Abstract
The sarA locus in Staphylococcus aureus controls the expression of many virulence genes. The sarA regulatory molecule, SarA, is a 14.7-kDa protein (124 residues) that binds to the promoter region of target genes. Here we report the 2.6 Å-resolution x-ray crystal structure of the dimeric winged helix SarA protein, which differs from the published SarA structure dramatically. In the crystal packing, multiple dimers of SarA form a scaffold, possibly via divalent cations. Mutations of individual residues within the DNA-binding helix–turn–helix and the winged region as well as within the metal-binding pocket implicate basic residues R84 and R90 within the winged region to be critical in DNA binding, whereas acidic residues D88 and E89 (wing), D8 and E11 (metal-binding pocket), and cysteine 9 are essential for SarA function. These data suggest that the winged region of the winged helix protein participates in DNA binding and activation, whereas the putative divalent cation binding pocket is only involved in gene function.
Staphylococcus aureus is a versatile bacterium capable of causing a wide spectrum of human infections, ranging from superficial abscesses to pneumonia, endocarditis, and sepsis (1, 2). This versatility may be attributable to the impressive array of extracellular and cell wall-associated virulence determinants coordinately expressed during the infectious process (3). Many of these virulence factors are either secreted or cell surface-associated proteins. Secreted proteins such as hemolysins, lipase, and proteolytic enzymes are responsible for invasion and tissue damage. Cell surface-associated proteins such as protein A and fibronectin-binding proteins mediate adhesion to host tissues (4). The coordinate expression of many virulence determinants in S. aureus has been shown to be regulated by global regulatory elements such as staphylococcal accessory regulator (sarA) and accessory global regulator (agr) (5, 6). These regulatory elements, in turn, control the transcription of a wide variety of unlinked genes, many of which have been implicated in pathogenesis. Transcription profiling study showed that an agr mutation could alter the expression of ≈138 genes, with 104 up-regulated and 34 down-regulated, whereas a sarA mutation leads to altered expression of ≈120 genes, with 76 and 44 genes up-regulated and down-regulated, respectively (7).
The sarA locus has been shown to promote the synthesis of selected extracellular and cell wall-associated proteins while repressing the transcription of the protein A and protease genes (8). The sarA locus is composed of three overlapping transcripts, sarA P1, sarA P3, and sarA P2, each encoding the major 372-bp sarA ORF that yields the 14.7-kDa SarA protein (5). DNase I footprinting studies revealed that the SarA protein binds to several target gene promoters (9), including those of agr, hla, spa, and fnbA, thus implicating SarA to be a DNA-binding protein that can modulate target gene transcription via both agr-dependent and agr-independent pathways (9–12).
SarA is the first member of the SarA protein family that has been found to play vital roles in the regulation of virulence genes. A search of the recently released S. aureus genomes revealed at least 10 additional SarA homologs, six of which have been partially characterized. These members include SarR, SarS, SarT, SarU, SarV, and MgrA (also called Rat) (12–16). Members of the SarA protein family also share sequence homology with the MarR protein or homologs in Gram-negative bacteria (13). Schumacher et al. (17) have reported the SarA structure as well as cocrystallization data with an 8-bp agr promoter sequence. Based on the reported structures, they proposed a “shortening” effect of DNA as a result of binding by the dimeric SarA protein, leading to gene activation (17). Independently, we reported the dimeric SarR structure to be a typical “winged-helix DNA-binding protein” in the absence DNA, with the helix–turn–helix (HTH) and the winged regions interacting with the major and minor grooves of target promoter DNA, respectively (18). We also proposed the two conserved acidic patches on the convex side of the SarR structure as possible functional motifs, whereas the concave surface, containing a tract of basic residues, is likely to be the DNA-binding side (18). Based on sequence alignment, we speculated SarR, SarA, SarS, and possibly other MarR homologs to have similar folds, which are strongly supported by the structure of SarS (13, 19); however, these structures diverge significantly from the reported SarA structure with a completely different topology. Additionally, despite low sequence homology, the reported MarR structure has similar topological folds as the SarR and SarS proteins, consistent with our structural and sequence alignment predictions (20). To resolve the discrepancy between the reported SarA and SarR structures and to understand the structure-function relationship of SarA, we independently determined the SarA structures in two crystal forms and found it to be also a winged helix protein. Mutagenesis analysis of single and selected double residues that are conserved within the SarA protein family indicated that the basic residues within the winged region are important for DNA binding, whereas the acidic residues within the wing and a putative divalent cation binding pocket are involved in SarA function. These findings confirm the structural prediction of the SarA crystal structure.
Results
Overall Structure of SarA.
