Abstract
The anaphase-promoting complex/cyclosome (APC/C) is a multisubunit ubiquitin-protein ligase that targets for degradation cell-cycle regulatory proteins during exit from mitosis and in the G1 phase of the cell cycle. The activity of APC/C in mitosis and in G1 requires interaction with the activator proteins Cdc20 and Cdh1, respectively. Substrates of APC/C–Cdc20 contain a recognition motif called the “destruction box” (D-box). The mode of the action of APC/C activators and their possible role in substrate binding remain poorly understood. Several investigators suggested that Cdc20 and Cdh1 mediate substrate recognition, whereas others proposed that substrates bind to APC/C or to APC/C–activator complexes. All these studies used binding assays, which do not necessarily indicate that substrate binding is functional and leads to product formation. In the present investigation we examined this problem by an “isotope-trapping” approach that directly demonstrates productive substrate binding. With this method we found that the simultaneous presence of both APC/C and Cdc20 is required for functional substrate binding. By contrast, with conventional binding assays we found that either Cdc20 or APC/C can bind substrate by itself, but only at low affinity and relaxed selectivity for D-box. Our results are consistent with models in which interaction of substrate with specific binding sites on both APC/C and Cdc20 is involved in selective and productive substrate binding.
Keywords: CDC20, ubiquitin, cell cycle
The anaphase-promoting complex/cyclosome (APC/C) is a large, multisubunit ubiquitin (Ub)-protein ligase that has important roles in the control of the eukaryotic cell division cycle. It targets for degradation essential cell-cycle regulatory proteins, such as mitotic cyclins and securin, an inhibitor of anaphase initiation. APC/C-mediated degradation of specific cell-cycle regulators is critical for proper exit from mitosis and for prevention of premature entry into the S-phase (reviewed in refs. 1–4). The activity of APC/C itself is tightly regulated in all stages of the cell cycle. It is inactive in the S-phase, G2 phase, and early mitosis, becomes active in late mitosis, and is converted back to an inactive form at the end of the G1 phase of the next cell cycle. This tight control is due to the action of a variety of positive and negative regulatory mechanisms. The activation of APC/C in late mitosis is initiated by the phosphorylation of several of its subunits by mitotic protein kinases (5, 6). These phosphorylation events allow the conversion of APC/C to an active form by binding to the WD40 repeat-containing activator protein Cdc20. After exit from mitosis, APC/C is dephosphorylated but is kept active in G1 by another related activator, Cdh1. In the G1-to-S-phase transition APC/C is inactivated by inhibitory phosphorylations of Cdh1 and by the rise in the level of the inhibitory protein Emi1 (reviewed in refs. 2–4).
Although the APC/C has been subject to intensive recent investigation, its mode of action and the mechanisms of its regulation remain poorly understood. It is not clear how Cdc20 and Cdh1 activate the APC/C. This is an important issue, because these activators are the target of several major regulatory systems that affect APC/C and cell-cycle progression. For example, Cdc20 is the target of the mitotic (or “spindle assembly”) checkpoint system, which is a surveillance mechanism that ensures that anaphase is initiated only after all chromosomes are correctly attached to the mitotic spindle (7, 8). Another important, partially related problem is how specific substrates of APC/C are recognized by this highly selective Ub ligase. All known substrates of APC/C–Cdc20 contain a 9-aa degenerate motif called the “destruction box” (D-box) (9), whereas substrates of APC/C–Cdh1 contain either D-box or KEN-box (10) recognition motifs. However, there are conflicting reports on the problem of how these motifs are recognized. Several investigators have reported that Cdc20 and Cdh1 bind substrates in the absence of APC/C 11–16 and suggested that these activators mediate substrate recognition for APC/C. However, the properties of substrate binding to Cdc20 or Cdh1 did not reflect the selectivity of their ubiquitylation in all cases (reviewed in ref. 17). Furthermore, the notion that activators are solely responsible for substrate binding was not compatible with the observation that yeast APC/C lacking Doc1 subunit binds activator but not substrate (18). In another study it was reported that the mitotic form of APC/C binds D-box-containing substrates in the absence of Cdc20 (17). In still another recent study it was reported that specific substrates bind only to a stoichiometric APC/C–Cdh1 complex (19). These authors suggested that both APC/C and its activator participate in substrate binding (19).
All of the above-described studies, yielding conflicting conclusions, used binding assays such as coimmunoprecipitation, pull-down, and native gel electrophoresis techniques. These assays did not necessarily indicate that this binding is functional, i.e., that it leads to product formation. In the present investigation we used a functional “isotope-trapping” method devised by Rose (20) to reexamine the roles of APC/C and Cdc20 in specific substrate binding. Furthermore, we compared the results with those obtained by conventional binding assays. We discuss models for specific and functional substrate binding that are compatible with all our findings, as well as with much of previously reported data of other investigators.
