Abstract
Cancers display a diverse set of cellular defects, which are thought to be elicited by multiple genetic mutations. In this study, we show that when a single adherens junction protein, α-catenin, is removed by conditional targeting, the entire skin epidermis systematically transforms to a hyperproliferative, invasive tissue replete with inflammation. Transcriptional profiling and biochemical analyses reveal that α-catenin ablation is accompanied by activation of NF-κB and its proinflammatory target genes, along with genes involved in proliferation, wound healing, angiogenesis, and metastasis. Many of these alterations occur in vitro and in the embryo, and thus seem at least partly to be intrinsic to the loss of α-catenin. We show that reductions in α-catenin, activation of NF-κB, and inflammation are common features of human squamous cell carcinomas of the skin.
Keywords: cancer, inflammation, E-cadherin
Adherens junctions (AJs) are intercellular junctions prominent in normal epithelia and reduced in cancers. The prototypic transmembrane core of AJs is composed of E-cadherin, whose cytoplasmic domain binds β-catenin, which in turn binds α-catenin. α-Catenin then regulates actin dynamics to integrate AJs with the cytoskeleton and promote intercellular adhesion (1). Throughout development, epithelia also use AJs to coordinate and maintain an intricate balance between intercellular adhesion and growth control. The best-studied mechanism is through Wnt signaling, which stabilizes additional β-catenin to serve as a transcription cofactor for the Tcf/Lef family of DNA binding proteins (2). Among the targets of Tcf/Lef/β-catenin complexes are cell-cycle-regulated genes.
Mutations in β-catenin, E-cadherin, and α-catenin genes have been found in some human cancers, and reduced E-cadherin and α-catenin are often prognostic markers of poor clinical outcome in squamous cell carcinomas (SCCs) (2, 3). It has been assumed that reductions in E-cadherin and α-catenin act by liberating β-catenin, freeing it to trigger Tcf/Lef-mediated transcription and causing uncontrolled tumor growth (4, 5). Thus it is curious that in skin, 2 days after conditional ablation of α-catenin, the epidermis loses polarity and becomes hyperproliferative, yielding large masses that resemble SCC in situ, a precancerous state (6). By contrast, activating mutations in β-catenin typically result in benign hair tumors (pilomatricomas) rather than epidermal SCCs (7, 8). Such findings cannot be explained by α-catenin acting simply as a tumor suppressor to sequester β-catenin.
This conundrum prompted us to explore how loss of α-catenin elicits this phenotype. Here we report that α-catenin conditional knockout (cKO) epidermis systematically progresses from a hyperpoliferative adherent tissue to a dermal infiltrate of nonadherent, dysplastic cells. We have also unraveled a link between α-catenin and NF-κB signaling that unexpectedly points to α-catenin at the nexus of additional pathways that go awry in cancers.
Results
Progression to Hyperproliferation and Invasion.
K14-Cre/α-catenin fl/fl mice display quantitative loss of α-catenin in skin and oral epithelium by embryonic day 17.5 (E17.5) and die shortly after birth (6). We explored the phenotypic consequences in greater detail by grafting E18.5 α-catenin cKO and control littermate (WT) skins onto the backs of thymus-defective (Nude) mice. Within 4–6 weeks, WT grafts appeared normal, replete with hairs. By contrast, cKO grafts lacked hairs and displayed tumor-like nodules (Fig. 1A).
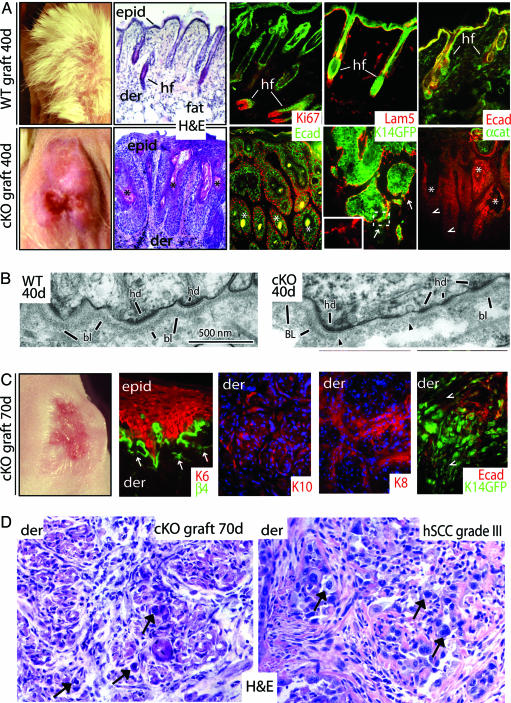
Fig. 1.