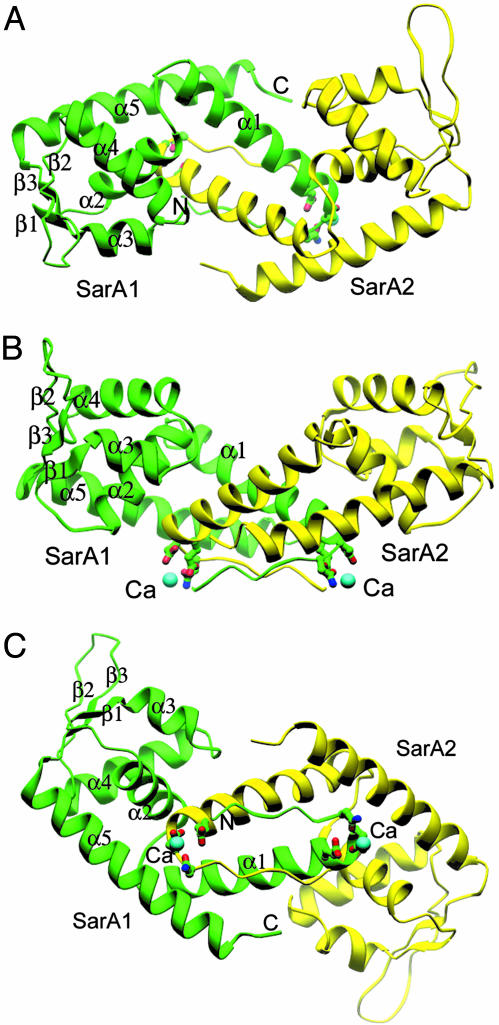
The structure of the SarA protein reported here has a similar topology to structures of SarR and SarS (Fig. 1; refs. 18 and 19). Briefly, there is one homodimer of SarA in one asymmetric unit, which is the assumed basic functional unit. The monomeric SarA structure consists mainly of five α-helices and three β-strands. α1-helices from both monomers are the major components that bring the two monomers together. The dimer association surface is highly hydrophobic and conserved among the family members. α3 and α4 build up the typical HTH motif, a widely existing protein scaffold for transcription factors accounting for DNA binding. Two of the three total antiparallel β-strands (β1, β2, and β3), β2 and β3, form the beta hairpin (the wing), which is predicted to interact with the minor groove of target DNA (18). The overall rms deviation between SarA and SarR is 2.5 Å. Individual comparison of the two winged helical motifs yields an even smaller difference (1.9 Å). The two α5-helices from each monomer are longer than those of the SarR in the C termini. Big differences are found at the two β-hairpin regions, which are the flexible pro-DNA binding regions of SarR (18) but are well defined in the SarA structure. Interestingly, the SarA structure reported here differs significantly from those of SarA reported by Schumacher et al. (ref. 17; with and without DNA bound, PDB ID codes 1FZN and 1FZP,, which are published as supporting information on the PNAS web site), except for three helical regions (α1, α4, and part of α5). Two helices (α2 and α3) are completely absent in the reported SarA structures and the β hairpin motif is missed in the complex structure of SarA and DNA (17). An additional extended helix exists in the C-terminal of the reported SarA structures (17).
Fig. 1.
Structures of the SarA dimer, side chains of residues that involve in Ca2+ binding are shown on the convex side of the SarA dimer. The assumed DNA-binding surface is opposite of Ca2+-binding sites. Two β-hairpins (β2 and β3 from monomer one, and those from monomer two) two HTH motifs (α3 and α4 from monomer one, and those from another monomer) build up the putative DNA-binding surface on the concave side of the SarA dimer. All structural model figures are made by program ribbons (21).
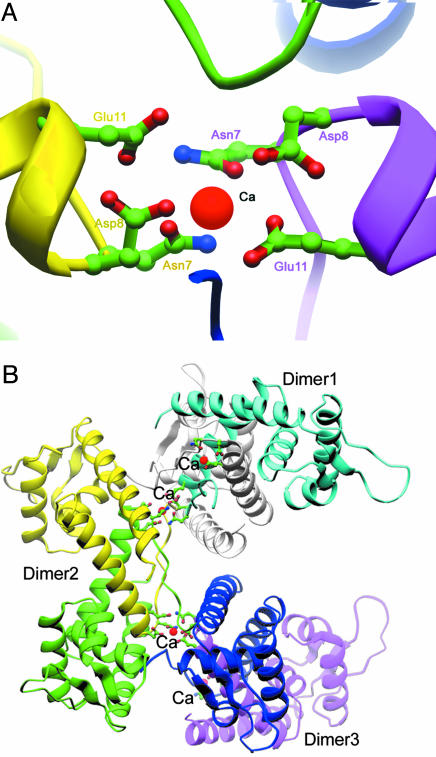
Two half-calcium ions were found at the N termini of the two SarA monomers, which are shown strongly on a 2Fc − Fo map with eight-sigma level (Fig. 8, which is published as supporting information on the PNAS web site). Three residues from one monomer of one dimer and three correspondent residues from another monomer of a neighboring dimer related by crystallographic symmetry are involved in the coordination for each calcium ion (Fig. 2A). Residue Glu-11 is located at the beginning of helix α 1. Residue Asp-8 is on the short coil that is antiparallel to the corresponding coil strand from the other monomer (Fig. 2A). The side chain of Asn-7 also takes part in the coordination of the calcium ion through the proper orientation of the carboxyl group. Six residues from two monomers build up a standard chelating site for one Ca2+ (Fig. 2A). Notably, each Ca2+ brings two SarA dimers together through a crystallographic symmetry relation (61) (Fig. 2B). Three or more SarA dimers stack up and generate a super helix with the putative DNA-binding surface located at the outer side and calcium binding sites inside (Fig. 2B). This type of oligomerization raises the question whether SarA functions as dimer alone or multiple dimers, with the aid of divalent cations. The prediction will be that mutation of Asp-8 and Glu-11 would render SarA nonfunctional in complementation (see below).
