Abstract
Tumor cells have the capacity to proliferate indefinitely that is qualified as replicative immortality. This ability contrasts with the intrinsic control of the number of cell divisions in human somatic tissues by a mechanism called replicative senescence. Replicative immortality is acquired by inactivation of p53 and p16INK4a genes and reactivation of hTERT gene expression. It is unknown whether the cancer cell replicative immortality is reversible. Here, we show the spontaneous induction of replicative senescence in p53-and p16INK4a-deficient hepatocellular carcinoma cells. This phenomenon is characterized with hTERT repression, telomere shortening, senescence arrest, and tumor suppression. SIP1 gene (ZFHX1B) is partly responsible for replicative senescence, because short hairpin RNA-mediated SIP1 inactivation released hTERT repression and rescued clonal hepatocellular carcinoma cells from senescence arrest.
Keywords: immortality, liver cancer, SIP1, telomerase, p53
Tumor cells are clonal (1), and tumorigenesis usually requires three to six independent mutations in the progeny of precancerous cells (2). For this to occur, preneoplastic somatic cells would need to breach the replicative senescence barriers. Replicative senescence is a telomere-dependent process that sets a limit to the successive rounds of cell division in human somatic cells (3). Progressive telomere shortening is observed in almost all dividing normal cells. This phenomenon is linked to the lack of efficient hTERT expression that is observed in most human somatic cells (3). Replicative senescence (permanent growth arrest also called M1 stage) is believed to be initiated by a DNA damage-type signal generated by critically shortened telomeres, or by the loss of telomere integrity, leading to the activation of cell cycle checkpoint pathways involving p53, p16INK4a, and/or retinoblastoma (pRb) proteins (4, 5). In the absence of functional p53 and p16INK4a/pRb pathway responses, telomeres continue to shorten resulting in crisis (also called M2 stage). Cells that bypass the M2 stage by reactivating hTERT expression gain the ability for indefinite cell proliferation, also called immortality (3, 4, 6). There is accumulating evidence that cancer cells undergo a similar process during carcinogenesis to acquire immortality. Telomerase activity associated with hTERT reexpression is observed in ≈80% of human tumors (7), and senescence controlling p53 and p16INK4A genes are commonly inactivated in the majority of human cancers (8). Moreover, experimental transformation of normal human cells to tumor cells requires hTERT-mediated immortalization, as well as inactivation of p53 and pRb genes (9).
Aberrant expression of hTERT, together with the loss of p53 and p16INK4a/pRb control mechanisms, suggests that the replicative immortality is a permanent and irreversible characteristic of cancer cells. Although some cancer cells may react to extrinsic factors by a senescence-like stress response, this response is immediate, telomere-independent, and cannot be qualified as replicative senescence (10). Experimental inactivation of telomerase activity in cancer cells mostly results in cell death (11), whereas ectopic expression of p53, p16INK4a, or pRb provokes an immediate senescence-like growth arrest or cell death (10). Thus, to date there is no experimental evidence for spontaneous reprogramming of replicative senescence in immortalized cancer cells. Using hepatocellular carcinoma (HCC)-derived Huh7 cells as a model system, here we show that cancer cells with replicative immortality are able to spontaneously generate progeny with replicative senescence. Thus, we provide preliminary evidence for the reversibility of cancer cell immortality. The replicative senescence of cancer cells shares many features with normal cell replicative senescence such as repression of hTERT expression, telomere shortening, and permanent growth arrest with morphological hallmarks of senescence. However, the p53 gene is mutated, whereas p16INK4a promoter is hypermethylated in these cells. Thus, we show that fully malignant and tumorigenic HCC cells that display aberrant hTERT expression and lack functional p53 and p16INK4a genes are able to revert from replicative immortality to replicative senescence by an intrinsic mechanism. Furthermore, we demonstrate that the SIP1 gene, encoding a zinc-finger homeodomain transcription factor protein involved in TGF-β signaling (12, 13) and hTERT regulation (14), serves as a molecular switch between replicative immortality and replicative senescence fates in HCC cells.