Progression to SCC in the absence of α-catenin. E18.5 WT and cKO skins were grafted onto Nude mice for the days indicated. Seventy-day WT grafts were analogous to 40 days and are not shown. (A, C, and D) Far left images in A and C are of grafts (note the absence of follicles in cKO grafts). Hematoxylin and eosin (H&E)-stained sections are from grafts, with the exception of D Lower Right, which is of a grade III hSCC (arrows denote atypical cells, classical morphological signs of SCC, and prevalence in the dermis of the 70-day cKO graft). Color coding of immunostained sections is according to secondary Abs; some sections are counterstained with DAPI (blue). Note that in C most sections are of invasive keratinocytes within the dermis as indicated in upper left of each frame. Arrows in A and C denote perturbations at dermo-epidermal borders (hemidesmosome/basement membrane), more extensive at 70 days than 40 days. Arrowheads denote epidermal cells displaying weak or no E-cadherin, which in C are also weak for K14-GFP+. epid, epidermis; der, dermis; hf, hair follicles; Lam5, laminin-5; Ecad, E-cadherin; αcat, α-catenin; K14GFP, transgenic epifluorescence expression of GFP; Ki67, a proliferating nuclear antigen; K, keratin. Asterisks denote keratinized pearls. (B) EM of ultrathin sections that correspond to boxed areas in Fig. 6 A′ and B′. Shown are the dermo-epidermal boundaries. Note that the basal lamina (bl) and hemidesmosomes (hd) are intact in WT but are perturbed in cKO (region flanked by arrowheads).
Histological analyses revealed massive invaginations throughout the entire epithelium of 10 of 10 40-day cKO skin grafts. These lesions were epidermal in nature, as judged by the presence of keratinized centers (∗ in Fig. 1A) and by immunofluorescence microscopy (Fig. 1A and Figs. 5 and 6, which are published as supporting information on the PNAS web site). Masses were positive for epidermal keratins and granular layer markers, but negative for hair follicle markers. Masses were also hyperproliferative, as judged by multiple external layers of mitotically active, anti-Ki67-positive cells and internal differentiating layers positive for hyperproliferation-associated keratins. Anti-E-cadherin staining was markedly diminished at the invaginating fronts of α-catenin cKO epidermis (arrowheads in Fig. 1A). Abs against hemidesmosomal and extracellular matrix ligands revealed signs of discontinuity in the basement membrane, which was confirmed by ultrastructural analyses (Figs. 1 A and B and 5). Similar dysplastic changes were observed in α-catenin ablated regions of cKO mice that survived because they were mosaic for the targeting event (Fig. 5). Overall, the morphological and biochemical perturbations resembled those of well differentiated (grade I) SCCs in human.
By 70 days, WT grafts remained unchanged, but cKO grafts resembled human grade III SCC with many atypical cells present (Fig. 1 C and D). Near the skin surface, the epithelium was hyperplastic and disorganized, and discontinuities in the basement membrane were prevalent. Keratinocytes invading the dermis were mostly atypical and fibroblast-like in appearance. We confirmed their origin by breeding cKO to K14-GFPactin mice (9) and repeating the grafts with GFP+ epidermis. The GFP+ dermal keratinocytes expressed both simple epithelial and epidermal keratins and showed reduced E-cadherin. Cells expressing lower K14-GFPactin exhibited little or no E-cadherin.
Signaling Pathways Changed by Loss of α-Catenin.
To understand how loss of α-catenin leads to such global effects, we first examined mRNA expression patterns in cKO and WT epidermis. For microarray profiling, we used epidermis from E18.5 embryos, i.e., shortly after quantitative loss of α-catenin. Analyzing embryonic skin also minimized secondary events that might occur if the epidermal barrier was not intact at birth.
High-stringency comparative analyses uncovered the distinguishing features that mark the early consequences of α-catenin ablation. Most of the genes scored as unchanged, reflective of epidermal character. A small group of genes scored as up- or down-regulated by ≥2× in cKO versus WT epidermis (Fig. 2A). We confirmed these differences by real-time PCR using mRNAs isolated from FACS populations of keratin-expressing basal cells of E18.5 epidermis and cultured keratinocytes (Fig. 2B).