Results
Examination of the Roles of APC/C and Cdc20 in Substrate Binding by a Functional Isotope-Trapping Assay.
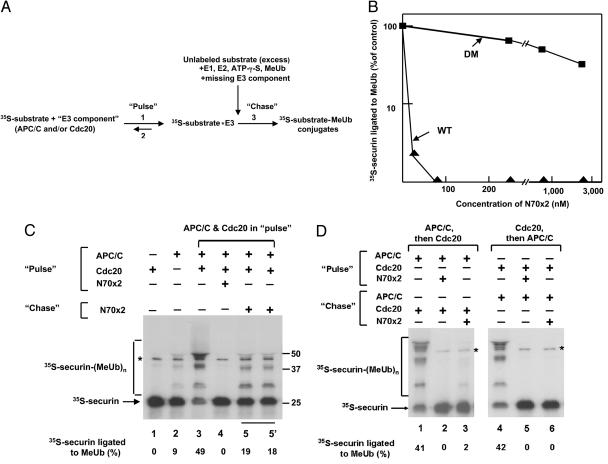
The Ub-protein ligase APC/C acts on cell-cycle regulatory proteins that contain D-box or KEN-box recognition motifs, but it is not clear how these motifs are recognized (see Introduction). Previous studies used binding assays, which do not necessarily indicate that binding is functional, i.e., that it leads to the conjugation of the protein substrate to Ub. We approached this problem by a functional isotope-trapping procedure. This method was devised by Rose (20), and we subsequently used it to show that E3α, the first Ub-protein ligase characterized, binds specific protein substrates (21). In this method, isotopically labeled substrate is first incubated with enzyme to form an enzyme–substrate complex (“pulse”). Subsequently, the sample is rapidly mixed with excess unlabeled substrate, added together with all further components required to complete the reaction (“chase”). In such assay, part of enzyme-bound labeled substrate is converted to labeled products, provided that product formation is faster than substrate dissociation. A scheme of the isotope-trapping procedure used in the present study is shown in Fig. 1A. In the pulse, 35S-labeled substrate was incubated with the tested substrate-binding “E3 component” (APC/C, Cdc20, or both). The chase mixture contained excess unlabeled substrate and all other components necessary for the formation of ubiquitylated products [E1, E2-C/UbcH10, adenosine-5′-O-(3′-thiotriphosphate (ATP-γ-S), and methylated Ub (MeUb)]. We used ATP-γ-S instead of ATP and MeUb instead of Ub for technical reasons (see Materials and Methods). After an additional, 1-min chase incubation, the reaction was quenched, and the conversion of 35S-labeled substrate to ubiquitylated derivatives was examined. We used 35S-securin as a high-affinity labeled substrate, and we used a bacterially expressed construct consisting of two copies of a 70-aa D-box-containing N-terminal fragment of Schizosaccharomyces pombe cyclin B (N70–2X; ref. 17) as the unlabeled substrate. As shown in Fig. 1B, sufficient excess of unlabeled substrate was used, because the prior addition of WT N70–2X completely prevented the formation of 35S-securin–MeUb conjugates at concentrations ≈30-fold lower than those used for isotope-trapping experiments. That this was indeed due to specific competition of unlabeled substrate on specific substrate-binding site(s) was indicated by the observation that a similar construct of N70–2X in which the RxxL sequences in the D-box motifs have been mutated to AxxA (DM-N70–2X; ref. 17) inhibited only slightly the formation of 35S-securin–MeUb conjugates even at high concentrations (Fig. 1B).
Fig. 1.
The simultaneous presence of mitotic APC/C and Cdc20 is required for productive substrate binding. (A) Outline of isotope-trapping (pulse–chase) procedure. Step 1, binding of labeled substrate to relevant E3 component(s); step 2, dissociation of 35S-substrate–enzyme complex; step 3, product formation from enzyme-bound labeled substrate upon the addition of a mixture of excess unlabeled substrate and all other components necessary for ubiquitylation. (B) Selective inhibition of ubiquitylation of 35S-securin by excess unlabeled N70–2X substrate. Experimental conditions were similar to those described for the pulse–chase assay, except that the indicated concentrations of WT or DM N70–2X proteins were added in the pulse (and not in the chase) incubation, together with APC/C and Cdc20. (C) Productive binding of labeled substrate for conjugate formation in the presence of both APC/C and Cdc20. Experimental conditions were as described in Materials and Methods, with both APC/C and Cdc20 added in the pulse phase. The isotope-trapping incubation (lanes 5 and 5′) was performed in duplicate to test that results were not affected by possible slight changes in mixing conditions upon the addition of the chase mixture. Lanes 1–4 show control incubations, in which the indicated components were added in the indicated phase of the pulse–chase incubation. The position of free 35S-securin and of 35S-securin–MeUb conjugates are indicated on the left. The asterisk indicates a contaminating protein band in the preparation of 35S-securin. Numbers on the right side indicate the positions of molecular mass marker proteins (kDa). The percentage of 35S-securin ligated to MeUb is indicated at the bottom of each lane. (D) The omission of either Cdc20 or APC/C from the pulse mixture abolished the binding of labeled substrate for product formation. Experimental conditions were as described in Materials and Methods, except that the indicated components were added at the indicated phases of the pulse–chase incubation.
