Abstract
Infections with hepatitis C virus (HCV) are marked by frequent viral persistence, chronic liver disease, and extraordinary viral genetic diversity. Although much has been learned about HCV since its discovery, progress has been slowed by a lack of permissive cell culture systems supporting its replication. Productive infections have been achieved recently with genotype 2a virus, but cirrhosis and liver cancer are typically associated with genotype 1 HCV, which is more prevalent and relatively resistant to IFN therapy. We describe production of infectious genotype 1a HCV in cells transfected with synthetic RNA derived from a prototype virus (H77-S). Viral proteins accumulated more slowly in H77-S transfected cells than in cells transfected with genotype 2a (JFH-1) RNA, but substantially more H77-S RNA was secreted into supernatant fluids. Most secreted RNA was noninfectious, banding in isopycnic gradients at a density of 1.04–1.07 gm∕cm3, but infectivity was associated with H77-S particles possessing a density of 1.13–1.14 gm∕cm3. The specific infectivity of H77-S particles (5.4 × 104 RNA copies per focus-forming unit) was significantly lower than JFH-1 virus (1.4 × 102 RNA copies per focus-forming unit). Infection with either virus was blocked by CD81 antibody. Sera from genotype 1a-infected individuals neutralized H77-S virus, but had little activity against genotype 2a virus, suggesting that these genotypes represent different serotypes. The ability of this genotype 1a virus to infect cultured cells will substantially benefit antiviral and vaccine discovery programs.
Keywords: bouyant density, CD81, cell culture, neutralizing antibody, serotype
Despite intensive research efforts, many gaps remain in our understanding of hepatitis C virus (HCV) and the mechanisms by which it causes chronic liver injury (1). To a large extent, this situation reflects the absence of tractable cell culture systems that are permissive for virus replication. Recent reports describing the efficient propagation of a genotype 2a strain of HCV, JFH-1, and a related, wholly genotype 2a chimera, FL-J6∕JFH, have thus stimulated much interest (2–4). The JFH-1 virus appears to be unique among strains of HCV in terms of its ability to undergo productive infection. Many aspects of the virus–host interaction, including viral entry, assembly, and release, that were previously inaccessible to experimental manipulation, can now be studied using the JFH-1 strain and its chimeric derivatives. However, the genotype 2a JFH-1 strain is not representative of the genotype 1 strains of HCV that are principally associated with liver disease in most regions of the world (5). There is thus an important need to develop systems supporting replication of other HCV genotypes in cell culture.
Like all positive-strand RNA viruses, HCV possesses an error-prone RNA replicase. Strains of HCV show extraordinary genetic diversity, both in terms of quasi-species variation within infected individuals, as well as genetic distances between viruses belonging to different genotypes (5, 6). Pairwise differences in the nucleotide sequences of the six HCV genotypes are on the order of 31–33%. This degree of variation approximates the genetic distance between members of the classical flavivirus serogroups, such as the four dengue viruses and members of the Japanese encephalitis serogroup, that represent serologically and genetically distinct viruses (7). The extent to which critical epitopes involved in antibody (Ab)-mediated neutralization varies among different HCV genotypes is not well understood. Neutralization studies using pseudotyped retrovirus particles suggest considerable relatedness among different HCV genotypes (8), but these conclusions need to be confirmed in neutralization studies using authentic HCV. There also may be important distinctions in the capacity of different genotypes of HCV to establish long-term persistence or to cause liver disease. Cirrhosis and liver cancer are typically associated with genotype 1 viruses, which are most prevalent (5). As important, there are marked differences in the therapeutic responses of different genotypes, with genotype 1 viruses being least likely (≈45%) and genotype 2 viruses most likely (≈85%) to respond to IFN-based therapy (9). Collectively, this marked genetic heterogeneity and the corresponding clinical outcome differences underscore the importance of developing genotype 1 replication systems.
Unmodified genomic RNA derived from the genotype 2a JFH-1 strain of HCV produces infectious virus particles after transfection into Huh7 hepatoma cells (2, 3). In contrast, the genome of the prototype genotype 1a virus, H77, although capable of efficient replication in chimpanzees, replicates poorly in cell culture (10, 11). Nonetheless, we recently reported the efficient replication of H77 genomic RNA containing five adaptive mutations (referred to herein as “H77-S”) in Huh7 hepatoma cells (12). These adaptive mutations are located within the NS3, NS4A, and NS5A proteins (see Fig. 1A). Here, we describe production of infectious HCV in cells transfected with this RNA. We compare the biophysical properties of genotype 1a and 2a particles produced in cell culture and show that these viruses can be readily distinguished serologically. Although possessing lower specific infectivity than JFH-1 virus produced in cell culture, the ability of this genotype 1a virus to infect cultured cells should substantially benefit antiviral and vaccine discovery programs.
