Abstract
Although mantle cell lymphoma (MCL) frequently harbors inactivated ataxia telangiectasia mutated (ATM) and p53 alleles, little is known about the molecular phenotypes caused by these genetic changes. We identified point mutations and genomic deletions in these genes in a series of cyclin D1-positive MCL cases and correlated genotype with gene expression profiles and overall survival. Mutated and/or deleted ATM and p53 alleles were found in 56% (40/72) and 26% (21/82) of the cases examined, respectively. Although MCL patients with inactive p53 alleles showed a significant reduction in median overall survival, aberrant ATM status did not predict for survival. Nevertheless, specific gene expression signatures indicative of the mutation and genomic deletion status of each gene were identified that were different from wild-type cases. These signatures were comprised of a select group of genes related to apoptosis, stress responses, and cell cycle regulation that are relevant to ATM or p53 function. Importantly, we found the molecular signatures are different between cases with mutations and deletions, because the latter are characterized by loss of genes colocalized in the same chromosome region of ATM or p53. This information on molecular phenotypes may provide new areas of investigation for ATM function or may be exploited by designing specific therapies for MCL cases with p53 aberrations.
Keywords: cancer, cell cycle, genetics, microarray, signature
Mantle cell lymphoma (MCL) is an aggressive tumor that accounts for ≈6% of all non-Hodgkin lymphoma cases in the U.S., with higher rates in North America (1, 2). Although the median survival of MCL patients is only 3 years, some individuals survive >10 years from the time of diagnosis (2, 3). There is considerable interest in defining the molecular basis for this clinical heterogeneity to develop better prognostic markers and more effective therapies.
MCL corresponds to B cells of the mantle zone of the lymphoid follicles that have acquired distinctive alterations in genes related to cell cycle control and apoptosis (4). The hallmark of these genetic alterations is the t(11;14)(q13;q32) translocation that juxtaposes the IGH locus near the CCND1 gene, resulting in the overexpression of cyclin D1 (5). A subset of MCL cases acquire p53 mutations, and these patients have a significantly shortened median survival relative to cases with wild-type p53 (6–8). Interestingly, the ataxia telangiectasia mutated (ATM) gene, whose product regulates some p53-dependent apoptosis pathways, is mutated or deleted in 25–40% of MCL cases (reviewed in refs. 9 and 10). Although preliminary studies suggest that ATM mutation status does not have a significant impact on patient survival (7, 11), they may have lacked the statistical power to identify more subtle effects on survival, such as the effect of functional subsets of mutations.
We determined the ATM and p53 genotypes in a large cohort of MCL cases with previous gene expression profiles to further elucidate the relationship between molecular phenotypes and clinical outcomes. Whereas p53 mutation status correlated with overall survival (OS), ATM mutation status did not. Nonetheless, there were gene expression signatures indicative of ATM and p53 mutation status. These results provide insights into the altered molecular processes in MCL harboring mutations in these genes and how they relate to clinical outcomes.
Results
ATM Aberrations.
Sequence variants were found throughout the coding region of the ATM gene (, which are published as supporting information on the PNAS web site). We placed these variants into three categories: deleterious mutations, unclassified missense changes, or neutral variants (Tables 3–5, which are published as supporting information on the PNAS web site). We define deleterious mutations as sequence changes that produce a truncated ATM protein or alter amino acids critical for ATM function. Unclassified missense changes, which are not present in public databases [i.e., database single nucleotide polymorphism (dbSNP)], refer to amino acid substitutions with an unknown effect on ATM activity. Neutral variants refer to previously reported sequence variants (i.e., SNPs in dbSNP, or to silent substitutions, regardless of allele frequency in the general population).
Deleterious point mutations in the ATM gene were found in 33.3% (24/72) of MCL cases. These included eight nonsense mutations and 13 insertions or deletions. There were six samples with missense mutations in the PI-3 kinase domain as well as six samples with missense changes outside the PI-3 kinase domain that altered conserved amino acids in mouse and/or the African clawed frog. The presence of wild-type alleles, presumably from normal cells, was detected upon sequencing analysis of all cases with deleterious mutations and/or missense variants.
Genomic deletions of the region containing ATM locus (11q22.3) were previously found in 30 of 85 (35.3%) MCL cases (4). A detailed mapping analysis of 11q deletions in MCL demonstrated they are typically >10 Mb in length (12). Of the 72 MCL cases analyzed for both ATM point mutations (i.e., any sequence change not present in dbSNP, excluding silent changes) and genomic deletions, 11 (15.3%) contained only genomic deletions, 16 (20.8%) contained only point mutations, and 13 (18.1%) contained a genomic deletion and a point mutation (see Table 6, which is published as supporting information on the PNAS web site). This last category represents samples that unambiguously have biallelic ATM mutations. The weak correlation between the occurrence of deleterious point mutations and deletions in the ATM gene (κ statistic = 0.2) indicated the need to use methods for detecting both mutations and deletions to identify all aberrations in the ATM gene.
p53 Aberrations.
A total of 82 cases were screened for hotspot point mutations, polymorphisms, and genomic deletions in the p53 gene (Tables 7–9, which are published as supporting information on the PNAS web site). We found that 16 of 82 (19.5%) MCL cases contained deleterious p53 mutations. The mutations included 10 missense mutations, three nonsense mutations, two deletions, and one altered splice site. This is similar to previous reports in MCL, in which missense mutations predominated (6, 8). Genomic deletions of the p53 locus were previously found in 9.8% (8/82) of MCL cases (4). Here, we find that 6.1% (5/82) had only genomic deletions, 15.9% (13/82) had only point mutations, and 3.7% (3/82) had a genomic deletion and a point mutation. The weak correlation between the occurrence of deleterious point mutations and deletions in the p53 gene (κ statistic = 0.14) demonstrates the need to use both methods to identify all aberrations in the p53 gene.