Fig. 2.
The positions of Ca2+ and their association with multiple dimers of the SarA. (A) Detailed side chains of residues that involve Ca2+ binding are shown from both dimer of SarA; calcium ions are colored purple. (B) The association of dimers of SarA through Ca2+; three SarA homodimers are involved through an x-ray axis (61).
Alignment of SarA Homologs and Mutagenesis Studies of Conserved Residues Within the Winged Helix Regions.
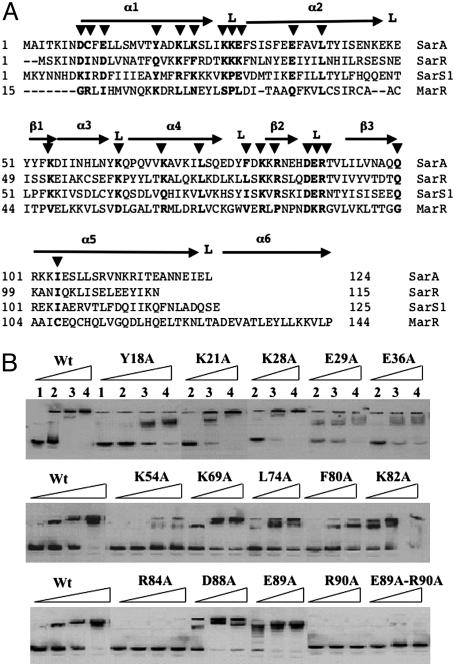
In superimposing the secondary structure of SarA with the alignment of several SarA homologs (SarA, SarR, SarT, MarR, and the N-terminal half of SarS), we have found the following: (i) conserved basic residues within the α3-α4 HTH; (ii) conserved basic and acid residues within β2-L-β3 winged region, with DER (residues 88–90) highly conserved among the SarA family members; (iii) semiconserved acidic residues (D8 and E11) at the divalent metal binding pocket; (iv) unique cysteine residue at position 9; and (v) conserved residues within the SarA homologs, including L40, L74, Q100, and I104 (Fig. 3A). To obtain mutated SarA protein with alanine substitutions at corresponding residues, the intact 375-bp gene was mutated, cloned in frame into the NdeI/BamHI sites of pET14b, expressed in BL21, and purified separately over a nickel resin affinity column. Most of the mutant proteins were successfully expressed and purified to homogeneity. A 160-bp promoter fragment of the protein A gene (spa), a known SarA binding target (11), was used in gel shift assays to determine the binding of the mutant SarA proteins. SarA proteins with K21A, K28A, K69A, K82A, D88A, and E89A mutations (Fig. 3B) and D8A, E11A, C9A, K23A, L40A, K27A, and K63A mutations (data not shown) were able to bind to the spa promoter in a manner similar to the wild-type protein. Mutations of C9, Y18, E29, E36A, K54, L74, and F80 to alanine reduced the binding of protein to the spa promoter (Fig. 3B). Remarkably, two mutant SarA proteins with R84A and R90A mutations failed completely to bind to the spa promoter, thus indicating the importance of these two residues within the winged region (Fig. 3A) in binding to the target promoter. Interestingly, among the highly conserved DER residues within the loop of the winged region (13), only R90 is essential to DNA binding. In addition, R84, residing within β2 strand that also constituted part of the wing, is also critical to binding. A double E89A-R90A mutation in SarA also failed to bind to the spa promoter, thus confirming that E89 likely did not contribute to binding.
Fig. 3.
Mutations and analyses of SarA residues conserved in the SarA protein family. (A) The primary sequence alignment of SarA, SarR, SarT, SarS1 (1–125 residues of SarS), and MarR superimposed with the secondary structures of SarR and SarA proteins of S. aureus (17). Numbers at the beginning of each line indicate amino acid positions relative to the start of each protein sequence. Helices are indicated by α, β-sheets are indicated by β, and loops and turns are marked by L. The residues (▾) altered by mutagenesis have been highlighted. (B) Autoradiogram of a nondenaturing 8% PAGE showing the binding of the wild-type and the mutant forms of SarA to a 160-bp spa promoter fragment. Lanes 1–4 correspond to 0 ng, 100 ng, 300 ng, and 500 ng of wild-type SarA or the mutant SarA as indicated on the lanes, with 0.1 ng of a γ-32P-labeled 160-bp DNA fragment used for each lane in the binding reaction.