We first examined whether the isotope-trapping technique can be used to examine substrate binding to APC/C–Cdc20. In the experiment shown in Fig. 1C, trapping of labeled substrate for product formation was tested with both APC/C and Cdc20 present in the pulse incubation. The experiment was accompanied by several controls. In Fig. 1C, lanes 1–3, we tested the activity of APC/C and Cdc20 preparations by an incubation pattern similar to that of the “pulse–chase” experiment (including rapid mixing and 1-min second incubation), except that unlabeled substrate was not added. A strong activity in the formation of 35S-securin–MeUb conjugates was observed when both APC/C and Cdc20 were present (Fig. 1C, lane 3), but none without APC/C (lane 1), and only slight activity was observed with APC/C in the absence of Cdc20 (lane 2). The slight activity seen in the absence of Cdc20 is due to the small amount of Cdc20 present in our purified preparations of mitotic APC/C (see Materials and Methods). Another control showed that the addition of unlabeled substrate in the pulse incubation completely prevented the formation of 35S-securin–MeUb conjugates (lane 4), confirming that the excess of unlabeled substrate was sufficient under the experimental conditions used. By contrast, when a similar concentration of unlabeled substrate was added in the chase phase of the isotope-trapping incubation, significant formation of labeled securin–MeUb conjugates could be seen (Fig. 1C, lanes 5). The amount of 35S-securin–MeUb conjugates formed in the single-turnover, isotope-trapping incubation was approximately one-third of that obtained in a 1-min control incubation carried out without unlabeled substrate (compare lane 5 with lane 3), indicating a high efficiency of productive substrate binding when both APC/C and Cdc20 are present in the pulse phase. It is also noteworthy that several higher-molecular-weight derivatives of 35S-securin were formed in the pulse–chase incubation, suggesting strong processivity in the ligation of several MeUb molecules to lysine residues of the substrate under these conditions. These data indicated that the isotope-trapping approach for the detection of productive substrate–enzyme binding is feasible for the case of APC/C–Cdc20.
We next used the isotope-trapping technique to examine the problem of whether the presence of APC/C, Cdc20, or both is necessary for productive substrate binding. In the experiment shown in Fig. 1D, lanes 1–3, 35S-securin was first incubated with APC/C alone, and Cdc20 was subsequently added in the chase mixture, together with all other components. A control incubation of similar design, but without unlabeled substrate, resulted in strong formation of 35S-securin–MeUb conjugates (Fig. 1D, lane 1), indicating that APC/C interacts rapidly with Cdc20 in the second 1-min incubation. Here again, the supplementation of unlabeled substrate in the pulse incubation completely prevented the formation of 35S-securin–MeUb conjugates (lane 2). In the corresponding pulse–chase incubation, no appreciable formation of 35S-securin–MeUb conjugates could be detected (Fig. 1D, lane 3). These results suggested that, without Cdc20, 35S-securin does not bind to APC/C in a manner that can lead to product formation.
In further experiments we examined whether the binding of substrate to Cdc20 before the addition of APC/C can lead to product formation (Fig. 1D, lanes 4–6). Here again, a control incubation demonstrated strong formation of ubiquitylated derivatives when 35S-securin was first incubated with Cdc20, and then APC/C was added for a 1-min second incubation without unlabeled substrate (Fig. 1D, lane 4). However, when unlabeled substrate was present in the chase incubation, there was no significant trapping of labeled substrate for the formation of ubiquitylated derivatives (Fig. 1D, lane 6). From these experiments we concluded that the presence of both APC/C and Cdc20 are necessary for high-affinity, productive binding of substrate.
Cdc20 Binds Substrate with Relaxed Selectivity for D-Box.