Correlation of ATM and p53 Aberrations.
Of the 70 cases analyzed for all aberrations (point mutations and/or genomic deletions) in both genes, 10 (14%) had only p53 aberrations, 33 (47%) had only ATM aberrations, 7 (10%) exhibited aberrations in both genes, and 20 (29%) were wild type for both genes (Table 10, which is published as supporting information on the PNAS web site). ATM and p53 mutations were mutually exclusive in 43 of 50 (86%) cases.
OS.
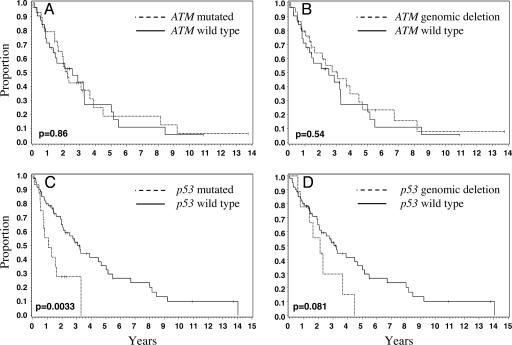
The differences in the median OS of patients with ATM point mutations (2.2 years) or patients with ATM genomic deletions (3.1 years) from those with only wild-type ATM alleles (2.5 years) were not significant (P = 0.86 and 0.54, respectively) (Fig. 1 A and B). The inability of ATM mutation status to predict for survival correlates with the mean proliferation signature value (4) of the wild-type versus the ATM mutated cases (i.e., 0.02 versus 0.06; see Table 6 and Fig. 4, which is published as supporting information on the PNAS web site). The hypothesis was considered that both alleles of the ATM gene may need to be inactivated to affect OS. However, there was no difference in the OS of the 13 cases (3.1 years), where both ATM alleles are indisputably inactivated (i.e., via both a point mutation and a genomic deletion) compared with the wild-type group (P = 0.73) (data not shown).
Fig. 1.
Overall survival by ATM and p53 mutational status. OS was estimated by the Kaplan–Meier method, and the log-rank test was used to estimate the difference in survival between groups. Here, mutated alleles refer to sequence changes not present in dbSNP, excluding silent changes. Solid black lines indicate cases with wild-type gene of interest, whereas dashed lines indicate cases with aberrations in the gene of interest. The median OS for each genotype is given in parentheses. (A) Mutated ATM (2.2 years) versus wild-type ATM (2.5 years); (B) deleted ATM (3.1 years) versus wild-type ATM; (C) mutated p53 (1.1 years) versus wild-type p53 (3.1 years); and (D) deleted p53 (2.1 years) versus wild-type p53.
In contrast to the results for ATM status, there was a significant difference (P = 0.0033) in the median OS of patients with p53 point mutations (1.1 years) compared with those with only wild-type p53 alleles (3.1 years) (Fig. 1C). There was also a trend for worse OS in patients with p53 genomic deletions (2.1 years) compared with those with only wild-type p53 alleles (3.1 years; P = 0.081) (Fig. 1D). The mean proliferation average value (see Table 9 and Fig. 5, which is published as supporting information on the PNAS web site) for tumors with mutant p53 was −0.1 versus 0.3 for the wild-type cases, which places these groups in two different survival quartiles described by Rosenwald et al. (4). The median survival for the seven patients with MCL that contained both ATM and p53 aberrations was 1.7 years, which shows no additive effect on survival.
Gene Expression Profiles in Tumors with ATM Aberrations.
ATM was among the 10 genes that were differentially expressed between cases with ATM point mutations relative to those containing two wild-type alleles (excluding cases containing genomic deletions) (Table 1 and Fig. 6, which is published as supporting information on the PNAS web site). Thirty-one genes were differentially expressed in the tumors with ATM genomic deletions relative to those containing wild-type alleles (excludes cases with ATM point mutations) (Table 1). They were nonrandomly distributed across the genome, with chromosome 11 showing a significant enrichment (see Fig. 7, which is published as supporting information on the PNAS web site). This was due to three genes (ATM, CASP4, and NPAT) in the 11q22–q23 genomic region that were underexpressed in tumors with ATM genomic deletions. Interestingly, ATM was the only gene in this region that was differentially expressed in tumors with ATM point mutations.
Table 1.