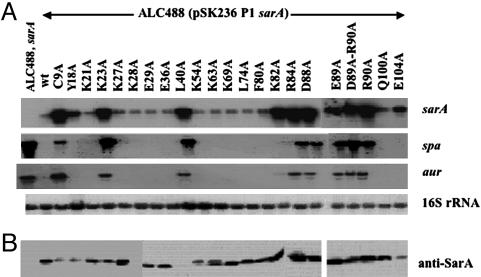
To ascertain the effect of these mutations on target genes, we chose to evaluate the transcription of sarA-regulated genes in sarA mutants containing shuttle plasmids (pSK236) with mutations at various codon positions. Because the sarA locus contains three transcripts, we elected to simplify our interpretation by using the strongest and the most proximal sarA P1 promoter to drive wild-type or mutant sarA gene. To ensure that the sarA mutant genes were expressed, we first examined the sarA P1 transcription in sarA mutant strains. As shown in Fig. 4A, all clones were able to express sarA P1 transcription. Immunoblot studies with anti-SarA monoclonal antibody 1D1 also confirmed that, with the exception of the L40A mutation, which likely rendered the SarA protein insoluble (unpublished observation), the sarA mutant clones, upon complementation, were able to express the mutant SarA proteins, whereas the sarA mutant negative control ALC488 did not (Fig. 4B). To evaluate sarA target genes, we chose to examine two genes, spa and aur (aureolysin gene), normally repressed by a functional SarA molecule. As expected, the transcription of spa and aur was elevated in the sarA mutant ALC488 but not in the mutant complemented with the wild type sarA gene. Interestingly, sarA mutant clones, expressing mutant SarA proteins (Y18, E29, E36A, K54, L74, and F80) that exhibited reduced binding to the spa promoter (Fig. 3), did not display any functional deficit with respect to spa and aur repression. For two constructs expressing mutant SarA proteins (R84A and R90A) that failed to bind to the spa promoter, these clones, expectedly, did not repress transcription of spa and aur. Of interest are the D88A and E89A mutations within the highly conserved DER motifs within the SarA and the MarR protein families (13); mutant clones expressing these proteins bound spa promoter normally (Fig. 3) but did not complement the sarA mutant phenotype with respect to spa and aur as compared with the nonmutated wild-type control. Besides examining D88A and E89A mutations, we also mutated the unique cysteine residue of SarA and the less-conserved lysine residue (K23) within the α1 helix to alanine. Although the C9A mutant SarA protein bound well to the spa promoter, the sarA fragment with this mutation did not restore repression of the spa and aur genes in the sarA mutant, thus indicating that the unique cysteine residue (9C), adjacent to the divalent cation binding pocket, plays a role in SarA function. Additionally, a K23A mutation that yielded normal binding protein also failed to complement the spa and aur genotype in a sarA mutant. Because the α1 helix is involved in dimerization of two SarA monomers, the possibility of a dimerization defect in the K23A mutant has not been defined.
Fig. 4.
Effects of sarA mutations at various codon positions in in vivo expression of target genes. (A) Northern blots of RNA isolated from isogenic sarA mutant ALC488, and ALC488 constructs containing shuttle plasmid with the wild-type and mutated sarA fragments at postexponential phases (OD650 ≈1.7 with 18-mm borosilicate glass tubes by using spectrophotometer Spectronic 20). A total of 10 μg of cellular RNA was loaded onto each lane. The blots were probed with 375-bp sarA (Top), 1,200-bp spa (Center Top), 1,500-bp aur (Center Bottom), and 724-bp of 16S rRNA (Bottom; nucleotides 777-1,500 of GenBank accession no. X68417) gene fragments. (B) Western blots of intracellular extract were probed with monoclonal anti-SarA antibody 1D1. Equivalent amounts of extracts (5 μg each) from the same growth conditions were used to detect SarA expression.
The Role of the Residues Within the Divalent Cation Binding Pocket in SarA Function.