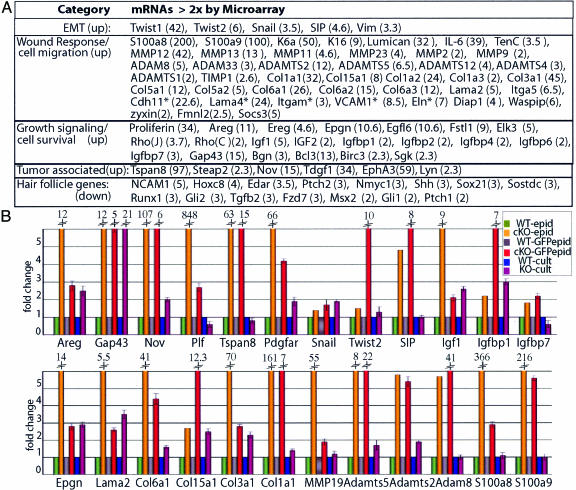
Fig. 2.
Changes in gene expression elicited by loss of α-catenin. (A) mRNAs in five relevant categories that scored by microarray analyses as up-regulated or down-regulated in E18.5 cKO vs. WT epidermis. Numbers in parentheses correspond to fold change. Asterisks refer to mRNAs not expressed by keratinocytes. EMT, epithelial-mesenchymal transition. (B) Real-time PCR of mRNAs from E18.5 epidermis (epid), FACS-purified keratinocytes from epidermis (GFPepid), and primary keratinocytes (cult) cultured in low calcium medium to prevent intercellular adhesion. Fold change is relative to WT-epid (=1).
The up-regulated genes within the α-catenin null molecular signature included cell survival and growth signaling genes, consistent with the elevated Ras-mitogen-activated protein kinase signaling that occurs in the cKO epidermis (6). Especially prominent were insulin growth factor signaling molecules. Insulin growth factors are potent stimulators of keratinocyte growth and survival, and this pathway is frequently perturbed in human SCCs (hSCCs) (10). Only a few established Wnt pathway members scored as up-regulated in cKO skin. By contrast, genes associated with hair follicle morphogenesis, a Wnt-regulated process, were uniformly down-regulated (Fig. 2), consistent with the absence of follicles. Additional evidence in Fig. 7, which is published as supporting information on the PNAS web site, fails to support the view that Wnt signaling underlies the α-catenin cKO phenotype in skin.
The up-regulation of Snail, Twist, and SIP transcriptional repressors was consistent with the down-regulation of E-cadherin at the edges of invaginating epithelial masses and with the epithelial to mesenchymal-like transition (11) seen in day-70 cKO grafts. Normally at the migrating front of hair germs (12), nuclear Snail1 was detected in E18.5 cKO epidermis (Fig. 8, which is published as supporting information on the PNAS web site). Also up-regulated in cKO epidermis were other migration, wound-healing, and invasion-associated genes, including those encoding tenascin C, biglycan, K6, K16, matrix metalloproteinases (MMPs), and other basement membrane remodeling factors. Increased MMP-2 expression was notable, because it is a measure of metastatic aggressiveness for hSSCs (13).
Many of these differences were also in cultured cKO versus WT keratinocytes, suggesting that they were not solely caused by loss of tissue integrity (Fig. 2B). Previously, we observed enhanced proliferation and migration in cultured α-catenin null keratinocytes (6).
NF-κB Activation upon α-Catenin Ablation.
Perhaps the most striking and unanticipated feature of the arrays was the large number of NF-κB target genes up-regulated early after α-catenin ablation (Fig. 3A). Of particular interest were wound-responsive and inflammatory response genes. NF-κB is a classic regulator of antiapoptotic and proinflammatory gene expression (14, 15). The best-characterized form of NF-κB is a heterodimer composed of p50 and p65 subunits, which in resting cells is sequestered in a cytoplasmic complex with inhibitory protein IκBα (16). NF-κB-activating signals lead to IκBα phosphorylation, freeing NF-κB for phosphorylation and nuclear translocation, enabling it to transactivate its target genes (15, 17).
