.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carpenter E. J., Price C. C. Science. 1976;191:1278–1280. doi: 10.1126/science.1257749. [DOI] [PubMed] [Google Scholar]
- 27.Schneegurt M. A., Sherman D. M., Nayar S., Sherman L. A. J. Bacteriol. 1994;176:1586–1597. doi: 10.1128/jb.176.6.1586-1597.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang T. C., Lin R. F., Chu M. K., Chen H. M. Microbiology. 1999;145:743–753. doi: 10.1099/13500872-145-3-743. [DOI] [PubMed] [Google Scholar]
- 29.Bateson M. M., Ward D. M. Appl. Environ. Microbiol. 1988;54:1738–1743. doi: 10.1128/aem.54.7.1738-1743.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nold S. C., Ward D. M. Appl. Environ. Microbiol. 1996;62:4598–4607. doi: 10.1128/aem.62.12.4598-4607.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Anderson K. L., Tayne T. A., Ward D. M. Appl. Environ. Microbiol. 1987;53:2343–2352. doi: 10.1128/aem.53.10.2343-2352.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van der Meer M. T. J., Schouten S., Bateson M. M., Nübel U., Wieland A., Kühl M., de Leeuw J. W., Sinninghe-Damsté J. S. S., Ward D. M. Appl. Environ. Microbiol. 2005;71:3978–3986. doi: 10.1128/AEM.71.7.3978-3986.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stal L. J., Moezelaar R. FEMS Microbiol. Rev. 1997;21:179–211. [Google Scholar]
- 34.Wickstrom C. E. Curr. Microbiol. 1984;10:275–280. [Google Scholar]
- 35.Steppe T. F., Pearl H. W. Microb. Ecol. 2005;49:1–10. doi: 10.1007/s00248-004-0245-x. [DOI] [PubMed] [Google Scholar]
- 36.Omoregie E. O., Crumbliss L. L., Bebout B. M., Zehr J. P. Appl. Environ. Microbiol. 2004;70:2119–2128. doi: 10.1128/AEM.70.4.2119-2128.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmitz R. A., Klopprogge K., Grabbe R. J. Mol. Microbiol. Biotechnol. 2002;4:235–242. [PubMed] [Google Scholar]
- 38.Flores E., Herrero A. Biochem. Soc. Trans. 2005;33:164–167. doi: 10.1042/BST0330164. [DOI] [PubMed] [Google Scholar]
- 39.Villbrandt M., Stal L. J., Krumbein W. E. FEMS Microbiol. Ecol. 1990;74:59–72. [Google Scholar]
- 40.Revsbech N. P. Limnol. Oceanogr. 1989;34:474–478. [Google Scholar]
- 41.Revsbech N. P., Ward D. M. Appl. Environ. Microbiol. 1984;48:270–275. doi: 10.1128/aem.48.2.270-275.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bhaya D., Vaulot D., Amin P., Takahashi A. W., Grossman A. R. J. Bacteriol. 2000;182:5692–5699. doi: 10.1128/jb.182.20.5692-5699.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Whelan J. A., Russell M. B., Whelan M. A. J. Immunol. Methods. 2003;278:261–269. doi: 10.1016/s0022-1759(03)00223-0. [DOI] [PubMed] [Google Scholar]
- 44.David K. A. V., Apte S. K., Banerji A., Thomas J. Appl. Environ. Microbiol. 1980;39:1078–1080. doi: 10.1128/aem.39.5.1078-1080.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.