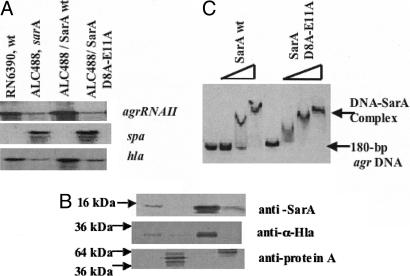
In the crystal structure of SarA, we have observed the involvement of divalent cations in linking SarA dimers. The three residues that are involved in Ca2+ binding (Asn-7, Asp-8, and Glu-11) were conserved among SarA, SarR, and part of SarS, with occasional conserved substitution of Glu-11 by Asp, but not in SarT and other members. This observation raises the question whether the two negatively charged residues that constitute the putative calcium-binding sites are required for SarA function. To address this question, two point mutations within SarA were generated by replacing residues Glu-11 and Asp-8 with alanines. The shuttle plasmid containing the wild-type or the mutated sarA gene was introduced into sarA mutant ALC488. Northern blots with a sarA probe revealed transcription of the native and mutated sarA genes (data not shown). Immunoblot analysis disclosed that both the intact and mutated SarA proteins were expressed in the sarA mutant construct (Fig. 5B). However, the amount of the mutated SarA protein was less than that of the nonmutated control but higher than the parental strain carrying a single copy of the sarA gene (Fig. 5B). DNA-binding assays with purified intact SarA and mutant SarA protein (D8A-E11A) and an 180-bp agr promoter fragment disclosed that the mutated SarA protein retained DNA-binding properties similar to that of the native protein (Fig. 5C). Transcriptional analysis disclosed that the mutated gene (D8A and E11A) did not complement the sarA genotype and phenotype in the sarA mutant with respect to restoring agr RNAII and α-hemolysin gene transcription and repressing spa expression, whereas the nonmutated sarA gene did (Fig. 5 A and B). These data confirmed that D8 and E11 within the putative divalent cation-binding pocket are essential to SarA function.
Fig. 5.
Effects of sarA mutations at codon positions 8 and 11 in in vivo and in vitro expression of target genes. (A) Northern blots of RNA isolated from S. aureus RN6390, isogenic sarA mutant ALC488, and ALC488 containing shuttle plasmid with the wild-type and mutated sarA fragments and probed with agr RNAII, spa or hla probe. (B) Western blots for intracellular, secreted spent supernatant and cell-wall associated protein extracts were probed with monoclonal anti-SarA antibody 1D1, polyclonal sheep anti-α-hemolysin antibody and affinity-purified chicken anti-protein A antibody, respectively. Equivalent amounts of extracts from the same growth conditions were used to detect SarA and protein A expression, whereas equal OD650 of spent cell culture supernatants were precipitated to detect α-hemolysin expression. Lanes are indentical to A. (C) Autoradiogram of an 8.0% nondenaturing gel showing gel-mobility shifts for increasing amounts (0, 50, 100, and 300 ng) of purified wild-type and mutated SarA proteins with a 180-bp agr P2-P3 promoter fragment (0.1 ng per reaction).
Discussion
The Discrepancy Between Two SarA Structures.
Several lines of evidence suggest that the SarA structure reported here represents the native form of SarA. First, sequence alignment of SarA, SarR, SarS, and other SarA family members indicated that all of these proteins likely belong to the same protein family. Second, structural conservation among SarA family is very consistent, based on the comparison of available structural information of several family members. Third, the HTH motif and β-hairpin (wing) showed that the SarA protein family belongs to the very well characterized superfamily of transcription factors called the “winged-helix” proteins. Based on potential interactions between the elongated β-hairpin and minor groove of their target DNAs, SarA and its homologs likely belong to a unique subfamily of winged-helix proteins (18–19). Finally, we discovered the same structure of SarA by using different crystallization condition (see below). Thus, it seems unlikely that dramatic conformational change occurs under unique crystallization conditions for the SarA protein as has been suggested by other authors (17).
The DNA-Binding Domains.
Members of the winged-helix protein family traditionally have very conserved DNA-binding motifs, including the HTH and the winged region constituted by two β-sheets and an intervening loop (13). Modeling studies of SarR, a SarA homolog, with promoter DNA also revealed that the HTH and potential β-hairpins (i.e., the winged region) on SarA are involved in DNA interactions (18). Notably, the previously reported structure of SarA with DNA by Schumacher et al. (17) showed an unusual mode of interaction between protein and DNA in which a bended 7-bp duplex DNA fragment with one nucleotide 5′ overhangs is held by residues that are not conserved among members of the SarA protein family. In contrast to the above studies, we did not detect any binding activity between the 7-bp duplex DNA (AGTTAAG) (17, 22, 23) and the SarA protein. To reexamine this issue, we mixed SarA protein with the 7-bp DNA fragment for crystallization trials by following the reported conditions in ref. 17. Crystals of the supposed complex were obtained and diffracted beyond 3.0 Å at an in-house x-ray resource with the same crystal forms as reported in ref. 17. The structure was solved by a partial model (we just used several helices that have correspondent ones in our SarA structures to calculate the difference fouries) from the reported complex structure in ref. 17. To our surprise, our SarA model has a good fit in the additional electron density. However, we did not observe any extra density of DNA fragment, with the assumed DNA-binding location actually occupied by the SarA protein itself. Accordingly, we have determined the same SarA structure with different crystallization conditions in the presence of a 7-bp DNA fragment, but the DNA fragment was absent in the crystal packing. Collectively, these data supported the notion that there is only one form of SarA structure, and that form is a winged-helix protein.