Gene expression changes in MCL with ATM aberrations
| Gene symbol | Gene description | Location | Fold change | P value |
|---|---|---|---|---|
| ATM point mutations versus wild-type cases | ||||
| EEF1A1 | Eukaryotic translation elongation factor 1 α 1 | 6q14.1 | −1.38 | 1.9 × 10−5 |
| CYP4B1 | Cytochrome P450, subfamily IVB, polypeptide 1 | 1p34-p12 | −1.21 | 4.1 × 10−4 |
| C7orf35 | DC32 (unknown function) | 7q11.23 | −1.30 | 5.4 × 10−4 |
| MAP3K4 | Mitogen-activated protein kinase kinase kinase 4 | 6q26 | −1.24 | 7.3 × 10−4 |
| BNlP1 | BCL2/adenovirus E1B 19-kDa interacting protein 1 | 5q33-q34 | −1.28 | 7.7 × 10−4 |
| ATM | Human ataxia-telangiectasia locus protein | 11q22-q23 | −1.44 | 8.7 × 10−4 |
| CCL20 | Chemokine (C-C motif) ligand 20 | 2q33-q37 | −1.52 | 8.7 × 10−4 |
| Hs.112482 | Similar to glutathione peroxidase | 11q14.3 | −1.36 | 8.9 × 10−4 |
| CDK10 | Cyclin-dependent kinase (CDC2-like) 10 | 16q24 | 1.27 | 6.8 × 10−4 |
| PARVG | Parvin, γ | 22q13.2-q13 | 1.49 | 7.7 × 10−4 |
| ATM genomic deletions versus wild-type cases | ||||
| ATM* | Human ataxia-telangiectasia locus protein | 11q22-q23 | −1.53 | 1.8 × 10−5 |
| CCL20 | Chemokine (C-C motif) ligand 20 | 2q33-q37 | −1.70† | 3.1 × 10−5 |
| NPAT* | Nuclear protein, ataxia-telangiectasia locus | 11q22-q23 | −1.49† | 3.6 × 10−5 |
| RGS16 | Regulator of G-protein signaling 16 | 1q25-q31 | −1.53 | 4.0 × 10−5 |
| EPHA2 | EPH receptor A2 | 1p36 | −1.19 | 1.5 × 10−4 |
| EEF1A1 | Eukaryotic translation elongation factor 1 alpha 1 | 6q14.1 | −1.33 | 3.0 × 10−4 |
| CXCL13 | Chemokine (C-X-C motif) ligand 13 | 4q21 | −1.26 | 4.5 × 10−4 |
| RPL5 | Ribosomal protein L5 | 1p22.1 | −1.35 | 5.4 × 10−4 |
| RPS20 | Ribosomal protein S20 | 8q12 | −1.25 | 6.1 × 10−4 |
| BNIP1 | BCL2/adenovirus E1B 19-kDa interacting protein 1 | 5q33-q34 | −1.25 | 6.7 × 10−4 |
| XPO1 | Exportin 1 | 2p16 | −1.26 | 6.7 × 10−4 |
| NOMO2 | NODAL modulator 2 | 16p12.3 | −1.22 | 6.8 × 10−4 |
| RPL11 | Ribosomal protein L11 | 1p36.1-p35 | −1.20 | 6.9 × 10−4 |
| PCDH1 | Protocadherin 1 (cadherin-like 1) | 5q32-q33 | −1.58 | 7.0 × 10−4 |
| CASP4* | Caspase 4, apoptosis-related cysteine protease | 11q22.2-q22.3 | −1.45† | 7.1 × 10−4 |
| CDKN3 | Cyclin-dependent kinase inhibitor 3 | 14q22 | −1.27 | 7.8 × 10−4 |
| RPS18 | Ribosomal protein S18 | 6p21.3 | −1.30† | 9.4 × 10−4 |
| LCK | Lymphocyte-specific protein tyrosine kinase | 1p34.3 | 1.64 | 2.4 × 10−5 |
| ZNF207 | Zinc finger protein 207 | 17q11.2 | 2.20 | 5.3 × 10−5 |
| ERCC2 | Excision repair, complementation group | 19q13.3 | 1.31 | 1.8 × 10−4 |
| ILT7 | Leukocyte immunoglobulin-like receptor | 19q13.4 | 1.93 | 1.9 × 10−4 |
| ALDH5A1 | Aldehyde dehydrogenase 5 family, member A1 | 6p22.2-p22.3 | 1.39 | 2.1 × 10−4 |
| STAT2 | Signal transducer and activator of transcription 2 | 12q13.3 | 1.31 | 2.3 × 10−4 |
| ITGB7 | Integrin, β 7 | 12q13.13 | 1.32 | 3.1 × 10−4 |
| LILRB1 | Leukocyte immunoglobulin-like receptor | 19q13.4 | 1.42 | 3.3 × 10−4 |
| LAIR1 | Leukocyte-associated Ig-like receptor 1 | 19q13.4 | 1.47 | 3.9 × 10−4 |
| SFRS7 | Splicing factor, arginine/serine-rich 7 | 2p22.1 | 1.48 | 4.1 × 10−4 |
| ST3GAL1 | ST3 β-galactoside α-2,3-sialytransferase 1 | 8q24.22 | 1.48 | 4.3 × 10−4 |
| CDK10 | Cyclin-dependent kinase (CDC2-like) 10 | 16q24 | 1.27 | 7.5 × 10−4 |
| LBH | Likely ortholog of mouse limb-bud and heart gene | 2p23.1 | 1.37 | 8.2 × 10−4 |
| CCM2 | Cerebral cavernous malformation 2 | 7p13 | 1.42 | 8.4 × 10−4 |
*Located in the 11q22-q23 ATM genomic region.
†Average of multiple values. The largest P value from random-variance t tests for multiple probe sets is provided.
Gene Expression Profiles in Tumors with p53 Aberrations.