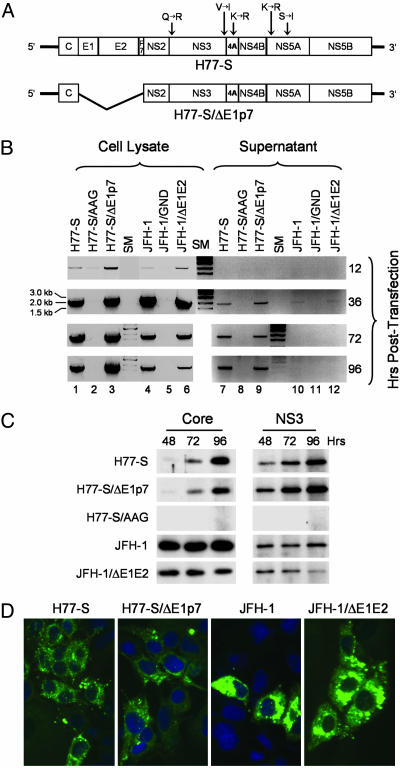
Fig. 1.
Replication of H77-S and JFH-1 RNAs in transfected Huh-7.5 cells. (A) Organization of the H77-S genomic RNA, showing location of the five adaptive mutations and the H77-S∕ΔE1p7 mutant in which sequence encoding the structural proteins was deleted. (B) Semiquantitative RT-PCR assays for HCV RNA in lysates (Left) and supernatant fluids (Right) of transfected Huh-7.5 cells. (C) Immunoblot detection of the HCV core and NS3 proteins in transfected Huh-7.5 cells. (D) Core antigen detected by indirect immunofluorescence 96 h after transfection of Huh-7.5 cells with the indicated RNA.
Results
Previous studies have shown that a combination of five cell culture-adaptive mutations provide for efficient replication of the genotype 1a H77-S RNA in transfected Huh7 cells (12). To assess the ability of this highly cell-culture-adapted RNA to produce infectious virus in transfected cells, we created a related mutant, H77-S∕ΔE1p7, with an in-frame deletion of sequence encoding the HCV structural proteins, E1, E2, and p7, that should eliminate virus particle formation but not impair viral RNA replication (Fig. 1A). Synthetic RNA transcribed from these two constructs, and an RNA replication-defective mutant containing an Ala–Ala–Gly substitution for the conserved Gly–Asp–Asp motif in the NS5B polymerase active site (H77-S∕AAG), were electroporated into Huh-7.5 cells. These cells are deficient in signaling virus activation of IFN-β synthesis through the intracellular retinoic acid-inducible gene I (RIG-I) pathway and are highly permissive for HCV RNA replication (13, 14). In parallel, Huh-7.5 cells were transfected with similar RNA transcripts derived from the JFH-1 virus and related mutants containing either a deletion of E1 and E2 or a Gly–Asn–Asp substitution in NS5B. Cells were monitored for replication of the transfected RNAs by a semiquantitative RT-PCR assay targeting an ≈1.9-kb segment of the NS3-coding region (see Materials and Methods). This assay is sensitive and specific for replication of synthetic viral RNAs after transfection.
Both the H77-S and JFH-1 RNAs replicated efficiently in Huh-7.5 cells, as did the related mutants in which the structural proteins were deleted (Fig. 1B, lanes 1, 3 and 4, 6). RNA synthesis was evident as early as 12 h after electroporation, with strong RT-PCR signals obtained from lysates prepared 36 h after transfection. These levels of RNA persisted in the H77-S and H77∕S-ΔE1p7 transfected cells through 96 h after transfection (lanes 1 and 3), whereas the RT-PCR signal was notably decreased in JFH-1 and JFH-1∕ΔE1E2-transfected cells by 72–96 h compared with that present at 36 h (lanes 4 and 6). In contrast, viral RNA was not detected in lysates of cells transfected with the NS5B mutants, indicating a failure of replication (Fig. 1B, lanes 2 and 5). These results were confirmed by immunoblot detection of the core and NS3 proteins in lysates of the transfected cells (Fig. 1C) and by immunofluorescence imaging of the core protein (Fig. 1D).
Several interesting differences were apparent in the expression of H77-S and JFH-1 proteins. Despite high abundance of viral RNA in H77-S transfected cells (Fig. 1B), JFH-1 transfected cells contained substantially more core protein, particularly at early time points (Fig. 1C). Core accumulated slowly in H77-S transfected cells, whereas its abundance was maximal at the earliest time point, 48 h, in JFH-1 transfected cells. Immunofluorescence staining was also substantially more intense for core antigen in the JFH-1 transfected cells (Fig. 1D). NS3 abundance also increased with time in H77-S transfected cells, whereas maximal levels were present at 48 h in JFH-1 transfected cells (Fig. 1C). The less intense NS3 signal in JFH-1 immunoblots likely reflects antigenic differences between JFH-1 and H77-S, as the NS3 Ab was raised to genotype 1a protein. In contrast, the anti-core monoclonal Ab (mAb) we used recognizes an epitope located between residues 21–40 of the protein, which is conserved in both viruses. These results suggest that JFH-1 RNA produces more core protein than H77-S RNA or that the JFH-1 core protein has greater stability than the H77-S protein in Huh-7.5 cells. However, a smaller proportion of cells expressed detectable core antigen in the JFH-1 transfected cultures 96 h after transfection (Fig. 1D). This difference suggests that JFH-1 RNA replication may be associated with greater cell death than H77-S.