If our structure is correct, we speculate that SarA likely interacts with DNA in a manner similar to other winged-helix proteins such as SarR (18), with the HTH interacting with the major groove and the “wing” with the minor groove of DNA. Using this finding as a guide, we mutated conserved and, in particular, basic residues within the HTH and the winged region (see Fig. 3). Mutations of individual conserved residues (K54, K63, K69, and L74) within HTH yielded proteins that were expressed. The K54A and L74A mutant proteins bound less well to the spa promoter, a target for SarA binding, than the K63A and K69A proteins (11). These data suggested that more than one residue in the HTH makes contact with the major groove of DNA, and mutation of a single residue would only reduce, but not abolish, binding to the target gene promoter. Whether cooperative binding occurs among these residues is not clear. Of interest is the finding that two basic residues within the HTH (K63 and K69) conserved among many SarA family members did not play a role in DNA binding.
Besides the HTH motif, we also mutated conserved and basic residues within the winged region (Fig. 3). In examining these and other conserved basic residues, it is interesting that only basic residues R84 and R90 are critical to binding to the spa promoter, whereas others, including K82A, have no effect. Importantly, R84 and R90 are highly conserved within the putative winged region among many SarA homologs (Fig. 3) (13). Collectively, these data suggested that R84 and R90 are important residues that make contact with target DNA among the SarA protein family.
Structure and Function.
Many of the mutations that led to reduced binding, albeit moderately, to the spa promoter (Fig. 3) was capable of complementing the sarA mutant, thus demonstrating that a mild reduction in binding to the target promoter in vitro would not translate to functional deficiency in vivo. As expected, mutations that eventuated in defective binding proteins (R84A and R90A) also failed to complement the sarA mutant (Fig. 4), thus validating our gel-shift assay. Of interest, the acidic residues, D88 and E89, adjacent to the basic DNA-binding residue R90 and highly conserved in the “wing” region among members of the winged helix family (13), displayed normal binding to the spa promoter but were not functional with respect to complementation. These data thus demonstrated the DER residues within SarA and possibly the winged helix family are critical for both DNA binding and activation. Further high-resolution structures of the protein–DNA complex may plausibly explain how three contiguous residues in the winged region could have important roles in both DNA binding and function.
As with the SarR protein, SarA also has a concave side and a convex side. Previous studies on SarR have suggested the DNA-binding surface to reside on the concave side, whereas the putative activation motifs are located on the convex side (18). Based on the location of the acidic residues on SarR, we predicted D8 and E11 of SarA to represent an activation domain. Curiously, the crystal structure of SarA also predicted the role of E8 and E11 in connecting two dimers via a divalent cation bridge (Fig. 2). The notion of a SarA multimer in binding to DNA was also supported by results of gel-retardation assays, showing a large mobility shift of the protein–DNA complex (9, 11, 12, 22–24). It will be of interest to understand how these multiple homodimers bind to their target DNA and their association with each other. The current SarA structure and their association with divalent cations seem to reveal a potential association of multiple homodimers. However, the exact number of dimers is not clear. The data by Rechtin et al. (23) indicated that three homodimers of SarA might be a reasonable estimate. To address the importance of the divalent cation-binding sites on SarA, D8 and E11 were both mutated to alanine. Contrary to the wild-type gene, the D8A-E11A mutant did not complement the sarA mutant phenotype (Fig. 4), even though the mutant D8A-E11A protein bound comparably to the target agr promoter vs. the wild-type protein. Thus, D8 and E11 are not involved in DNA binding but are necessary for activation. Combining all these available data, one potential regulation scenario would be that, in the absence of divalent cations, multiple SarA dimers (with the major site occupied by the first SarA dimer, which could cooperatively bring more SarA into minor binding sites) bind to the promoter region of the target gene, but the repulsive force conferred by the negative charge of two acidic residues (Asp-8 and Glu-11) coerce the SarA dimers to work independently to bend and shorten the target DNA to position the RNA polymerase into a proper initiation state. However, the introduction of divalent cations at the proper site will facilitate the juxtaposition of two SarA homodimers. This cation-mediated interaction could encourage shortening of the promoter DNA and, hence, proper engagement/disengagement of RNA polymerase on the promoter. An additional but untested possibility is that SarA may stabilize some RNA transcripts and potentially promote specific gene expression.
Methods
Protein Expression, Purification, and Crystallization.
Protein expression and purification followed the procedure as described in ref. 11. Crystals for SarA protein (15 mg/ml) was crystallized by vapor diffusion against a solution of 200 mM calcium acetate/100 mM sodium cacodylate, pH 6.5)/20% PEG 8000 at room temperature. Heavy-atom derivatives were prepared by soaking crystals for 24 h in 1 mM HgCl2, which was dissolved in the crystallization solution. Crystals of SarA were transferred to crystallization buffer with 20% glycerol/25% PEG 8000 for a half hour before flash-freezing. The second crystal of the SarA protein in the presence of a short DNA fragment was obtained as reported in ref. 17.