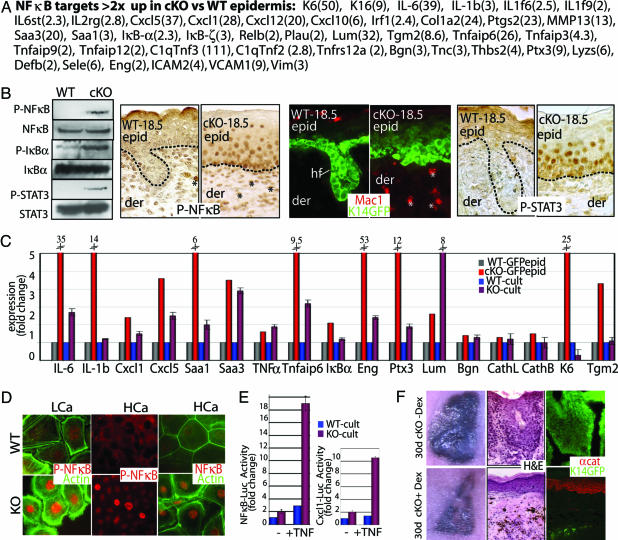
Fig. 3.
Early activation of the NF-κB pathway in α-catenin null epidermis. (A) Putative NF-κB target genes whose mRNAs were scored by microarray as up-regulated in E18.5 cKO vs. WT epidermis. Numbers in parentheses correspond to fold change. (B) NF-κB activation and inflammation in E18.5 cKO skin. (Left) Immunoblot with panel and phospho-specific Abs: P-NF-κB (activated p65 subunit), P-IκBα (inhibited state), and P-STAT3 (activated). (Right) Immunostainings with Abs against P-NF-κB, Mac1 (macrophage-specific), and P-STAT3. Asterisks denote positive dermal nuclei; additional identification of inflammatory cells is in Fig. 9. (C) Real-time PCR of NF-κB target genes. mRNAs were from FACS-purified GFP-positive epidermal cells and cultured keratinocytes (low calcium). Fold change was relative to WT-GFPepid (=1). (D) Anti-NF-κB and P-NF-κB immunofluorescence of KO and WT keratinocytes that are also GFPactin transgenic. Cells were cultured in low-calcium medium (LCa) and high-calcium medium (HCa). (E) NF-κB-luciferase and Cxcl1-luciferase reporter gene assays on cultured keratinocytes (LCa) ± TNF-α. Values shown are relative to Renilla luciferase levels. ∗, t test statistical P ≤ 0.005. (F) Nude mice harboring K14-GFP-expressing E18.5 WT and cKO skin grafts were treated ± Dex to suppress NF-κB activation. Grafts were processed at 30 days for hematoxylin and eosin (H&E) staining, and immunomicroscopy was performed with Ab against α-catenin (αcat). Pigmented melanocytes from grafted skin (C57/Bl6) are black.
As judged by anti-phospho-p65 immunoblot analyses, E18.5 cKO nuclei displayed activated NF-κB and phosphorylated (inactive) IκBα (Fig. 3B). Immunohistochemistry verified that NF-κB was activated and nuclear in cKO epidermis. A few dermal cells near the epidermal/dermal border were also positive (∗ in Fig. 3B), and further investigation verified that these were macrophages, granulocytes, mast cells, and lymphocytes (Fig. 3B and Fig. 9, which is published as supporting information on the PNAS web site). A few of these cells were even seen in the epidermis, explaining some discrepancies between the in vivo array and in vitro real-time PCR data.
The proinflammatory response transpired shortly after α-catenin ablation, i.e., when the immune repertoire is still developing. The few “escaper” mosaic-Cre cKO mice that reached adulthood displayed signs of chronic inflammation. Inflammation even arose in cKO (but not WT) epidermis grafted onto adult Nude mice, defective in thymic and hair development (Fig. 9 and data not shown).
We verified that the up-regulated NF-κB array genes were not simply caused by stray immune cells in cKO epidermis by conducting real-time PCR on our mRNAs isolated from GFP-purified E18.5 epidermal cells (Fig. 3C). We used in vitro studies to demonstrate that the NF-κB cascade was partially an intrinsic response of the α-catenin null state (Fig. 3C). Although cultured cKO cells displayed elevated nuclear phosphorylated NF-κB (Fig. 3D), the real-time PCR differences were always more pronounced in vivo, suggesting that immune cells may exacerbate the response. Support for an extrinsic effect came from NF-κB luciferase reporter gene assays, which were sensitive to TNF-α, expressed by activated immune cells (Fig. 3E). TNF-α sustains elevated kinase activity necessary for IκB phosphorylation (18). Notably, both epidermal and immune cells possess surface receptors for TNF-α and other NF-κB targets, e.g., interleukin receptors. cKO epidermal interleukin receptors appeared to be activated, as judged by phosphorylation of their downstream effector, STAT3 (19) (Fig. 3B).