Because Huh7 cells transfected with the JFH-1 RNA are known to release infectious virus particles (2, 3), we compared the secretion of viral RNA from these cells into the supernatant culture fluids. Substantially more viral RNA was released into supernatant fluids from H77-S transfected cells than from JFH-1 transfected cells (Fig. 1B, compare lanes 7 and 10), with the abundance of H77-S RNA released increasing between 36 and 96 h. In contrast, JFH-1 RNA release was minimal and detected only at 36 h after transfection in this assay. However, the release of either of these RNAs into the supernatant fluids was not dependent on expression of the envelope proteins (lanes 9 and 12), indicating that the presence of viral RNA in the culture media is not indicative of viral particle assembly.
Clarified supernatant culture fluids collected 24–96 h after transfection were tested for the presence of infectious virus by inoculation onto naïve Huh-7.5 cells, followed by fixation and staining for the presence of core antigen 96 h later. Core was present in numerous cells inoculated with the JFH-1 supernatant fluids (Fig. 2A Right), consistent with infection with JFH-1 virus (2, 3). Importantly, we also observed core antigen in a smaller number of cells inoculated with the H77-S supernatant fluids (Fig. 2. Left). As described previously for the JFH-1 virus (2), H77-S infected cells were grouped in small clusters. These clusters appear to result from division of a single infected cell during the 96-h incubation period or possibly by cell-to-cell spread of virus. Thus, in subsequent experiments, we measured virus infectivity in terms of “focus-forming units” (FFU). Importantly, the intensity of antigen staining in these cells mirrored that in the transfected Huh-7.5 cells, with less intense staining of core antigen in H77-S infected cells. Supernatant fluids remained infectious after passage through a 0.2-μm filter, consistent with cell-free virus.
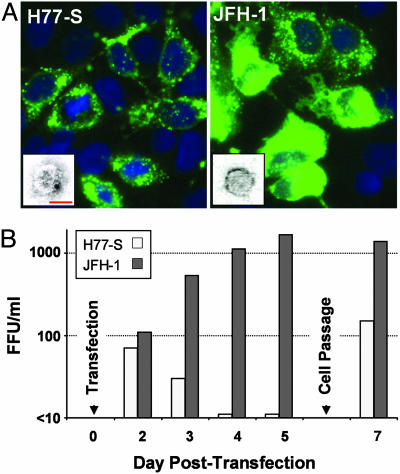
Fig. 2.
Infection of Huh-7.5 cells with H77-S and JFH-1 virus released into supernatant fluids of transfected Huh-7.5 cells. (A) HCV core antigen expression in cells infected with H77-S (Left) or JFH-1 (Right) virus. Left Inset shows a particle with immunogold labeling indicating recognition by the AP33 mAb to E2. (Bar: 50 nm.) Right Inset shows a typical JFH-1 particle for comparison. (See also Fig. 5.) (B) Time course of infectious H77-S (open bars) and JFH-1 (filled bars) virus released into supernatant fluids of RNA-transfected Huh-7.5 cells. H77-S release was greatest 24–48 h after transfection or passage of cells.
Interestingly, the release of infectious virus from H77-S transfected cells was not continuous, but was greatest 24–48 h after transfection and reduced subsequently (Fig. 2B). Trypsin treatment of the cell monolayer, followed by a 1:3 split and reseeding of the cells at a lower density, resulted in a reproducible burst of virus production. These results are consistent with previous observations indicating that HCV RNA replication is tightly coupled to host cell proliferation and that viral RNA synthesis is enhanced during the S phase of the cell cycle (15, 16). In contrast, the release of JFH-1 virus was continuous and increased with time (Fig. 2B).
Electron microscopic examination of supernatant fluids from both H77-S- and JFH-1-transfected cultures revealed the presence of occasional virus-like particles measuring 44–64 nm in diameter (Fig. 2A Insets; see also Fig. 5, which is published as supporting information on the PNAS web site). Some particles in the H77-S infectious material bound gold-labeled mAbs to the E2 glycoprotein of HCV (Fig. 2A Left Inset). Importantly, neither viral antigen expression in inoculated cells nor virus-like particles were observed with supernatant fluids taken from cells transfected with the H77-S∕ΔE1p7 mutant, despite equivalently robust replication of that RNA in transfected cells (Fig. 1B, lanes 3 and 9). Together, these results provide strong evidence for the production of cell culture-infectious H77-S virus in transfected Huh-7.5 cells. However, the lower number of antigen-positive cells obtained with inoculation of the H77-S harvest compared with the JFH-1 harvest suggests that the production of infectious virus is 10- to 100-fold less efficient with H77-S.