Twenty genes were differentially expressed in the tumors containing p53 point mutations relative to those containing double wild-type alleles (excludes cases with genomic deletions) (Table 2 and Fig. 8, which is published as supporting information on the PNAS web site). Twenty-seven genes were differentially expressed in the tumors with p53 genomic deletions relative to those containing wild-type alleles (excludes cases with p53 point mutations) (Table 2). Interestingly, they were nonrandomly distributed across the genome, with chromosome 17 showing a significant enrichment seven genes in the p53 genomic region (SPAG7, ATP2A3, DVL2, DPH2L1, CENTB1, ITGAE, and MAP2K4) (see Fig. 9, which is published as supporting information on the PNAS web site). All seven were underexpressed in tumors with p53 genomic deletions but not in tumors with p53 point mutations. However, p53 was not differentially expressed in cases with either p53 point mutations or genomic deletions.
Table 2.
Gene expression changes in MCL with p53 aberrations
| Gene symbol | Gene description | Location | Fold change | P value |
|---|---|---|---|---|
| p53 point mutations versus wild-type cases | ||||
| XPC | Xeroderma pigmentosum, complementation group C | 3p25 | −1.40* | 3.3 × 10−5 |
| HRB2 | HIV-1 rcv-binding protein 2 | 12q21.1 | −1.45 | 7.3 × 10−5 |
| PVT1 | Pvt1 oncogene homolog, MYC activator (mouse) | 8q24 | −1.55 | 3.6 × 10−4 |
| GBE1 | Glucan (1,4-α-), branching enzyme 1 | 3p12.3 | −1.27 | 5.8 × 10−4 |
| STK38 | Serine/threonine kinase 38 | 6p21 | −1.47 | 7.6 × 10−4 |
| GLIPR1 | GLI pathogenesis-related 1 (glioma) | 12q21.1 | −1.41 | 8.2 × 10−4 |
| CLK2 | CDC-like kinase 2 | 1q21 | −1.24 | 8.7 × 10−4 |
| SPAP1 | SH2 domain containing phosphatase anchor protein 1 | 1q21 | −1.35 | 9.1 × 10−4 |
| PTPN22 | Protein tyrosine phosphatase, nonreceptor type 22 | 1p13.3-p13.1 | −1.47 | 9.5 × 10−4 |
| MEF2B | MADS box transcription enhancer factor 2 | 19p12 | −1.68 | 9.9 × 10−4 |
| BIRC5 | Baculoviral IAP repeat-containing 5 (survivin) | 17q25 | 1.68 | 3.2 × 10−4 |
| MTHFD2 | Methylenetetrahydrofolate dehydrogenase 2 | 2p13.1 | 1.39 | 4.6 × 10−4 |
| KIT | V-kit Hardy–Zuckerman 4 feline sarcoma oncogene | 4q11-q12 | 1.95 | 4.6 × 10−4 |
| CASP7 | Caspase 7, apoptosis-related cysteine protease | 10q25 | 1.42 | 4.7 × 10−4 |
| CEP2 | Centrosomal protein 2 | 20q11.22-q12 | 1.44 | 5.3 × 10−4 |
| HBZ | Hemoglobin, ζ | 16p13.3 | 1.93 | 5.7 × 10−4 |
| PLK1 | Polo-like kinase 1 (Drosophila) | 16p12.1 | 1.62 | 6.9 × 10−4 |
| DAD1 | Defender against cell death 1 | 14q11-q12 | 1.18 | 8.2 × 10−4 |
| JUP | Junction plakoglobin | 17q21 | 1.77 | 8.7 × 10−4 |
| ASPM | Asp (abnormal spindle)-like | 1q31 | 1.51 | 9.8 × 10−4 |
| p53 genomic deletions versus wild-type cases | ||||
| MAP2K4† | Mitogen-activated protein kinase kinase 4 | 17p11.2 | −1.70 | 2.0 × 10−7 |
| SPAG7† | Sperm-associated antigen 7 | 17p13.2 | −1.63 | 1.2 × 10−5 |
| ATP2A3† | ATPase, Ca++ transporting, ubiquitous | 17p13.3 | −1.61 | 1.2 × 10−4 |
| TAL2 | T-cell acute lymphocytic leukemia 2 | 9q32 | −1.47 | 1.4 × 10−4 |
| DVL2† | Dishevelled, dsh homolog 2 (Drosophila) | 17p13.2 | −1.60 | 1.8 × 10−4 |
| XPC | Xeroderma pigmentosum, complementation group C | 3p25 | −1.44 | 1.8 × 10−4 |
| NFATC2 | Nuclear factor of activated T cells | 20q13.2–3 | −1.41 | 2.6 × 10−4 |
| DPH2L1† | Candidate tumor suppressor in ovarian cancer 2 | 17p13.3 | −1.41 | 3.9 × 10−4 |
| CENTB1† | Centaurin, β 1 | 17p13.1 | −1.47 | 5.3 × 10−4 |
| ITGAE† | Integrin, α E | 17p13 | −1.39 | 6.4 × 10−4 |
| ZFP161 | Zinc finger protein 161 homolog (mouse) | 18pter-p11.2 | −1.38 | 7.1 × 10−4 |
| RGS13 | Regulator of G protein signaling 13 | 1q31.2 | −2.73 | 7.7 × 10−4 |
| TERT | Telomerase reverse transcriptase | 5p15.33 | −1.39 | 7.8 × 10−4 |
| SELL | Selectin L (lymphocyte adhesion molecule 1) | 1q23-q25 | −2.10 | 8.3 × 10−4 |
| POLD1 | Polymerase, delta 1, catalytic subunit 125 kDa | 19q13.3 | 1.52 | 6.5 × 10−5 |
| CDT1 | DNA replication factor | 16q24.3 | 1.62 | 1.2 × 10−4 |
| PPP5C | Protein phosphatase 5, catalytic subunit | 19q13.3 | 1.43 | 1.6 × 10−4 |
| PLK1 | Polo-like kinase 1 (Drosophila) | 16p12.1 | 2.00 | 7.5 × 10−4 |
| PTP4A3 | Protein tyrosine phosphatase type IVA, member 3 | 8q24.3 | 1.64 | 4.0 × 10−4 |
| BIRC5 | Baculoviral IAP repeat-containing 5 (survivin) | 17q25 | 1.90 | 5.1 × 10−4 |
| UQCRB | Ubiquinol-cytochrome c reductase-binding protein | 8q22 | 1.39 | 6.1 × 10−4 |
| CKS2 | CDC28 protein kinase regulatory subunit 2 | 9q22 | 1.61* | 8.0 × 10−4 |
| MYC | V-myc myelocytomatosis viral oncogene homolog | 8q24.12-.13 | 2.03 | 7.4 × 10−4 |
| PRKAR1B | Protein kinase, cAMP-dependent, reg., type I, β | 7pter-p22 | 1.32 | 8.3 × 10−4 |