Finally, we used dexamethasone (Dex) to evaluate the extent to which NF-κB activation in epidermal and inflammatory cells contributed to the progressive dysplasia in α-catenin cKO, K14-GFP-actin skin grafts. Although Dex is not exclusive to NF-κB repression, it did not have an obvious effect on WT grafts (data not shown). In contrast, within 30 days of Dex treatment, most GFP+ cells from four of five treated cKO grafts had degenerated and been replaced by α-catenin+, Nude epidermis (Fig. 3F). These studies suggest that α-catenin null epidermal cells may be more sensitive to apoptosis than their WT counterpart, but that this sensitivity is masked by their activated NF-κB.
Relevance to SCCs in Humans.
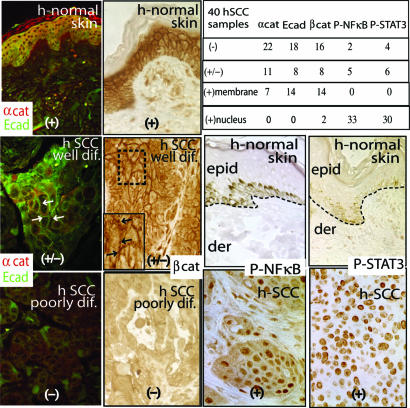
The progressive events leading to pervasive transformation of cKO skin are hallmarks of the natural transition from benign hyperplasia or SCC in situ, to a noninvasive SCC (grade III) and finally an invasive SCC with poor prognosis (grade IV) (20, 21). To explore the clinical significance of our findings, we analyzed 40 hSCCs for changes in α-catenin, NF-κB activation, and inflammation. Of these, 33 exhibited reduced (±) or undetectable (−) intercellular border staining for α-catenin, and most of these also showed reductions in E-cadherin and β-catenin (Fig. 4). Similar to our mouse skin grafts, even well differentiated hSCCs often displayed reduced and often no signs of α-catenin expression. Notably, only 2 of the 40 hSCCs displayed signs of nuclear β-catenin, so prevalent in human and mouse pilomatricomas (7, 8), but not detected in our mouse SCCs.
Fig. 4.
Status of α-catenin (αcat) expression, β-catenin (βcat) expression, E-cadherin (Ecad) expression, and NF-κB/STAT activity in hSCCs. Immunostainings of hSCCs ranged from well to poorly differentiated (dif). Normal human skin is from unaffected areas of hSCC sections. Arrows point to cell–cell border staining. Chart summarizes immunodata from 40 hSCCs (+, present; +/−, weak; −, not detected). Note that some background staining was detected in the anti-β-catenin immunohistochemistry, which was comparable to that seen in the poorly differentiated hSCC sample shown. epid, epidermis; der, dermis.
Increasing evidence suggests that secondary inflammatory responses could be responsible for invasion of cancer cells (14, 15, 22). Although NF-κB inhibition correlates with reduced tumorigenicity in most cell types, the epidermis has been an exception, where oncogenic Ras-initiated keratinocytes progress to a cancerous state if their NF-κB activity is blocked (23). It was thus important to evaluate the status of the p65 subunit of NF-κB in the 33 hSCCs with reduced α-catenin staining. As judged by immunohistochemistry, 33 displayed signs of nuclear, activated NF-κB (Fig. 4), which was accompanied by inflammation and nuclear phosphorylated STAT3, previously implicated in hyperproliferative skin disorders (24, 25). Together, these findings suggest that the specific gene defective in a tumor may also determine the impact of sustained NF-κB activity on cancer progression.