To compare the physical properties of infectious H77-S and JFH-1 particles, aliquots of concentrated posttransfection supernatant fluids were passed through a 0.2-μm filter and then layered onto a preformed 10–40% iodixanol gradient, which was centrifuged to equilibrium. Fractions collected from this isopycnic gradient were tested for viral RNA by the semiquantitative RT-PCR assay described above (Fig. 3A). Much of the H77-S RNA was present in fractions 2 to 3, near the top of the gradient, but a major fraction of the RNA banded discreetly at a higher density (≈1.13–1.14 g∕cm3) in fractions 5 to 6. Importantly, this second RNA peak was absent in gradients loaded with concentrated supernatant fluids from cells transfected with the H77-S∕ΔE1p7 mutant (Fig. 3A). Although a small amount of JFH-1 RNA was detectable in fraction 3, most was present in fractions 5 to 6 (Fig. 3A). These results were confirmed by real-time quantitative RT-PCR assays targeting a small, conserved segment of the 5′ nontranslated RNA (Fig. 3 B and C). When fractions were inoculated onto naïve Huh-7.5 cells, H77-S infectivity was found only in fractions 4 and 5, whereas the maximum JFH-1 infectivity was present in fraction 5 (Fig. 3B). No infectious virus was present in supernatant fluids from the H77-S∕ΔE1p7 transfected cells. The intensity of core antigen staining in cells infected with the H77-S and JFH-1 virus from these gradients showed the characteristic difference in staining intensity described above (Fig. 3, compare D and E). These results indicate that infectious H77-S virus has a buoyant density similar to that of JFH-1 virus: ≈1.13–1.14 g∕cm3.
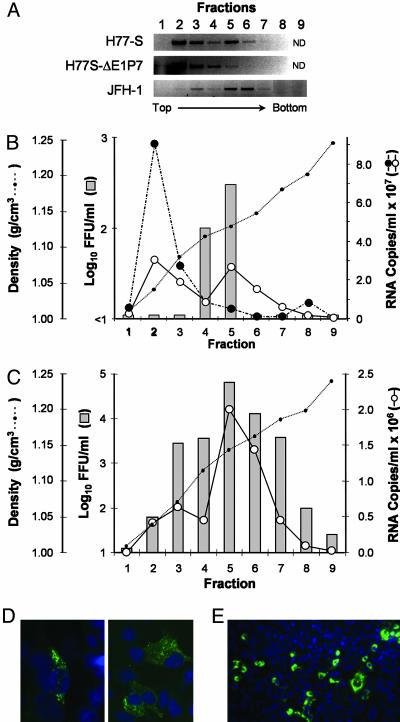
Fig. 3.
Equilibrium ultracentrifugation of H77-S and JFH-1 particles in isopycnic iodixanol gradients. (A) Semiquantitative RT-PCR detection of HCV RNA in fractions of gradients loaded with concentrated, filtered (0.2 μm) supernatant fluids from cells transfected with the indicated RNAs. (B) Results of infectivity assays (bars) and quantitative TaqMan RT-PCR assays in fractions from gradients loaded with concentrated supernatant fluids from cells transfected with H77-S (solid line with open circles) or H77-S∕ΔE1p7 (dashed line with filled circles) RNA. H77-S∕ΔE1p7 supernatant fluids contained no infectious virus. (C) Results of similar assays using fractions from gradients loaded with concentrated JFH-1 supernatant fluids. (D and E) HCV core antigen detected by indirect immunofluorescence in cells inoculated with fraction 5 of gradients loaded with H77-S (D) or JFH-1 (E) material (lower magnification).
We estimated the specific infectivity of the JFH-1 and H77-S viruses banding at this density by comparing the abundance of viral RNA in these fractions (determined by real-time RT-PCR) with the infectious titer. These results suggested a specific infectivity of 140 ± 13 S.E. RNA copies per FFU for the JFH-1 virus, compared with 54,000 ± 18,000 copies per FFU for the H77 virus (based on analysis of three fractions in two independent experiments). Thus, the JFH-1 virus has ≈400-fold greater specific infectivity than the H77-S virus in these fractions. The magnitude of this difference cannot be explained by differences in the ability of the RT-PCR assay to detect JFH-1 vs. H77-S RNA, nor is it likely to be due to contamination of the H77-S virus in fractions 4 and 5 with the RNA peaking in fraction 2 of the gradient that was not associated with infectious virus (Fig. 3B).
Because synthetic RNA transcripts banded at a higher density than infectious virus in these gradients (1.15–1.17 g∕cm3; data not shown), it is likely that the noninfectious H77-S RNA in fractions 2–3 is present in membranous complexes. The possibility that this material might be derived from viral replicase complexes is suggested by the presence of NS3 and NS5B within these fractions (see Fig. 6, which is published as supporting information on the PNAS web site). The abundance of E2 and core protein was not sufficient in fractions containing infectious virus for reproducible detection by immunoblot.