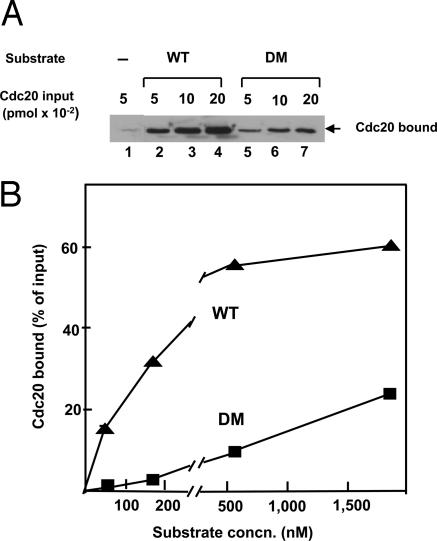
Our results showing that both APC/C and Cdc20 are required for functional substrate binding are at variance with conclusions of other investigators, which were based on binding experiments (11–17). We therefore reexamined this problem by conventional binding assays, with the preparations used in the present investigation. Because the N70–2X substrates are expressed as a GST fusion proteins (17), the binding of Cdc20 or APC/C to substrate could be conveniently estimated by GST pull-down, followed by immunoblotting with appropriate antibody. We first examined the binding of Cdc20 to substrate in the absence of APC/C. In the experiment shown in Fig. 2A, increasing amounts of recombinant purified Cdc20 were incubated with a constant, relatively low concentration (180 nM) of either WT or D-box mutant (DM) substrates. As expected from previous reports (11–16), considerable binding of Cdc20 to WT substrate was observed. However, we also observed less, but significant, binding of Cdc20 to the DM substrate. Quantitation of the immunoblot data showed that, under these experimental conditions, the binding of the DM substrate to Cdc20 was ≈15% that obtained with WT substrate. These findings suggest that Cdc20 binds substrate in the absence of APC/C, but at reduced selectivity for D-box.
Fig. 2.
Cdc20 binds substrate with relaxed selectivity for D-box.(A) The binding of Cdc20, added at the indicated amounts, to WT or DM N70–2X substrate (180 nM) was estimated as described in Materials and Methods. (B) Effect of substrate concentration on the binding of Cdc20. The binding of Cdc20 (0.1 pmol) to WT or DM substrates at the indicated concentrations was estimated by quantitative immunoblotting.
We further examined the affinity of the binding of Cdc20 to WT and DM substrate. The effect of substrate concentration on Cdc20 binding is shown in Fig. 2B. The binding of Cdc20 to WT substrate increased until a concentration of ≈500 nM, which is much higher than the concentrations required for inhibition of the ubiquitylation of 35S-securin by APC/C–Cdc20 (Fig. 1B), suggesting that substrate binds to Cdc20 at low affinity. As may be expected, the affinity of the binding of the DM substrate to Cdc20 is even lower, as indicated by the observation that Cdc20 binding continued to increase even at very high concentrations (1,800 nM) of the mutant substrate (Fig. 2B). We concluded that Cdc20 binds substrate in the absence APC/C, as reported by other investigators. However, this binding cannot account for the entire role of Cdc20 in APC/C-mediated ubiquitylation of specific substrates, because substrate binding to CDC20 is of less stringent specificity and of lower affinity than those of the ubiquitylation reaction.
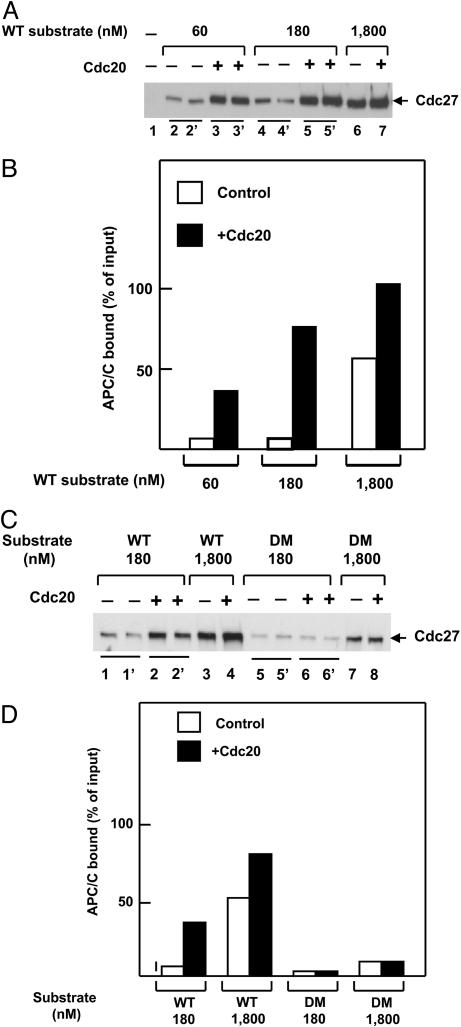
APC/C Binds Substrate at Low Affinity in the Absence of Cdc20 and at Higher Affinity in Its Presence.