| HRAS | V-Ha-ras Harvey rat sarcoma viral oncogene | 11p15.5 | 1.37 | 9.4 × 10−4 |
| AKAP1 | A kinase (PRKA) anchor protein 1 | 17q21-q23 | 1.45 | 9.6 × 10−4 |
| HN1 | Hematological and neurological expressed 1 | 17q25.1 | 1.62 | 9.9 × 10−4 |
*Average of multiple values. The largest P value from random-variance t tests for multiple probe sets is provided.
†Located in the 17p11–17p13 p53 genomic region.
Discussion
In agreement with previous studies (7, 9, 11, 13), deleterious or unclassified missense changes in the ATM gene were found in 40.3% of MCL cases. Furthermore, we identified deletions in the 11q22-q23 genomic region that covers the ATM gene in 24 of 72 (33.3%) cases (4). This is somewhat lower than previous studies showing genomic deletion of 11q22-q23 in 50–60% of MCL cases (11–16). This could be due to the sensitivity of our genomic qPCR approach (e.g., given a tumor cell content of 70% in a sample, >30% of the tumor cells in the sample must carry ATM genomic deletions in order for the event to be scored).
Because carriers of germ-line ATM mutations have not been reported to have a higher incidence of MCL (17), we sought to identify those MCL tumors in which it is certain that both copies of the ATM gene were inactivated. Surprisingly, this simultaneous occurrence was lower than expected. Assuming that these ATM aberrations are not primarily random events, incurring no selective advantage for cancer cell proliferation and/or survival, there are several possible explanations for the preponderance of single-copy ATM mutations. For example, DNA methylation could inactivate expression of the wild-type ATM allele, ATM may be haploinsufficient, or dominant-negative ATM mutations may occur. However, it is unlikely that epigenetic mechanisms silence ATM gene expression in all cases, because there are significant differences in ATM expression among the wild-type cases and those with ATM aberrations. Furthermore, ATM promoter hypermethylation was not detected in a study of lymph node biopsies from MCL patients (18). Last, our assays may have failed to detect sequence changes due to their nature (19) or location (e.g., introns or promoter region) in the ATM gene.
In addition, we uncovered point mutations and genomic deletions at the p53 locus at the expected frequencies. There was a weak correlation between p53 mutation and genomic deletion status. However, epigenetic phenomenon or false negatives in each analysis may account for a subset of cases in which aberrations affecting both alleles were not found.
Given the interrelationship of ATM and p53 gene function in regulating apoptosis and the cell cycle, we sought to determine whether specific tumors contained at least one inactivated allele (point mutation or genomic deletion) in both genes. In agreement with previous studies, we found no preferential association between p53 and ATM aberrations (7, 11). This indicates there is no strong selective advantage for the acquisition of both ATM and p53 aberrations in MCL.
Consistent with previous reports (7, 11), there was no significant difference in the median OS of patients with tumors having wild-type ATM and those with ATM point mutations (Fig. 1A) or ATM genomic deletions (Fig. 1B). Interestingly, patients having two mutated ATM alleles (one point mutation and one deletion) also showed no significant difference in overall survival relative to patients with only wild-type ATM alleles (data not shown). In contrast, patients with aberrant p53 alleles showed a worse OS relative to wild-type cases (Fig. 1 C and D), in keeping with our previous observations (6). Furthermore, p53 mutation status correlates with a higher-proliferation signature average value than wild-type cases, which was determined by expression profiling.
Because ATM mutations do not affect the overall survival of MCL patients, it is tempting to question their relevance in MCL development and progression. One could speculate that random point mutations accumulate in the ATM gene due to genomic instability. If this were the case, one would predict that missense changes would be randomly scattered throughout the coding region. However, 53.3% of missense changes were located in the last 10% of the ATM coding region that contains the highly conserved PI-3 kinase domain. However, there was no difference in OS for this subset of mutations versus mutations located in the rest of the ATM sequence (data not shown). This suggests that the point mutations, if somatic in origin, are selected for during lymphomogenesis and are not primarily the consequence of genomic instability in tumors.