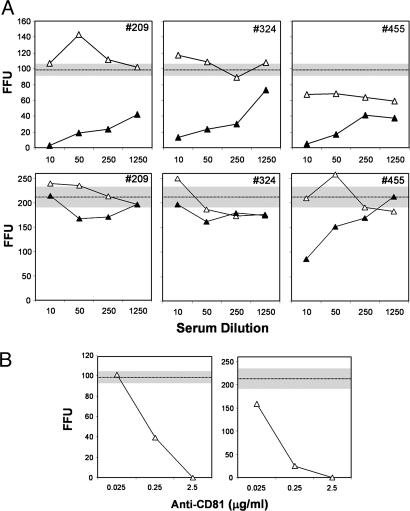
To characterize further the infectious particles produced in H77-S transfected cells, we carried out neutralization assays using serum samples collected prospectively from injection drug users who had experienced acute genotype 1 HCV infection (17). Dilutions of each serum sample were mixed with a fixed amount of H77-S, incubated for 60 min, then inoculated onto naïve Huh-7.5 cells. A >50% reduction in FFU was considered indicative of virus neutralizing Abs. We tested paired sera from three subjects, collected before and 8–14 months after initial serologic evidence of HCV infection. As shown in Fig. 4A Upper), none of the preinfection sera neutralized >50% of the H77-S inoculum, at any dilution, whereas a 1:10 dilution of each of the postinfection sera resulted in an 87–97% reduction in FFU relative to either the paired preinfection specimen, or a no serum control (98 ± 7 FFU). Fifty percent neutralization endpoints for the postinfection sera ranged from 1:700 to >1:1250. The specific neutralization of H77-S infectivity with postinfection human sera confirms that the infectious particles produced by H77-S transfected cells are antigenically related to those produced during human infections with wild-type virus. We also tested these sera for the ability to neutralize JFH-1 virus (Fig. 4A Lower). Only one of the three postinfection sera (patient no. 455) was capable of neutralizing JFH-1 virus (titer = 1:40), indicating substantial serologic differences between these two viruses.
Fig. 4.
Neutralization of cell culture-produced virus infectivity by antibody to HCV or CD81. (A) Neutralization of H77-S (Upper) and JFH-1 (Lower) viruses by paired preinfection (▵) or after seroconversion (▴) sera from three individuals sustaining infection with genotype 1a HCV. The horizontal lines and shaded zones indicate the mean and range of infectious foci obtained with each viral inoculum in the absence of any serum. (B) Anti-CD81 Ab, added to virus before its inoculation onto Huh-7.5 cells, prevents infection with either H77-S (Left) or JFH-1 (Right) inocula.
CD81 is expressed on the surface of many cells and has been shown to interact with the E2 protein of HCV (18). Abs to CD81 block the infection of Huh7 cells with HCV pseudotyped retroviral particles, as well as the genotype 2a JFH-1 virus (2, 3, 19). Moreover, a soluble CD81 fragment blocks infection of hepatoma cells with the FL-J6∕JFH chimera (4). We found that anti-CD81 Ab efficiently blocked the infection of Huh-7.5 cells with H77-S virus (Fig. 4B Left). These data suggest that H77-S and JFH-1 particles bind to and enter Huh-7.5 cells by similar mechanisms, most likely involving virus recognition of the CD81 molecule on the cell surface.
Discussion
Until recently, efforts to study HCV and its interactions with host cells have been impeded by the absence of cell culture systems that are capable of supporting all stages of the virus life cycle. The development of efficiently replicating subgenomic RNA replicons and genome-length selectable RNAs has been helpful in providing model systems that recapitulate events in viral polyprotein expression, processing, replicase assembly, and viral RNA synthesis (20–22). However, these experimental systems are not capable of providing insights into interactions of the virus with host-cell receptors, the process of viral entry, or assembly and release from the cell. The recent recognition that the genotype 2a virus, JFH-1, is capable of very efficient RNA replication as well as the production of fully infectious virus particles in transfected cells thus represents a major breakthrough for the hepatitis C field.
Our demonstration here of the ability of the H77-S virus to undergo the complete viral life cycle in Huh7.5 cells represents another important step in the development of useful cell culture systems for HCV. Unlike the genotype 2a JFH-1 virus, the genotype 1a H77-S virus is representative of the most prevalent HCV genotypes causing liver disease within the United States, as well as many other countries (5, 6). It carries five defined cell culture-adaptive mutations that distinguish it from the prototype Hutchinson strain (H77C) virus that is highly infectious for chimpanzees and that has been used in many early studies characterizing HCV (10, 12, 23). The adaptive mutations in H77-S that promote efficient viral RNA replication are located within the NS3∕4A protease complex and the NS5A protein, a nonstructural phosphoprotein (12). Both of these proteins appear to be essential components of the viral RNA replicase, but both proteins also play important roles in confounding innate cellular antiviral defenses (24, 25). How these five adaptive mutations modulate these viral functions to promote HCV RNA replication remains unknown, as is their impact, if any, on viral assembly and release. It will be interesting to determine whether these mutations reduce the ability of the virus to infect chimpanzees; previous studies with a genotype 1b virus suggest mutations that promote RNA replication in cultured cells reduce the ability of the virus to infect chimpanzees (26).
Although the lower quantities of infectious H77-S virus released from transfected cells correlates well with the lower abundance of viral proteins expressed, compared with JFH-1 transfected cells (Fig. 1C), the production of infectious virus does not appear to be determined only by the cellular abundance of HCV RNA and∕or its proteins. In other studies, we could not detect release of infectious virus from cells transfected with a highly cell culture-adapted, genotype 1b RNA derived from HCV-N (22), despite the expression of viral RNA and proteins roughly comparable with that observed with H77-S (M.Y. and S.M.L., unpublished data).