We next examined whether substrate can bind to APC/C in the absence of Cdc20, as suggested by Yamano et al. (17). In the experiment shown in Fig. 3A, the binding of purified mitotic APC/C to N70–2X–GST substrate containing WT D-box sequences was estimated in the absence or presence of Cdc20, by GST pull-down followed by immunoblotting with an antibody directed against the Cdc27 subunit of APC/C. Quantitation of the results is shown in Fig. 3B. In the absence of Cdc20 and at low substrate concentrations (60–180 nM), ≈5–10% of APC/C supplemented was bound to substrate, whereas at high substrate concentrations (1,800 nM), >50% of input APC/C was bound to substrate. This finding indicates that APC/C can bind substrate directly in the absence of Cdc20 at high substrate concentrations. It should be noted that the experiments of Yamano et al. (17), which showed direct binding of APC/C to substrates, were carried out with very high concentrations of N70–2X substrate. The binding of APC/C to substrate at low concentrations was greatly increased by the addition of Cdc20 (Fig. 3 A and B). We concluded that APC/C can bind substrate in the absence of Cdc20, but only with low affinity. High-affinity binding of APC/C at low substrate concentrations requires the presence of Cdc20.
Fig. 3.
Binding of APC/C to substrate in the absence or presence of Cdc20. (A) Influence of substrate concentration. The binding of APC/C (0.015 pmol) to WT N70–2X substrate, added at the concentrations specified, was estimated as described in Materials and Methods. Where indicated, 0.15 pmol Cdc20 was added jointly with APC/C. The immunoblots in lanes 2–5 were done in duplicate for better accuracy of quantitation. (B) Quantitation of the experiment shown in A. There was no significant binding of APC/C to glutathione beads in the absence of substrate, so such correction was not necessary in this case. (C) Influence of Cdc20 (0.05 pmol) on the binding of APC/C (0.015 pmol) to WT or DM substrates. The immunoblots in lanes 1, 2, 5, and 6 were done in duplicate. (D) Quantitation of the experiment shown in C.
The selectivity for D-box of the binding of APC/C to substrate in the presence or absence of Cdc20 was examined in the experiment shown in Fig. 3 C and D. In the absence of Cdc20, we observed low but significant binding of APC/C to the DM substrate, especially at high substrate concentrations. This finding suggests that the low-affinity binding of APC/C to substrate in the absence of Cdc20 has somewhat relaxed D-box selectivity. As seen above, Cdc20 markedly stimulated the binding of APC/C to WT substrate at low concentrations. By contrast, no significant influence of Cdc20 on the binding of APC/C to the DM could be detected (Fig. 3 C and D). These data suggest that both the affinity and the selectivity of the binding of substrate to APC/C were increased by Cdc20.
Discussion
In this study, we used an isotope-trapping method to examine the problem of which components of the APC/C–Cdc20 Ub ligase complex are required for productive substrate binding. As opposed to conventional binding assays, which do not necessarily reflect productive binding of substrate to enzyme, the isotope-trapping technique directly demonstrates functional substrate binding. We observed significant trapping of 35S-securin for product formation when both APC/C and Cdc20 were present in the pulse phase (Fig. 1C), but not when one of these was omitted from the pulse and added in the chase (Fig. 1D). These findings show that both APC/C and Cdc20 are involved in productive substrate binding.
Although conventional binding assays do not indicate productive substrate binding, they may yield partial information on the mode of the action of the system. We therefore reexamined the properties of direct substrate binding to Cdc20 or to APC/C, with preparations similar to those used in the functional assays. We found that Cdc20 binds substrate, but at relaxed selectivity for D-box and at reduced affinity (Fig. 2). This result was not surprising, because some previously reported data showed that properties of substrate binding to activators are not exactly similar to those of APC/C-mediated ubiquitylation and degradation. Thus, it was reported that, although the degradation of yeast B-type cyclins in vivo required D-box motifs, their binding to Cdh1 was independent of D-box sequences (12). Similarly, the APC/C-mediated degradation of the yeast protein kinase Hsl1p required both D-box and KEN-box sequences, but only the KEN box was necessary for its binding to Cdh1 in vitro (14). Our further binding experiments showed that APC/C can also bind substrate in the absence of added Cdc20, but only at high substrate concentrations (Fig. 3). The binding of APC/C to substrate could not be due to residual endogenous Cdc20, because the extent of APC/C binding at high substrate concentrations (>50%) greatly exceeded that of Cdc20 in our preparations of APC/C (≈5–10%). Thus, we confirmed the findings of Yamano et al. (17) on the existence of a substrate-binding site on APC/C, but we note the low affinity of this binding. We also observed that the binding of substrate to APC/C in the absence of Cdc20 has somewhat relaxed selectivity for D-box (Fig. 3D). High-affinity binding of APC/C to substrate that is also more selective for D-box was observed in the presence of Cdc20 (Fig. 3).