Results
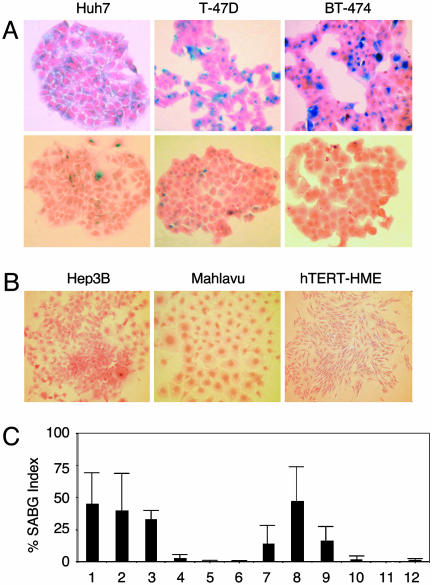
When analyzing clones from established cancer cell lines, we observed that some clones change morphology and cease proliferation at late passages with features reminiscent of cellular senescence (data not shown). We reasoned that this could be an indication for generation of progeny programmed for replicative senescence. We surveyed a panel of HCC and breast carcinoma cell lines and hTERT-immortalized human mammary epithelial cells (hTERT-HME). Plated at low clonogenic density, cells were maintained in culture until they performed 6–10 population doublings (PD), and tested for senescence-associated β-galactosidase (SABG) activity (15). Different cancer cell lines generated progeny with greatly contrasting SABG staining patterns. The first group, represented here by HCC-derived Huh7 and breast cancer-derived T-47D and BT-474 cell lines, generated heterogeneously staining colonies. Cells of some colonies were mostly positive for SABG, but others displayed significantly diminished or complete lack of staining (Fig. 1A). The second group, represented by HCC-derived Hep3B and Mahlavu, and hTERT-HME generated only SABG-negative colonies (Fig. 1B). Manual counting of randomly selected colonies demonstrated that mean SABG-labeling indexes for Huh7, T-47D and BT-474 progenies were 45 ± 23%, 40 ± 29%, and 33 ± 7%, respectively (Fig. 1C, lanes 1–3). In contrast, Hep3B, Mahlavu, and hTERT-HME progenies displayed <3 ± 3% mean SABG-labeling indexes (Fig. 1C, lanes 4–6). Clones from representative cell lines were expanded and subjected to the same analysis. SABG-staining patterns of all clones tested were closely similar to the patterns of their respective parental cell lines. For example, mean SABG staining indexes of Huh7-derived clones were 14 ± 15%, 47 ± 27%, and 17 ± 11% (Fig. 1C, lanes 7–9), whereas Hep3B-derived clones generated <2 ± 3% SABG-positive progenies (Fig. 1C, lanes 10–12). We speculated that the first group of cell lines comprised progenies in different stages of replicative senescence process at the time of analysis, whereas the second group of cell lines were composed mostly of immortal cells. The results obtained with the first group were unexpected. These cell lines have been established >20 years ago (16–18) and expanded in culture over many years, with PD well beyond the known senescence barriers for normal human cells (3), but they were still capable of generating presumably senescent progeny.
Fig. 1.
Established human cancer cell lines generate senescence-associated β-galactosidase (SABG)-expressing progeny. (A) Representative pictures of HCC (Huh7) and breast cancer (T-47D and BT-474) cell lines that generate both SABG-positive (Upper) and SABG-negative (Lower) colonies. (B) Representative pictures of HCC (Hep3B and Mahlavu) and telomerase-immortalized mammary epithelial (hTERT-HME) cell lines that generate only SABG-negative colonies. Cells were plated at clonogenic density to generate colonies with 6–10 population doublings, and stained for SABG activity (blue), followed by eosin counterstaining (red). (C) Quantification of SABG-positive cells in colonies. Randomly selected colonies (n ≥ 10) obtained from parental (lanes 1–6) cell lines and expanded clones (lanes 7–12) were counted to calculate the average % SABG positive cells per colony (% SABG index). Lanes 1–6 designate Huh7, T-47D, BT-474, Hep3B, Mahlavu, and hTERT-HME, respectively. Lanes 7–9 are Huh7-derived C1, C3, and C11 clones, and lanes 10–12 are Hep3B-derived 3B-C6, 3B-C11, and 3B-C13 clones. Error bars indicate SD.