Quantitatively more viral RNA was released from H77-S transfected cells than JFH-1 transfected cells, but most of this RNA banded at a very low density in iodixanol gradients (≈1.03–1.07 g∕ml) (Fig. 3B). This RNA was not naked viral RNA, which possesses a significantly higher density (1.15–1.17 g∕ml) (data not shown). The nature of the low-density RNA is uncertain. It may represent only membrane-bound RNA associated with replication complexes released from dying cells, as suggested by the presence of NS3 and NS5B in these fractions (Fig. 6). It is interesting to note, however, that some circulating HCV RNA molecules present in human sera are found at a density of ≈1.06 gm∕cm3 after equilibrium ultracentrifugation (27), suggesting that the low-density RNA released from H77-S transfected cells may have possible physiologic relevance.
The much lower specific infectivity of the H77-S particles banding at 1.13–1.14 gm∕cm3, compared with JFH-1 particles with the same density, also remains to be explained. Both viral RNAs appear to replicate efficiently in transfected cells (Fig. 1B), but the structural and nonstructural proteins accumulate more slowly in H77-S transfected cells (Fig. 1C). To document the presence of core antigen in H77-S infected cells, we were required to incubate cells for 96 h after inoculation with virus. In contrast, abundant core antigen was present in JFH-1 infected cells by 48 h. It is tempting to speculate that this difference might explain, at least in part, the 400-fold difference we observed in the specific infectivity of JFH-1 and H77-S particles. However, a specific defect in virus entry or uncoating cannot be excluded. Widely different cell entry efficiencies have been observed with pseudotyped retroviruses bearing envelope glycoproteins from different HCV strains (19).
Although further work will be required to answer these and many other questions, the availability of a genotype 1a virus that is capable of undergoing the complete viral cycle in cultured cells should be a major asset to the hepatitis C field. The widely divergent neutralizing Ab activities we found against genotype 1a and 2a viruses in human sera (Fig. 4A) suggest that these viruses may represent distinct serotypes, an observation that has significant implications for vaccine development.
Materials and Methods
Plasmids.
The H77-S virus was derived from the chimpanzee-infectious genotype 1a pCV-H77C cDNA clone (GenBank accession no.AF011751) (10). It contains five cell culture-adaptive mutations, two within NS3 (Q1067R, V1651I), one in NS4A (K1691R), and two in NS5A (K2040R, S2204I) (12). Construction of pH77-S, formerly called pH77c∕QR∕VI∕KR∕KR5A∕SI, as well as the related replication-defective NS5B mutant, H77-S∕AAG, has been described (12). pH77-S∕ΔE1p7 contains an in-frame deletion spanning the E1-p7 coding (see Supporting Materials and Methods, which is published as supporting information on the PNAS web site). The JFH-1, JFH-1∕GND, and JFH-1∕ΔE1E2 plasmids are described in ref. 3.
Cells.
The Huh7 cell subline, Huh-7.5, was kindly provided by Charles Rice (Rockefeller University, New York) (13). Cells were cultured as described in ref. 12.
Abs.
H53 mAb to E2 was a gift from J. Dubuisson (Institut de Biologie de Lille∕Institut Pasteur de Lille, Lille, France) (28); the AP33 mAb was kindly provided by A. Patel (Medical Research Council Virology Unit, University of Glasgow, Glasgow, U.K.) (29). Human sera were provided by D. Netski (The Johns Hopkins University, Baltimore) (17). Commercial Abs included anti-core C7–50 (Affinity BioReagents, Golden, CO), anti-NS3 BDI371 (BioDesign, Saco, ME), and anti-CD81 JS-81 (BD Pharmingen).
HCV RNA Transfection and Virus Production.
HCV RNAs were transcribed in vitro and electroporated into cells as described in ref. 12. In brief, 10 μg of in vitro synthesized HCV RNA was mixed with 5 × 106 Huh-7.5 cells in a 2-mm cuvette and pulsed twice at 1.4 kV and 25 μF. Cells were seeded into 12-well plates for RNA analysis or 6-well plates for protein analysis. For virus production, transfected cells were seeded into 75 cm2 flasks and fed with medium containing 10% FCS. These cells were passaged with a 3:1 split at 3–4 days after transfection. Twenty-four hours later, the medium was replaced with serum-free medium, which was collected 24 h later as the virus harvest. Virus harvests were clarified by low-speed centrifugation and, where indicated, passed through a 0.2-μm filter before stabilization by addition of 20% FCS and freezing at −80°C.
Infectivity Assays.
Huh-7.5 cells were seeded at 2 × 104 cells∕well in 8-well chamber slides (Nalge Nunc, Rochester, NY) 24 h before inoculation with 80–100 μl of culture medium or gradient fractions (see below). Cells were tested for the presence of intracellular core antigen by immunofluorescence 96 h later (48 h for JFH-1 virus), as described below. Clusters of infected cells identified by staining for core antigen were considered to constitute a single infectious focus, and virus titers were calculated accordingly in terms of FFU∕ml.