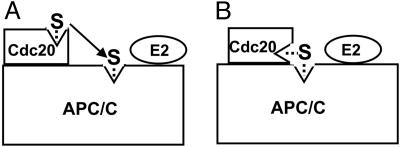
Any model for the roles of APC/C and Cdc20 in substrate binding should account for the observations that both APC/C and Cdc20 have substrate-binding sites, but productive and highly selective substrate binding requires the presence of both APC/C and Cdc20. One possibility, discussed by Yamano et al. (17), is that Cdc20 may have an “ushering” role, so that substrate first binds to Cdc20 and then is transferred to the substrate-binding site of APC/C (Fig. 4A). Such a sequential recognition mechanism would account for the higher selectivity of the APC/C–Cdc20 complex, because the substrate would be “tested” twice at the two binding sites. However, this mechanism does not account for increased affinity, so this model has to include a further assumption that the interaction of Cdc20 with APC/C induces a conformational change in one or both components that increases binding affinity. A putative Cdc20-induced conformational change in APC/C may also account for the observation that increasing concentrations of Cdc20 increase the Vmax of ubiquitylation, suggesting that Cdc20 stimulates not only substrate binding but also catalytic efficiency (Y.M. and A.H., unpublished observations). An alternative possible model is that, in the APC/C–Cdc20 complex, the substrate-binding site of Cdc20 is adjacent (or contiguous) to that of APC/C, so that an extended substrate-binding site is formed (Fig. 4B). This model would account for both high selectivity and increased affinity, assuming that different regions of the substrate interact with different parts of the composite binding site. In both models A and B, it appears reasonable to assume that the ultimate binding site on the APC/C–Cdc20 complex is in close proximity to E2 that is bound to APC/C, so that Ub transfer from E2 to the substrate can take place.
Fig. 4.
Possible models to account for the synergistic action of APC/C and Cdc20 in functional and specific binding of substrate. (A) Sequential transfer of substrate from Cdc20 to APC/C. (B) Composite APC/C–Cdc20 substrate binding site. See the text for details. S, substrate.
While this work was in progress, Passmore and Barford (19) reported that substrates bind preferentially to a stoichiometric APC/C–Cdh1 complex. The authors suggested that both APC/C and Cdh1 contribute to recognition sites with substrate. Our study, which used different and functional experimental approaches, yielded similar conclusions. However, the details of the molecular interactions involved remain unknown. Peptides or short fragments that contain D-box or KEN-box sequences have much lower affinity than longer protein substrates (15, 16, 22), implying that other, as-yet-unidentified regions in APC/C substrates are also important for substrate binding. It is also possible that regions in substrates that do not have canonical D-box sequences may interact with D-box receptors by virtue of a similar interaction surface. In the case of the model shown in Fig. 4B, such a possibility may account for the binding of APC/C substrates that have only a single canonical D-box motif. Much more remains to be learned about the interactions between APC/C–activator complexes and their substrates and about further possible mechanisms by which Cdc20 and Cdh1 activate the APC/C Ub protein ligase.
Materials and Methods
Reagents.
Ub and BSA (catalog no. A3294) were obtained from Sigma. ATP-γ-S and okadaic acid were purchased from Roche. E1 from human erythrocytes (23), recombinant E2-C/UbcH10 (24), MeUb (25), and Ub aldehyde (26) were prepared as described. pET-16pb–GST plasmids for the expression of a substrate for APC/C consisting of two tandem copies of the N-terminal 70 amino acid residues of fission yeast cyclin B fused to GST and its DM (N70–2X and DM-N70–2X, respectively; ref. 17) were kindly provided by H. Yamano (Marie Curie Research Institute, Surrey, U.K.). These proteins were expressed in bacteria and purified by chromatography on glutathione-Sepharose. Human securin cDNA was kindly provided by J. Pines (Cancer Research UK) and was subcloned into pcDNA3. This plasmid was used for the production of 35S-securin by in vitro transcription–translation with a TnT T7 Quick kit (Promega) and [35S]methionine (Amersham Pharmacia). Recombinant human his6–Cdc20 was expressed in baculovirus-infected 5B insect cells and was purified by chromatography on nickel-agarose. APC/C was purified from mitotically arrested HeLa cells by affinity chromatography on Suc1-Sepharose (27), followed by FPLC on MonoQ (6). This purified preparation still contained some Cdc20, detected either by residual Ub ligation activity in the absence of Cdc20 or by immunoblotting with anti-Cdc20 antibody. We found, however, that fractions of the APC/C peak eluted from MonoQ at higher salt concentrations contained less Cdc20 than did those eluted at lower salt concentrations. We therefore used for this study only the last two to three fractions of the peak of APC/C from MonoQ (approximately one-third of the total peak) eluted at the higher salt concentration. In this preparation, Ub ligation activity in the absence of Cdc20 was 5–10% of that obtained in the presence of Cdc20, and the molar amount of Cdc20 relative to APC/C (estimated by quantitative immunoblotting of Cd20 and the Cdc27 subunit of APC/C) was ≈10%.