Structure Determination and Refinement.
SarA crystals belong to space group P6122 with cell dimension of 65.34 Å × 65.34 Å × 237.29 Å and one dimer per asymmetric unit. The native data set was collected at CHESS. The HgCl2 derivative data were collected at Advanced Light Source (Table 1, which is published as supporting information on the PNAS web site). All data were processed with denzo and scalepack (25). Initial single isomorphous replacement (SIR) phases were derived from solve (26) and improved by solomon (27). Model building was carried out in o (28) with the help of SarR model (18). The electron density at “W1” regions was poor from the initial SIR and solvent flattening map, most of these regions showed up in the 2Fc − Fo and Fc − Fo omit maps, nevertheless, some side chains are partially ordered. The model was refined by cns (29) constrained with one noncrystallographic symmetry operator. Two half Ca2+ showed up in the Fc − Fo map with eight sigma peaks and were in the final refinement (Fig. 8). No water has been added at current resolution. The final model contains all residues of SarA. The coordinates have been submitted to the Brookhaven Protein Data Bank as entry 2FRH. The structure of the second crystal of the SarA protein was determined by the Difference Fouriers by using three α-helices from the published SarA structure (17) as initial model. A refined model SarA from this crystal form also has been deposited with PDB ID 2FNP.
Site-Directed Mutagenesis and Northern and Western Blots Analysis.
Site-directed mutagenesis was performed with the Muta-Gene in vitro mutagenesis kit (Bio-Rad) to introduce mutations into the sarA gene as described in the manufacturer’s insert. A 650-bp sarA P1 fragment containing the sarA gene was used as the template for mutation. Various primers were designed to introduce mutation at D8, C9, E11, Y18, K21, K23, K27, K28, E29, E36, L40, K54, K63, K69, L74, F80, K82, R84, D88, E89, R90, Q100, and I104 codon positions of SarA to yield various SarA mutant proteins with alanine at corresponding positions. The fragments containing mutations were gel-purified, ligated into shuttle plasmid pSK236 (ampr for E. coli and Cmr for S. aureus), and transformed E. coli XL-1 Blue strain. The authenticity of the recombinant constructs was confirmed by DNA sequencing. S. aureus RN4220 was used as the initial recipient for the correct recombinant constructs. Plasmid constructs from RN4220 were reintroduced into sarA mutant (ALC488) as described in ref. 30.
Northern blot analysis was performed as described in ref. 30 to assess the transcription of sarA and sarA-regulated target genes during exponential and postexponential phases in mutant strains containing the sarA gene with mutations at the various codon positions as described earlier. Western blots analysis of cell extract proteins from postexponentially grown cells was used to detect SarA expression. Five micrograms of total cell protein extracts from each sample were electrophoresed and blotted onto nitrocellulose membrane. The immunoblots were probed with monoclonal anti-SarA antibody 1D1 to detect the expression of SarA (30).
Supplementary Material
Acknowledgments
We thank John Kappler for discussions and advice; Jianye Zang for the refinement of the second crystal structure; Shaodong Dai for computing assistance; Howard Hughes Medical Institute, Zuckerman/Canyon Ranch, and Allen Laporte for support of our x-ray and computing facility; Jay Nix and Keith Henderson for assistance at 5.0.2 and 4.2.2 of Advanced Light Source; and MacCHESS for high-resolution data. G.Z. is a Pew Scholar in the Biomedical Sciences. The work is supported by National Institutes of Health Grant AI50678 (to A.L.C. and G.Z.).
Glossary
Abbreviations:
- agr
accessory global regulator
- HTH
helix–turn–helix
- sarA
staphylococcal accessory regulator.
Footnotes
Conflict of interest statement: No conflicts declared.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 2FRH and 2FNP).