Immunofluorescence Detection of Intracellular HCV Antigen.
Cells were fixed in methanol:acetone (1:1) at room temperature for 9 min, then stained with mAb C7–50 to the core protein diluted 1:300, followed by extensive washing and staining with FITC-conjugated goat anti-mouse IgG Ab (1010-02, Southern Biotech, Birmingham, AL) at a 1:100 dilution. Nuclei were counterstained with Bisbenzimide H (Hoechst, Frankfurt am Main, Germany), and slides were examined with a Zeiss LSM 510 laser scanning confocal microscope.
Neutralization Assay.
Virus stock containing ≈2 × 103 FFU∕ml virus was mixed with an equal volume of serial dilutions of heat-inactivated (56°C for 30 min) human sera and incubated at 37°C for 1 h before inoculation onto Huh-7.5 cells in 8-well chamber slides, as described above. After incubation of the cultures for 48 (JFH-1) or 96 (H77-S) h, cells were fixed and stained for the presence of HCV core antigen by indirect immunofluorescence (see above), and the foci of antigen-positive cells were enumerated. A >50% reduction in FFU (compared with virus incubated with no serum) was considered indicative of neutralizing Ab; endpoint 50% neutralization titers were estimated by using the least-squares method.
Equilibrium Ultracentrifugation.
Filtered supernatant fluids collected from transfected cell cultures (no FCS) were concentrated 20- to 50-fold by using a Centricon PBHK Centrifugal Plus-20 Filter Unit with Ultracel PL membrane (100-kDa exclusion) (Millipore), then layered on top of a preformed continuous 10–40% iodixanol (OptiPrep, Sigma-Aldrich) gradient in Hanks’ balanced salt solution (HBSS; Invitrogen). Gradients were centrifuged in a SW60 rotor (Beckman Coulter) at 45,000 rpm for 16 h at 4°C, and fractions (500 μl each) were collected from the top of the tube. The density of each fraction was estimated by weighing a 100-μl drop from fractions of a gradient run in parallel but loaded with HBSS.
Quantitation of HCV RNA.
Both semiquantitative and quantitative real-time RT-PCR assays were used to determine the abundance of viral RNA in transfected cells and virus harvests. For details, see Supporting Materials and Methods.
Immunoblot Analysis.
Blots were incubated with Abs to core (C7–50, 1:30,000) or NS3 (BDI371, 1:20,000), followed by horseradish peroxidase-conjugated anti-mouse IgG (1030-05, Southern Biotech) (1:30,000). Proteins were visualized by chemiluminesence using reagents provided with the ECL Advance kit (Amersham Pharmacia Biosciences).
Supplementary Material
Acknowledgments
We thank Robert Purcell and Jens Bukh (both of the National Institutes of Health, Bethesda) for the pCV-H77C plasmid; Charles Rice for Huh7.5 cells; and Dale Netski, Jean Dubuisson, and Arvind Patel for human sera and mAbs. We thank Vsevolod Popov for technical advice, Jeremy Yates and Francis Bodola for excellent technical assistance, and Annette Martin for critically reviewing the manuscript. This work was supported by National Institute of Allergy and Infectious Diseases Grants U19-AI40035, R21-AI063451, and N01-AI25488.
Glossary
Abbreviations:
- FFU
focus-forming unit
- HCV
hepatitis C virus.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Chisari F. V. Nature. 2005;436:930–932. doi: 10.1038/nature04076. [DOI] [PubMed] [Google Scholar]
- 2.Zhong J., Gastaminza P., Cheng G., Kapadia S., Kato T., Burton D. R., Wieland S. F., Uprichard S. L., Wakita T., Chisari F. V. Proc. Natl. Acad. Sci. USA. 2005;102:9294–9299. doi: 10.1073/pnas.0503596102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wakita T., Pietschmann T., Kato T., Date T., Miyamoto M., Zhao Z., Murthy K., Habermann A., Krauslich H.-G., Mizokami M., et al. Nat. Med. 2005;11:791–796. doi: 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lindenbach B. D., Evans M. J., Syder A. J., Wolk B., Tellinghuisen T. L., Liu C. C., Maruyama T., Hynes R. O., Burton D. R., McKeating J. A., et al. Science. 2005;309:623–626. doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- 5.Zein N. N. Clin. Microbiol. Rev. 2000;13:223–235. doi: 10.1128/cmr.13.2.223-235.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Simmonds P., Bukh J., Combet C., Deleage G., Enomoto N., Feinstone S., Halfon P., Inchauspe G., Kuiken C., Maertens G., et al. Hepatology. 2005;42:962–973. doi: 10.1002/hep.20819. [DOI] [PubMed] [Google Scholar]