Isotope-Trapping Pulse–Chase Incubations.
The first (pulse) incubation contained (in a volume of 7 μl) 30 mM Tris-HCl (pH 7.6), 5% (vol/vol) glycerol, 2 μg/μl BSA, 1 mM okadaic acid, 0.1 μl of 35S-securin, and 0.015 pmol purified mitotic APC/C or 0.06 pmol purified recombinant Cdc20, as indicated. After incubation at 23°C for 5 min, the sample was rapidly mixed with 5 μl of chase mixture that contained 30 mM Tris-HCl (pH 7.6), 2 μg/μl BSA, 2 μg/μl MeUb, 0.1 μg/μl Ub aldehyde, 10 mM MgCl2, 4 mM ATP-γ-S, 0.02 μg/μl E1, 0.2 μg/μl E2-C/UbcH10, and 2 μM WT unlabeled N70–2X. When APC/C or Cdc20 were not added to the pulse incubation, the missing component was included in the chase mixture. We used MeUb instead of native Ub for ease of detection of distinct low-molecular-weight derivatives (25). ATP-γ-S was used instead of ATP to prevent the possible action of the small amount of 26S proteasome in reticulocyte lysate added with 35S-securin. Rapid mixing was done by a Vortex mixer at intermediate speed for 1–2 seconds. After the second (chase) incubation for 1 min at room temperature, the reaction was quenched by the addition of SDS electrophoresis sample buffer. The samples were subjected to electrophoresis on a 12.5% polyacrylamide/SDS gel. Results were quantified by phosphorimager analysis.
Binding Assays.
To measure the binding of Cdc20 or of APC/C to N70–2X–GST substrates, glutathione-Sepharose beads (Amersham Pharmacia Biosciences) were first equilibrated with buffer A (50 mM Tris-HCl, pH 7.2/10% glycerol/1 mM DTT/2 mg/ml BSA), by rotation at room temperature for 1 h. This treatment was necessary to minimize nonspecific adsorption of Cdc20 to beads. Twenty-microliter samples of glutathione beads were incubated with the amounts of N70–2X proteins indicated in the figures in 30 μl of buffer A that contained 4 mg/ml BSA. After rotation at room temperature for 1 h, the beads were washed twice with 0.4 ml of buffer A and then resuspended in 30 μl of buffer A that contained 1 μM okadaic acid and Cdc20 or APC/C as indicated. After rotation for further 1 h at room temperature, the beads were washed three times with 1-ml portions of buffer B that contained 50 mM Tris-HCl (pH 8.0), 150 mM NaCl, 0.5% Nonidet P-40, 50 mM NaF, 60 mM β-glycerophosphate, 5 mM EDTA, and 0.1 mM sodium vanadate (17). Bound proteins were eluted from beads with 30 μl of electrophoresis sample buffer that also contained 5 mg/ml BSA. The samples were separated by electrophoresis on 8% polyacrylamide/SDS gels, transferred to nitrocellulose, and blotted with monoclonal antibodies directed against Cdc20 (Santa Cruz Biotechnology, sc-13162, 1:200 dilution) or the Cdc27 subunit of APC/C (BD Transduction, catalog no. 610455, 1:500 dilution). Proteins were visualized with SuperSignal chemiluminescence reagent (Pierce) and were quantified with ImageMaster VSD-CL (Rhenium, Jerusalem). Results were expressed as the percentage of Cdc20 or APC/C (Cdc27 subunit) bound to substrate, estimated by input samples included in the same immunoblot.
Note.
While this work was in progress, Passmore and Barford (19) reported that substrates bind preferentially to a stoichiometric APC/C–Cdh1 complex.
Acknowledgments
We thank J. Pines for plasmid containing human securin cDNA and H. Yamano for pET-16pb–GST expression vectors for N70–2X substrates. This work was supported by grants from the Israel Cancer Research Fund and the Gruss Lipper Foundation.