Next, to identify the molecular phenotype of ATM mutations in MCL, we correlated mutation status with gene expression profiles previously obtained by cDNA microarray analysis (4). Unlike prior analyses, where profiles of mutant ATM or p53 were indistinguishable from profiles of wild-type chronic lymphocytic leukemia (20), we have shown that different profiles are present in mutant cases of MCL. This may be secondary to the larger numbers of cases analyzed and the larger number of genes with usable data in our study. MCL with ATM point mutations showed distinct gene expression profiles relative to cases with only wild-type ATM alleles. ATM was one of 10 genes differentially expressed between these two groups. Several of these genes have functions that are consistent with ATM biology. For example, both CDK10, a CDC2-related protein kinase involved in regulating the G2/M phase of the cell cycle, and the proapoptotic BNIP1 transcript are up-regulated in cases with ATM point mutations. Consistent with previous hypotheses of ATM function (21, 22), ATM-deficient MCL cases showed differential expression of genes related to oxygen stress response (e.g., a gene related to glutathione peroxidase, CYP4B1, and MAP3K4). Finally, the EEF1A1 gene is down-regulated in ATM mutant cases. This is consistent with the repression of a family member (EEF1D) in ATM-null fibroblasts (23).
Genomic deletions at the ATM locus yielded gene expression profiles that could be related to gene dosage. In addition to ATM, two other genes in the same 11q22–23 genomic region (NPAT and CASP4) were down-regulated in these cases. This finding is similar to observations made with U95 Affymetrix arrays in chronic lymphocyte leukemia cases with cytogenetic deletion of 11q22-q23, where seven genes colocalized with ATM were down-regulated (24). This confounds our analysis, because it is difficult to determine whether the other genes that are differentially expressed in the cases with ATM genomic deletions are due to the loss of ATM or its neighbors. Thus, point mutations in and genomic deletions encompassing tumor suppressor genes do not necessarily confer the same molecular phenotypes. Here, only ATM, EEF1A1, and CDK10 are differentially expressed in both categories of ATM genetic aberrations (point mutations and genomic deletions) relative to cases with only wild-type ATM alleles. These form a minimal core group of genes whose expression may be directly related to ATM functional integrity.
Next, we determined gene expression profiles related to p53 mutation status. Interestingly, only a few known p53 responsive genes (XPC, PLK1, BIRC5, and GLIPR1) were differentially expressed in cases with p53 point mutations relative to cases with two wild-type alleles. XPC, involved in nucleotide excision repair, is down-regulated in tumors with p53 mutations. Thus, this may contribute to the increase in secondary chromosomal abnormalities seen in lymphomas with p53 mutations. The overexpression of the PLK1 gene, an important regulator of cell-cycle-related events, is consistent with the decreased OS of p53-mutated MCL cases. Elevated PLK1 expression has been proposed as a prognostic marker and therapeutic target for large numbers of cancers. BIRC5 (also known as Survivin) is overexpressed in p53-mutated MCL and strongly associated with the proliferative activity of the tumors and the survival of the MCL patients (25). GLIPR1, a tumor suppressor gene that induces apoptosis in human prostate cancer cell lines, was down-regulated in p53-mutated cases.
p53 mutated MCL also showed the differential expression of multiple genes relevant to cancer biology. For example, KIT and JUP are overexpressed in numerous cancers and in p53-mutated MCL. ASPM, part of a proliferation gene signature related to OS in MCL patients (4), is up-regulated in cases with p53 point mutations. The induction of the proapoptotic CASP7 gene and repression of the DAD1 gene, a negative regulator of apoptosis, could help abrogate the prosurvival effect of p53 mutations in MCL.
Similar to ATM, genomic deletions of the p53 locus (17p13.1 genomic region) yielded gene expression profiles that could be affected by gene dosage. Strikingly, seven genes in or proximal to 17p11–13 were down-regulated, even though p53 was not differentially expressed. Similar to ATM, the molecular signature of MCL with a deleted p53 gene differs from that of MCL with p53 point mutations. Interestingly, expression analyses of p53-deleted chronic lymphocyte leukemia (CLL) cases also showed colocalized reductions of expression for 13 genes on the p53 chromosome segment by using U95 Affymetrix arrays (24). Here, only the XPC, BIRC5, and PLK1 genes are differentially expressed in cases with p53 genomic deletions and/or point mutations relative to those with only wild-type p53 alleles. These form a minimal core group of genes whose expression may be directly related to p53 functional integrity in MCL. XPC, an important p53-regulated gene that is induced by fludaribine therapy of CLL, may be a potential target that could be exploited in MCL (4).
The appearance of distinct clinical phenotypes in MCL cases with p53 mutations, but not in those with ATM mutations, is puzzling, because both genes are important regulators of cell cycle checkpoints and apoptosis. Nevertheless, the gene expression profiles of ATM-mutated MCL suggest they have altered biological properties relative to wild-type ATM MCL. Indeed, MCL with inactivated ATM alleles are associated with increasing numbers of chromosomal imbalances (11). ATM aberrations could also play roles in different stages of lymphomogenesis that are not directly related to later clinical manifestations.
Materials and Methods
Patient Materials.
Diagnostic biopsies from 92 patients with MCL from the Lymphoma Leukemia Molecular Profiling Project were studied after Institutional Review Board approval (4). Germline status for ATM and p53 is not known for these patients. Cases were chosen that were cyclin D1-positive and where tumor cells comprised the majority (>70%) of cells present on histology review. RNA and DNA were extracted simultaneously from the same piece of tissue. We made use of previously reported gene expression analyses of these tissues by using cDNA microarrays composed of 12,196 clones that interrogate genes that are expressed in lymphoid cells and/or are relevant to cancer or immune function (4).