- 7.Zanotto P. M., Gould E. A., Gao G. F., Harvey P. H., Holmes E. C. Proc. Natl. Acad. Sci. USA. 1996;93:548–553. doi: 10.1073/pnas.93.2.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Logvinoff C., Major M. E., Oldach D., Heyward S., Talal A., Balfe P., Feinstone S. M., Alter H., Rice C. M., McKeating J. A. Proc. Natl. Acad. Sci. USA. 2004;101:10149–10154. doi: 10.1073/pnas.0403519101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.National Institutes of Health Consensus Development Panel. NIH Consensus Statement on Management of Hepatitis C, NIH Consensus State Science Statements. Vol. 19. Bethesda: Natl. Inst. of Health; 2002. pp. 1–46. [Google Scholar]
- 10.Yanagi M., Purcell R. H., Emerson S. U., Bukh J. Proc. Natl. Acad. Sci. USA. 1997;97:8738–8743. doi: 10.1073/pnas.94.16.8738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blight K. J., McKeating J. A., Marcotrigiano J., Rice C. M. J. Virol. 2003;77:3181–3190. doi: 10.1128/JVI.77.5.3181-3190.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yi M., Lemon S. M. J. Virol. 2004;78:7904–7915. doi: 10.1128/JVI.78.15.7904-7915.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blight K. J., McKeating J. A., Rice C. M. J. Virol. 2002;76:13001–13014. doi: 10.1128/JVI.76.24.13001-13014.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sumpter R., Jr, Loo M. Y., Foy E., Li K., Yoneyama M., Fujita T., Lemon S. M., Gale M. J., Jr J. Virol. 2005;79:2689–2699. doi: 10.1128/JVI.79.5.2689-2699.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pietschmann T., Lohmann V., Rutter G., Kurpanek K., Bartenschlager R. J. Virol. 2001;75:1252–1264. doi: 10.1128/JVI.75.3.1252-1264.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scholle F., Li K., Bodola F., Ikeda M., Luxon B. A., Lemon S. M. J. Virol. 2004;78:1513–1524. doi: 10.1128/JVI.78.3.1513-1524.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Netski D. M., Mosbruger T., Depla E., Maertens G., Ray S. C., Hamilton R. G., Roundtree S., Thomas D. L., McKeating J., Cox A. Clin. Infect. Dis. 2005;41:667–675. doi: 10.1086/432478. [DOI] [PubMed] [Google Scholar]
- 18.Pileri P., Uematsu Y., Campagnoli S., Galli G., Falugi F., Petracca R., Weiner A. J., Houghton M., Rosa D., Grandi G., et al. Science. 1998;282:938–941. doi: 10.1126/science.282.5390.938. [DOI] [PubMed] [Google Scholar]
- 19.McKeating J. A., Zhang L. Q., Logvinoff C., Flint M., Zhang J., Yu J., Butera D., Ho D. D., Dustin L. B., Rice C. M., et al. J. Virol. 2004;78:8496–8505. doi: 10.1128/JVI.78.16.8496-8505.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lohmann V., Korner F., Koch J., Herian U., Theilmann L., Bartenschlager R. Science. 1999;285:110–113. doi: 10.1126/science.285.5424.110. [DOI] [PubMed] [Google Scholar]
- 21.Blight K. J., Kolykhalov A. A., Rice C. M. Science. 2000;290:1972–1974. doi: 10.1126/science.290.5498.1972. [DOI] [PubMed] [Google Scholar]
- 22.Ikeda M., Yi M., Li K., Lemon S. M. J. Virol. 2002;76:2997–3006. doi: 10.1128/JVI.76.6.2997-3006.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feinstone S. M., Alter H. J., Dienes H. P., Shimizu Y., Popper H., Blackmore D., Sly D., London W. T., Purcell R. H. J. Infect. Dis. 1981;144:588–598. doi: 10.1093/infdis/144.6.588. [DOI] [PubMed] [Google Scholar]
- 24.Lindenbach B. D., Rice C. M. Nature. 2005;436:933–938. doi: 10.1038/nature04077. [DOI] [PubMed] [Google Scholar]
- 25.Gale M., Jr, Foy E. M. Nature. 2005;436:939–945. doi: 10.1038/nature04078. [DOI] [PubMed] [Google Scholar]
- 26.Bukh J., Pietschmann T., Lohmann V., Krieger N., Faulk K., Engle R. E., Govindarajan S., Shapiro M., St Claire M., Bartenschlager R. Proc. Natl. Acad. Sci. USA. 2002;99:14416–14421. doi: 10.1073/pnas.212532699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hijikata M., Shimizu Y. K., Kato H., Iwamoto A., Shih J. W., Alter H. J., Purcell R. H., Yoshikura H. J. Virol. 1993;67:1953–1958. doi: 10.1128/jvi.67.4.1953-1958.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Op De Beeck A., Voisset C., Bartosch B., Ciczora Y., Cocquerel L., Keck Z., Foung S., Cosset F. L., Dubuisson J. J. Virol. 2004;78:2994–3002. doi: 10.1128/JVI.78.6.2994-3002.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Owsianka A., Tarr A. W., Juttla V. S., Lavillette D., Bartosch B., Cosset F. L., Ball J. K., Patel A. H. J. Virol. 2005;79:11095–11104. doi: 10.1128/JVI.79.17.11095-11104.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.