Glossary
Abbreviations:
- APC/C
anaphase-promoting complex/cyclosome
- Ub
ubiquitin
- MeUb
methylated Ub
- ATP-γ-S
adenosine-5′-O-(3′-thiotriphosphate
- D-box
destruction box
- DM
D-box mutant.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Zachariae W., Nasmyth K. Genes Dev. 1999;13:2039–2058. doi: 10.1101/gad.13.16.2039. [DOI] [PubMed] [Google Scholar]
- 2.Harper J. W., Burton J. L., Solomon M. J. Genes Dev. 2002;16:2179–2206. doi: 10.1101/gad.1013102. [DOI] [PubMed] [Google Scholar]
- 3.Peters J. M. Mol. Cell. 2002;9:931–943. doi: 10.1016/s1097-2765(02)00540-3. [DOI] [PubMed] [Google Scholar]
- 4.Castro A., Bernis C., Vigneron S., Labbe J. C., Lorca T. Oncogene. 2005;24:314–325. doi: 10.1038/sj.onc.1207973. [DOI] [PubMed] [Google Scholar]
- 5.Lahav-Baratz S., Sudakin V., Ruderman J. V., Hershko A. Proc. Natl. Acad. Sci. USA. 1995;92:9303–9307. doi: 10.1073/pnas.92.20.9303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Golan A., Yudkovsky Y., Hershko A. J. Biol. Chem. 2002;277:15552–15557. doi: 10.1074/jbc.M111476200. [DOI] [PubMed] [Google Scholar]
- 7.Shah J. V., Cleveland D. W. Cell. 2000;103:997–1000. doi: 10.1016/s0092-8674(00)00202-6. [DOI] [PubMed] [Google Scholar]
- 8.Bharadwaj R., Yu H. Oncogene. 2004;23:2016–2027. doi: 10.1038/sj.onc.1207374. [DOI] [PubMed] [Google Scholar]
- 9.Glotzer M., Murray A., Kirschner M. W. Nature. 1991;349:132–138. doi: 10.1038/349132a0. [DOI] [PubMed] [Google Scholar]
- 10.Pfleger C. M, Kirschner M. W. Genes Dev. 2000;14:655–665. [PMC free article] [PubMed] [Google Scholar]
- 11.Pfleger C. M., Lee E., Kirschner M. W. Genes Dev. 2001;15:2396–2407. doi: 10.1101/gad.918201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schwab M., Neutzner M., Mocker D., Seufert W. EMBO J. 2001;20:5165–5175. doi: 10.1093/emboj/20.18.5165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hilioti Z., Chung Y.-S., Mochizuki Y., Hardy C. F., Cohen-Fix O. Curr. Biol. 2001;11:1347–1352. doi: 10.1016/s0960-9822(01)00399-2. [DOI] [PubMed] [Google Scholar]
- 14.Burton J. L., Salomon M. J. Genes Dev. 2001;15:2381–2395. doi: 10.1101/gad.917901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burton J. L., Tsakraklides V., Salomon M. J. Mol. Cell. 2005;18:533–542. doi: 10.1016/j.molcel.2005.04.022. [DOI] [PubMed] [Google Scholar]
- 16.Kraft C., Vodermaier H. C., Maurer-Stroh S., Eisenhaber F., Peters J. M. Mol. Cell. 2005;18:543–553. doi: 10.1016/j.molcel.2005.04.023. [DOI] [PubMed] [Google Scholar]
- 17.Yamano H., Gannon J., Mahbubani H., Hunt T. Mol. Cell. 2004;13:137–147. doi: 10.1016/s1097-2765(03)00480-5. [DOI] [PubMed] [Google Scholar]
- 18.Passmore L. A., McCormack E. A., Au S. W., Paul A., Willison K. R., Harper J. W., Barford D. EMBO J. 2003;22:786–796. doi: 10.1093/emboj/cdg084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Passmore L. A., Barford D. EMBO Rep. 2005;6:873–878. doi: 10.1038/sj.embor.7400482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rose I. A. Methods Enzymol. 1980;64:47–59. doi: 10.1016/s0076-6879(80)64004-x. [DOI] [PubMed] [Google Scholar]
- 21.Hershko A., Heller H., Eytan E., Reiss Y. J. Biol. Chem. 1986;261:11992–11999. [PubMed] [Google Scholar]
- 22.Yamano H., Tsurumi C., Gannon J., Hunt T. EMBO J. 1998;17:5670–5678. doi: 10.1093/emboj/17.19.5670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hershko A., Heller H., Elias S., Ciechanover A. J. Biol. Chem. 1983;258:8206–8214. [PubMed] [Google Scholar]
- 24.Aristarkhov A., Eytan E., Moghe A., Admon A., Hershko A., Ruderman J. V. Proc. Natl. Acad. Sci. USA. 1996;93:4294–4299. doi: 10.1073/pnas.93.9.4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hershko A., Heller H. Biochem. Biophys. Res. Commun. 1985;128:1079–1086. doi: 10.1016/0006-291x(85)91050-2. [DOI] [PubMed] [Google Scholar]
- 26.Mayer A. N., Wilkinson K. D. Biochemistry. 1989;28:166–172. doi: 10.1021/bi00427a024. [DOI] [PubMed] [Google Scholar]
- 27.Hershko A. Methods Enzymol. 2005;398:170–175. doi: 10.1016/S0076-6879(05)98015-4. [DOI] [PubMed] [Google Scholar]