Discussion
Although loss of α-catenin at intercellular borders is a frequent occurrence in SCCs, it is extraordinary that this one defect accounts for so many diverse changes in tissue architecture and function. Initial clues that α-catenin’s effects extend beyond intercellular adhesion came when direct links were uncovered between the α-catenin null state and sustained activation of Ras-mitogen-activated protein kinase regulatory networks (6). Additionally, α-catenin is essential for directing the plane of asymmetric cell divisions in stratified squamous epithelial tissues (26), which may contribute to the multiple layers of dividing cells, loss of cell polarity, and differentiation defects when absent (6). Its ability to influence NF-κB activity and EMT genes now adds cell survival and proinflammatory and invasive responses to this repertoire. Although further investigations are necessary, we did not detect metastases in cKO mosaic or grafted mice.
The correlation between cancer and inflammation has been recognized for decades, but only in recent years has evidence begun to suggest that the inflammation is a prerequisite rather than a consequence of tumorigenesis (14, 15, 22). Our data now expose loss of intercellular adhesion and α-catenin as unanticipated instigators of a proinflammatory and cell survival response. Moreover, because α-catenin null epidermal cells not only intrinsically activate NF-κB, but also have the surface receptors to respond to its downstream targets, this activation not only protects the cells against apoptosis but also triggers a vicious proinflammatory signaling circuit through infiltrating immune cells. When accompanied by defects in intercellular adhesion and polarity, the α-catenin null epidermal barrier is also exposed to infection, further fueling this cascade. How loss of α-catenin leads to the initial difference in NF-κB activity is an intriguing question, beyond the scope of the present study.
In closing, it is notable that reduced α-catenin at intercellular borders and activation of NF-κB are facets of many cancers, raising the possibility that the mechanisms that we have uncovered may extend beyond skin SCCs. These features are also emblamatic of wound healing, where normal epidermal cells must transiently down-regulate cell–cell contact, increase migration, activate an inflammatory response, and remodel extracellular matrix (27, 28). It will be interesting in the future to evaluate whether the perturbations uncovered here might intersect with a pathway that is used during wound repair of normal epidermis. If a temporary modification, down-regulation, or sequestering of α-catenin is a natural response to injury, this could explain why chronic wound conditions ranging from ulcers to leprosy render patients susceptible to cancer (28).
Methods
Mice, Cultures, and Engraftments.
α-Catenin (fl/fl)/K14-Cre/K14-actinGFP cKO mice were as described (6, 9). Primary epidermal keratinocytes were isolated by floating E18.5 skins on dispase (Roche Diagnostics) 12 h, 4°C, followed by 0.1% trypsin (GIBCO) and filtration (40 μM, Falcon). Cells were grown in medium containing 0.05 or 1.5 mM calcium as described (9).
Engraftments onto the backs of Nude mice were performed by using E18.5 cKO back skin (29). Protective bandages were placed over grafts for a 10-day healing period. Where indicated, Dex (50 μg/kg in PBS) was injected i.p. for 20 days after recovery.
RNA Isolation and Analyses.
RNAs were purified by using the Absolutely RNA Microprep kit (Stratagene) and fluorometrically quantified (Ribogreen, Molecular Probes). Quality was assessed by RNA 6000 Pico Assay (Agilent Technologies, Palo Alto, CA), and 800 ng was primed with oligo(dT)-T7 primer and reverse-transcribed (Superscript III cDNA synthesis kit; Invitrogen). One round of amplification/labeling was performed to obtain biotinylated cRNA (MessageAmp aRNA kit, Ambion), and 10 μg of labeled cRNA was hybridized at 45°C for 16 h to mouse genome array MO-E430plus (Affymetrix, Santa Clara, CA). Processed chips were read by an argon-ion laser confocal scanner (Genomics Core Facility, Memorial Sloan-Kettering Institute, New York).
Scanned microarray images were imported into gene chip operating software (gcos, Affymetrix) to generate signal values and absent/present calls for each probe set by using the mas 5.0 statistical expression algorithm (chp files). Each array was scaled to a target signal of 500 by using all probe sets and default analysis parameters. For comparisons, raw data and .chp files were imported into genetraffic 3.8 (Iobion Informatics, La Jolla, CA), and replicate microarrays were grouped and compared by using the robust multichip analysis algorithm. Gene lists were compiled and annotated as described (29). Real-time PCRs were used to verify differential expression patterns (LightCycler system, Roche Diagnostics) as described (29).
Microscopy, Digital Photography, and Image Processing.