Statistical Analysis.
κ statistics were used to evaluate the agreement between deletion and mutation status in ATM and p53.A κ statistic >0.75 indicates excellent agreement (26). Survival times were calculated as the time from diagnosis to death or date of last contact. OS distributions were estimated by using the Kaplan–Meier method, and the distributions were compared by using the log-rank test (27, 28).
Differentially expressed genes were identified by using brb arraytools (15). Random-variance t tests were used to find genes that differed significantly between groups (Tables 1 and 2). Genes were considered statistically significant if their P value was <0.001. To confirm these results, permutation P values for significant genes were computed based on 1,000 random permutations. Only the genes that passed this permutation analysis are listed in Tables 1 and 2 (with all permutation P values <0.004 and data not shown). The chromosomal location of the differentially expressed genes was noted, and a χ2 goodness of fit test was used to test whether the differentially expressed genes were randomly distributed across the chromosomes. Because the absolute values of the fold changes observed in our comparisons typically range from 1.2 to 1.5, caution should be taken when considering the biological significance of a differentially expressed gene.
ATM Mutations.
A total of 72 cyclin D1-positive MCL cases were analyzed for mutations on previously described oligonucleotide microarrays (Affymetrix) (7, 29). Six cyclin D1-positive MCL cases previously screened for ATM sequence variation by using this platform were included in this study for completeness (7). In principle, the two-color cohybridization experiments and loss of hybridization signal analysis used in this study (see Fig. 2) allowed us to screen for all possible sequence variations in the 62 coding ATM exons and their splice junctions (7, 29, 30).
p53 Mutations.
A total of 82 cyclin D1-positive MCL cases were screened for mutations in p53 exons 5–8 by denaturing high-performance liquid chromatography using previously described GC clamped primers (6). The optimal assay temperatures were calculated for each exon by usingwavemaker software (Transgenomic, Omaha, NE). PCR products with heteroduplex formation were reanalyzed to collect the heteroduplex and homozygous mutant peaks for confirmatory sequencing as described (6).
Genomic Deletions.
A total of 85 MCL DNAs were previously analyzed for genomic deletions spanning the ATM and p53 genes using a real-time quantitative PCR technique (4).
Sequencing.
Sample exons were amplified, and unincorporated primers and dNTPs were inactivated before sequencing (31) by using the Big Dye Terminator Kit (Applied Biosystems) and halfBD sequencing reagent (Genetix, Boston). Reactions were analyzed on a PRISM 3100 Genetic Analyzer (Applied Biosystems), and sequencher software (GeneCodes, Ann Arbor, MI) was used to screen for sequence variation. ATM amplicons were subcloned and sequenced to identify mutations.
Supplementary Material
Acknowledgments
We thank A. Levine and J. Reichardt at the University of Southern California (Los Angeles) for thoughtful discussion. This study was funded, in part, by the Margaret E. Early and L. K. Whittier Foundations (to J.G.H.); the V Foundation for Cancer Research (to J.G.H.); National Cancer Institute Grants CA84967 and CA36727 (to W.C.C., T.C.G., J.O.A., J.V., and D.D.W.); the Lymphoma Research Foundation Mantle Cell Grant program (to T.C.G.); and the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research (to L.M.S.).
Glossary
Abbreviations:
- MCL
mantle cell lymphoma
- ATM
ataxia telangiectasia mutated
- OS
overall survival.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Anderson J. R., Armitage J. O., Weisenburger D. D. Ann. Oncol. 1998;9:717–720. doi: 10.1023/a:1008265532487. [DOI] [PubMed] [Google Scholar]
- 2.Lenz G., Dreyling M., Hiddemann W. Ann. Hematol. 2004;83:71–77. doi: 10.1007/s00277-003-0774-2. [DOI] [PubMed] [Google Scholar]
- 3.Weisenburger D. D., Vose J. M., Greiner T. C., Lynch J. C., Chan W. C., Bierman P. J., Dave B. J., Sanger W. G., Armitage J. O. Am. J. Hematol. 2000;64:190–196. doi: 10.1002/1096-8652(200007)64:3<190::aid-ajh9>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 4.Rosenwald A., Wright G., Wiestner A., Chan W. C., Connors J. M., Campo E., Gascoyne R. D., Grogan T. M., Muller-Hermelink H. K., Smeland E. B., et al. Cancer Cell. 2003;3:185–197. doi: 10.1016/s1535-6108(03)00028-x. [DOI] [PubMed] [Google Scholar]
- 5.Swerdlow S. H., Williams M. E. Hum. Pathol. 2002;33:7–20. doi: 10.1053/hupa.2002.30221. [DOI] [PubMed] [Google Scholar]