References
- 1.Bayer M. G., Heinrichs J. H., Cheung A. L. J. Bacteriol. 1996;178:4563–4570. doi: 10.1128/jb.178.15.4563-4570.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boyce J. M. In: The Staphylococci in Human Disease. Crossley K. B., Archer G. L., editors. New York: Churchill Livingstone; 1997. pp. 309–329. [Google Scholar]
- 3.Projan S. J., Novick R. P. In: The Staphylococci in Human Disease. Crossley K. B., Archer G. L., editors. New York: Churchill Livingstone; 1997. pp. 55–82. [Google Scholar]
- 4.Lowy F. N. Eng. J. Med. 1998;339:520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 5.Cheung A. L., Koomey J. M., Butler C. A., Projan S. J., Fischetti V. A. Proc. Natl. Acad. Sci. USA. 1992;89:6462–6466. doi: 10.1073/pnas.89.14.6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kornblum J., Kreiswirth B., Projan S. J., Ross H., Novick R. P. In: Molecular Biology of the Staphylococci. Novick R. P., editor. New York: VCH; 1990. pp. 373–402. [Google Scholar]
- 7.Dunman P. M., Murphy E., Haney S., Palacios D., Tucker-Kellogg G., Wu S., Brown E. L., Zagursky R. J., Shlaes D., Projan S. J. J. Bacteriol. 2001;183:7341–7353. doi: 10.1128/JB.183.24.7341-7353.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheung A. L., Eberhardt K., Heinrichs J. H. Infect. Immun. 1997;65:2243–2249. doi: 10.1128/iai.65.6.2243-2249.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chien Y. T., Manna A. C., Cheung A. L. Mol. Microbiol. 1998;31:991–1001. doi: 10.1046/j.1365-2958.1998.01126.x. [DOI] [PubMed] [Google Scholar]
- 9.Chan P. F., Foster S. J. J. Bacteriol. 1998;180:6232–6241. doi: 10.1128/jb.180.23.6232-6241.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chien Y. T., Manna A. C., Projan S. J., Cheung A. L. J. Biol. Chem. 1999;274:37169–37176. doi: 10.1074/jbc.274.52.37169. [DOI] [PubMed] [Google Scholar]
- 12.Manna A. C., Ingavale S. S., Maloney M., van Wamel W., Cheung A. L. J. Bacteriol. 2004;186:5267–5280. doi: 10.1128/JB.186.16.5267-5280.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheung A. L., Bayer A. S, Zhang G., Gresham H., Xiong Y. Q. FEMS Immunol. Med. Microbiol. 2004;40:1–9. doi: 10.1016/S0928-8244(03)00309-2. [DOI] [PubMed] [Google Scholar]
- 14.Luong T. T., Newell S. W., Lee C. Y. J. Bacteriol. 2003;185:3703–3710. doi: 10.1128/JB.185.13.3703-3710.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ingavale S. S., Van Wamel W., Cheung A. L. Mol. Microbiol. 2003;48:1451–1466. doi: 10.1046/j.1365-2958.2003.03503.x. [DOI] [PubMed] [Google Scholar]
- 16.Schmidt K. A., Manna A. C., Gill S., Cheung A. L. Infect. Immun. 2001;69:4749–4758. doi: 10.1128/IAI.69.8.4749-4758.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schumacher M. A., Hurlburt B. K., Brennan R. G. Nature 409, 215–219, and erratum (2001) 2001;414:85. doi: 10.1038/35051623. [DOI] [PubMed] [Google Scholar]
- 18.Liu Y., Manna A., Li R., Martin W. E., Murphy R. C., Cheung A. L., Zhang G. Proc. Natl. Acad. Sci. USA. 2001;98:6877–6882. doi: 10.1073/pnas.121013398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li R., Manna A. C., Dai S., Cheung A. L., Zhang G. J. Bacteriol. 2003;185:4219–4225. doi: 10.1128/JB.185.14.4219-4225.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alekshun M. N., Levy S. B., Mealy T. R., Seaton B. A., Head J. F. Nat. Struct. Biol. 2001;8:710. doi: 10.1038/90429. [DOI] [PubMed] [Google Scholar]
- 21.Carson M. J. Mol. Graphics. 1987;5:103. [Google Scholar]
- 22.Morfeldt E., Tegmark K., Arvidson S. Mol. Microbiol. 1996;21:1227–1237. doi: 10.1046/j.1365-2958.1996.751447.x. [DOI] [PubMed] [Google Scholar]
- 23.Rechtin T. M., Gillaspy A. F., Schumacher M. A., Brennan R. G., Smeltzer M. S., Hurlburt B. K. Mol. Microbiol. 1999;33:307–316. doi: 10.1046/j.1365-2958.1999.01474.x. [DOI] [PubMed] [Google Scholar]
- 24.Gao J., Stewart G. C. J. Bacteriol. 2004;186:3738–3748. doi: 10.1128/JB.186.12.3738-3748.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Otwinowski Z., Minor W. Methods Enzymol. 1997;276:307. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 26.Terwilliger T. C., Berendzen J. Acta Crystallogr. D. 1999;55:849. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sheldrick G. M. In: Isomorphous Replacement and Anomalous Scattering, Proc. CCP4 St. W. Wolf W., Evans P. R., Leslie A. G. W., editors. Warrington, U.K.: Daresbury Laboratory; 1987. pp. 23–38. [Google Scholar]
- 28.Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard Acta Crystallogr. A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 29.Brunger A. T., Adams P. D., Clore G. M., Delano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J.-S., Kuszewsk J., Nilges N., Pannu N. S. Acta Crystallogr. D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 30.Manna A., Cheung A. L. Infect. Immun. 2001;69:885–896. doi: 10.1128/IAI.69.2.885-896.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.