Light and fluorescent microscopy images were obtained by using a Zeiss Axioskop microscope driven by metamorph (Molecular Devices). OCT sections (10 μM) of frozen skins were fixed (10 min, 4% paraformaldehyde). hSCCs were from US Biomax, Rockville, MD. The MOM basic kit (Vector Laboratories) was used for mouse mAbs. Otherwise, sections were preblocked 1–4 h (2.5% normal goat serum, 2.5% normal donkey serum, 1% BSA, 0.1% Triton X-100), incubated with primary Ab (12 h, 4°C), treated with either fluorescence-conjugated secondary Abs (The Jackson Laboratory) (40 min, 25°C), or horseradish peroxidase-conjugated secondary Abs (Vector Nova-Red substrate) (Vector Laboratories). Abs were as in Vasioukhin et al. (6), except for AE13 (1:50, gift from T. T. Sun, New York School of Medicine, New York), AE15 (1:20, gift from T. T. Sun), Lef1 (1:300, lab generated), NF-κB (Santa Cruz Biotechnology), P-NF-κB (Cell Signaling Technology, Beverly, MA), and P-IκB (Imgenex, San Diego).
For transmission EM, samples were fixed and stained as described (6). Images were taken with a Tecnai G2-12 transmission electron microscope (FEI, Hillsboro, OR) equipped with an Advanced Microscopy Techniques (Danvers, MA) digital camera.
NF-κB and Cxcl1 Luciferase Reporter Assays.
A total of 250 × 103 cells per T24 well were plated and transfected the next day with 200 ng of NF-κB-luc (Stratagene) or Cxcl1-luc (gift from A. Richmond, Vanderbilt University School of Medicine, Nashville, TN) reporter constructs and 200 ng of Renilla luciferase plasmid (Promega). Twenty four hours later, cells were processed by using the Dual-Luciferase reporter assay system (Promega). Normalized firefly luciferase values are reported.
Supplementary Material
Acknowledgments
We thank H. A. Pasolli for conducting transmission electron microscopy; L. Polak and K. Kobielak for assistance with some of the engraftment experiments; A. North and the Bioimaging Facility, F. Quimby and the Animal Resource Facility, and S. Mazel and the FACS Facility at The Rockefeller University; and our colleagues cited in the text for providing antibodies and reagents. The microarray hybridizations were performed at the Genomics Facility at the Memorial Sloan-Kettering Institute. E.F. is an Investigator of the Howard Hughes Medical Institute. This work was supported in part by National Institutes of Health Grant NIH AR27883.
Glossary
Abbreviations:
- cKO
conditional knockout
- SCC
squamous cell carcinoma
- hSCC
human SCC
- En
embryonic day n
- Dex
dexamethasone.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Kobielak A., Fuchs E. Nat. Rev. Mol. Cell. Biol. 2004;5:614–625. doi: 10.1038/nrm1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nelson W. J., Nusse R. Science. 2004;303:1483–1487. doi: 10.1126/science.1094291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reya T., Clevers H. Nature. 2005;434:843–850. doi: 10.1038/nature03319. [DOI] [PubMed] [Google Scholar]
- 4.Gottardi C. J., Gumbiner B. M. J. Cell Biol. 2004;167:339–349. doi: 10.1083/jcb.200402153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kang Y., Massague J. Cell. 2004;118:277–279. doi: 10.1016/j.cell.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 6.Vasioukhin V., Bauer C., Degenstein L., Wise B., Fuchs E. Cell. 2001;104:605–617. doi: 10.1016/s0092-8674(01)00246-x. [DOI] [PubMed] [Google Scholar]
- 7.Chan E. F., Gat U., McNiff J. M., Fuchs E. Nat. Genet. 1999;21:410–413. doi: 10.1038/7747. [DOI] [PubMed] [Google Scholar]