- 6.Greiner T. C., Moynihan M. J., Chan W. C., Lytle D. M., Pedersen A., Anderson J. R., Weisenburger D. D. Blood. 1996;87:4302–4310. [PubMed] [Google Scholar]
- 7.Fang N. Y., Greiner T. C., Weisenburger D. D., Chan W. C., Vose J. M., Smith L. M., Armitage J. O., Mayer R. A., Pike B. L., Collins F. S., et al. Proc. Natl. Acad. Sci. USA. 2003;100:5372–5377. doi: 10.1073/pnas.0831102100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hernandez L., Fest T., Cazorla M., Teruya-Feldstein J., Bosch F., Peinado M. A., Piris M. A., Montserrat E., Cardesa A., Jaffe E. S., et al. Blood. 1996;87:3351–3359. [PubMed] [Google Scholar]
- 9.Boultwood J. J. Clin. Pathol. 2001;54:512–516. doi: 10.1136/jcp.54.7.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stankovi T., Stewart G. S., Byrd P., Fegan C., Moss P. A., Taylor A. M. Leuk. Lymphoma. 2002;43:1563–1571. doi: 10.1080/1042819021000002884. [DOI] [PubMed] [Google Scholar]
- 11.Camacho E., Hernandez L., Hernandez S., Tort F., Bellosillo B., Bea S., Bosch F., Montserrat E., Cardesa A., Fernandez P. L., et al. Blood. 2002;99:238–244. doi: 10.1182/blood.v99.1.238. [DOI] [PubMed] [Google Scholar]
- 12.Stilgenbauer S., Winkler D., Ott G., Schaffner C., Leupolt E., Bentz M., Moller P., Muller-Hermelink H. K., James M. R., Lichter P., et al. Blood. 1999;94:3262–3264. [PubMed] [Google Scholar]
- 13.Schaffner C., Idler I., Stilgenbauer S., Dohner H., Lichter P. Proc. Natl. Acad. Sci. USA. 2000;97:2773–2778. doi: 10.1073/pnas.050400997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Monni O., Knuutila S. Leuk. Lymphoma. 2001;40:259–266. doi: 10.3109/10428190109057924. [DOI] [PubMed] [Google Scholar]
- 15.Schraders M., Pfundt R., Straatman H. M., Janssen I. M., van Kessel A. G., Schoenmakers E. F., van Krieken J. H., Groenen P. J. Blood. 2005;105:1686–1693. doi: 10.1182/blood-2004-07-2730. [DOI] [PubMed] [Google Scholar]
- 16.Tagawa H., Karnan S., Suzuki R., Matsuo K., Zhang X., Ota A., Morishima Y., Nakamura S., Seto M. Oncogene. 2005;24:1348–1358. doi: 10.1038/sj.onc.1208300. [DOI] [PubMed] [Google Scholar]
- 17.Tort F., Camacho E., Bosch F., Harris N. L., Montserrat E., Campo E. Haematologica. 2004;89:314–319. [PubMed] [Google Scholar]
- 18.Chim C. S., Wong K. Y., Loong F., Srivastava G. Leukemia. 2005;19:880–882. doi: 10.1038/sj.leu.2403676. [DOI] [PubMed] [Google Scholar]
- 19.Hacia J. G., Edgemon K., Fang N., Mayer R. A., Sudano D., Hunt N., Collins F. S. Hum. Mutat. 2000;16:354–363. doi: 10.1002/1098-1004(200010)16:4<354::AID-HUMU8>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 20.Stankovic T., Hubank M., Cronin D., Stewart G. S., Fletcher D., Bignell C. R., Alvi A. J., Austen B., Weston V. J., Fegan C., et al. Blood. 2004;103:291–300. doi: 10.1182/blood-2003-04-1161. [DOI] [PubMed] [Google Scholar]
- 21.Weizman N., Shiloh Y., Barzilai A. J. Biol. Chem. 2003;278:6741–6747. doi: 10.1074/jbc.M211168200. [DOI] [PubMed] [Google Scholar]
- 22.Barzilai A., Rotman G., Shiloh Y. DNA Repair (Amsterdam) 2002;1:3–25. doi: 10.1016/s1568-7864(01)00007-6. [DOI] [PubMed] [Google Scholar]
- 23.Jang E. R., Lee J. H., Lim D. S., Lee J. S. J. Cancer Res. Clin. Oncol. 2004;130:225–234. doi: 10.1007/s00432-003-0522-y. [DOI] [PubMed] [Google Scholar]
- 24.Haslinger C., Schweifer N., Stilgenbauer S., Dohner H., Lichter P., Kraut N., Stratowa C., Abseher R. J. Clin. Oncol. 2004;22:3937–3949. doi: 10.1200/JCO.2004.12.133. [DOI] [PubMed] [Google Scholar]
- 25.Martinez A., Bellosillo B., Bosch F., Ferrer A., Marce S., Villamor N., Ott G., Montserrat E., Campo E., Colomer D. Am. J. Pathol. 2004;164:501–510. doi: 10.1016/S0002-9440(10)63140-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Le C. T. Applied Categorical Data Analysis. New York: Wiley; 1998. [Google Scholar]
- 27.Kaplan E. L., Meier P. J. Am. Stat. Assoc. 1958;53:457–481. [Google Scholar]
- 28.Peto R., Pike M. C., Armitage P., Breslow N. E., Cox D. R., Howard S. V., Mantel N., McPherson K., Peto J., Smith P. G. Br. J. Cancer. 1977;35:1–39. doi: 10.1038/bjc.1977.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karaman M. W., Groshen S., Lee C. C., Pike B. L., Hacia J. G. Nucleic Acids Res. 2005;33:e33. doi: 10.1093/nar/gni034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hacia J. G., Sun B., Hunt N., Edgemon K., Mosbrook D., Robbins C., Fodor S. P., Tagle D. A., Collins F. S. Genome Res. 1998;8:1245–1258. doi: 10.1101/gr.8.12.1245. [DOI] [PubMed] [Google Scholar]
- 31.Nickerson D. A., Tobe V. O., Taylor S. L. Nucleic Acids Res. 1997;25:2745–2751. doi: 10.1093/nar/25.14.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.