- 8.Gat U., DasGupta R., Degenstein L., Fuchs E. Cell. 1998;95:605–614. doi: 10.1016/s0092-8674(00)81631-1. [DOI] [PubMed] [Google Scholar]
- 9.Vaezi A., Bauer C., Vasioukhin V., Fuchs E. Dev. Cell. 2002;3:367–381. doi: 10.1016/s1534-5807(02)00259-9. [DOI] [PubMed] [Google Scholar]
- 10.Neely E. K., Morhenn V. B., Hintz R. L., Wilson D. M., Rosenfeld R. G. J. Invest. Dermatol. 1991;96:104–110. doi: 10.1111/1523-1747.ep12515914. [DOI] [PubMed] [Google Scholar]
- 11.Thiery J. P. Nat. Rev. Cancer. 2002;2:442–454. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- 12.Jamora C., Lee P., Kocieniewski P., Azhar M., Hosokawa R., Chai Y., Fuchs E. PLoS Biol. 2005;3:e11. doi: 10.1371/journal.pbio.0030011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fundyler O., Khanna M., Smoller B. R. Mod. Pathol. 2004;17:496–502. doi: 10.1038/modpathol.3800066. [DOI] [PubMed] [Google Scholar]
- 14.Balkwill F., Coussens L. M. Nature. 2004;431:405–406. doi: 10.1038/431405a. [DOI] [PubMed] [Google Scholar]
- 15.Greten F. R., Eckmann L., Greten T. F., Park J. M., Li Z. W., Egan L. J., Kagnoff M. F., Karin M. Cell. 2004;118:285–296. doi: 10.1016/j.cell.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 16.Lawrence T., Bebien M., Liu G. Y., Nizet V., Karin M. Nature. 2005;434:1138–1143. doi: 10.1038/nature03491. [DOI] [PubMed] [Google Scholar]
- 17.Beg A. A., Ruben S. M., Scheinman R. I., Haskill S., Rosen C. A., Baldwin A. S., Jr. Genes Dev. 1992;6:1899–1913. doi: 10.1101/gad.6.10.1899. [DOI] [PubMed] [Google Scholar]
- 18.Nidai Ozes O., Mayo L. D., Gustin J. A., Pfeffer S. R., Pfeffer L. M., Donner D. B. Nature. 1999;401:82–85. doi: 10.1038/43466. [DOI] [PubMed] [Google Scholar]
- 19.Zhong Z., Wen Z., Darnell J. E., Jr. Science. 1994;264:95–98. doi: 10.1126/science.8140422. [DOI] [PubMed] [Google Scholar]
- 20.Kato Y., Hirano T., Yoshida K., Yashima K., Akimoto S., Tsuji K., Ohira T., Tsuboi M., Ikeda N., Ebihara Y., Kato H. Lung Cancer. 2005;48:323–330. doi: 10.1016/j.lungcan.2004.11.012. [DOI] [PubMed] [Google Scholar]
- 21.Lyakhovitsky A., Barzilai A., Fogel M., Trau H., Huszar M. Am. J. Dermatopathol. 2004;26:372–378. doi: 10.1097/00000372-200410000-00005. [DOI] [PubMed] [Google Scholar]
- 22.Mantovani A. Nature. 2005;435:752–753. doi: 10.1038/435752a. [DOI] [PubMed] [Google Scholar]
- 23.Dajee M., Lazarov M., Zhang J. Y., Cai T., Green C. L., Russell A. J., Marinkovich M. P., Tao S., Lin Q., Kubo Y., Khavari P. A. Nature. 2003;421:639–643. doi: 10.1038/nature01283. [DOI] [PubMed] [Google Scholar]
- 24.Chan K. S., Sano S., Kiguchi K., Anders J., Komazawa N., Takeda J., DiGiovanni J. J. Clin. Invest. 2004;114:720–728. doi: 10.1172/JCI21032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sano S., Chan K. S., Carbajal S., Clifford J., Peavey M., Kiguchi K., Itami S., Nickoloff B. J., DiGiovanni J. Nat. Med. 2005;11:43–49. doi: 10.1038/nm1162. [DOI] [PubMed] [Google Scholar]
- 26.Kim J. H., Kim B., Cai L., Choi H. J., Ohgi K. A., Tran C., Chen C., Chung C. H., Huber O., Rose D. W., et al. Nature. 2005;434:921–926. doi: 10.1038/nature03452. [DOI] [PubMed] [Google Scholar]
- 27.Redd M. J., Cooper L., Wood W., Stramer B., Martin P. Philos. Trans. R. Soc. London B. 2004;359:777–784. doi: 10.1098/rstb.2004.1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trent J. T., Kirsner R. S. Adv. Skin Wound Care. 2003;16:31–34. doi: 10.1097/00129334-200301000-00014. [DOI] [PubMed] [Google Scholar]
- 29.Rendl M., Lewis L., Fuchs E. PLoS Biol. 2005;3:e331. doi: 10.1371/journal.pbio.0030331. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.