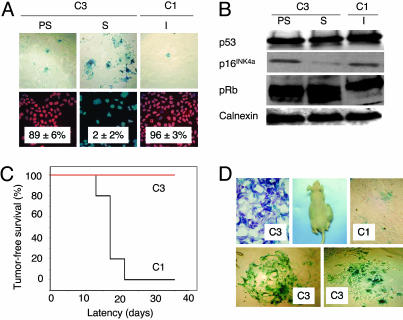
The study of a potentially active replicative senescence program in the progeny of immortal cancer cell lines requires the long-term follow up of single cell-derived clones. To this end, we chose to focus our investigations on Huh7 cell line. We expanded different Huh7-derived clones in long-term culture and examined their potential to undergo replicative senescence. Some clones performed >100 PD in culture with stable proliferation rates and heterogeneous SABG staining, whereas others sustained a limited number of PD, then entered a growth arrest phase with full SABG staining patterns. For example, C3 clone performed only 80 PD, whereas C1 clone replicated >150 PD. Permanently arrested C3 cells (PD 80) displayed enlarged size, flattened shape, and fully positive SABG staining, whereas early passage C3 (PD 57) and C1 (PD 179) cells displayed normal morphology with heterogeneous SABG staining (Fig. 2A Upper). Normal human cells at replicative senescence (M1) are refractory to mitotic stimulation and display <5% BrdUrd index (19). Growth-arrested C3 cells displayed very low BrdUrd staining (2 ± 2%), in contrast to early passage C3 and late passage C1 cells, which exhibited 89 ± 6%, and 96 ± 3% BrdUrd indexes, respectively (Fig. 2A Lower). Senescent C3 cells remained growth arrested, but alive when maintained in culture for at least 3 months, with no emergence of immortal clones (data not shown).
Fig. 2.
p53-and p16INK4a-deficient Huh7 cells generate progeny that undergo in vitro and in vivo replicative senescence resulting in loss of tumorigenicity. (A) Huh7-derived clones C3 and C1 were tested for replicative senescence arrest by SABG and BrdUrd staining at different passages. Presenescent C3 and immortal C1 cells display low SABG staining (Upper) and high BrdUrd incorporation (Lower), whereas senescent C3 cells are fully positive for SABG (Upper) and fail to incorporate BrdUrd into DNA after mitogenic stimuli (Lower). (B) p53 and p16INK4a protein levels show no increase in senescent C3 cells, compared to presenescent C3 and immortal C1 cells, but senescent C3 cells display partial hypophosphorylation of pRb. Calnexin was used as a loading control. Proteins were tested by Western blotting. PS, presenescent (PD 57); S, senescent (PD 80); I, immortal (PD 179). (C) C1 cells (black line) were fully tumorigenic, but C3 cells (red line) were not in nude mice. (D) C1 tumors displayed low SABG staining (Upper Right), whereas implanted C3 cells remaining at the injection site are fully positive for SABG in situ (Upper Left), as well as after short-term in vitro selection (Lower). Animals were injected with presenescent C3 (PD 59) and immortal C1 (PD 119) cells, and tumors and nontumorigenic cell samples were collected at day 35 and analyzed.
Biological mechanisms of replicative senescence observed here are of particular interest, because senescence-regulatory p53 is inactivated (20–22) and p16INK4a promoter is hypermethylated (23) in Huh7 cells. Accordingly, there was no change in p53 levels, whereas the low level p16INK4a expression did not increase, but decreased in senescent C3 (PD 80) cells, when compared to presenescent C3 (PD 57) or immortal C1 (PD 179) cells. Retinoblastoma protein (pRb) displayed partial hypophosphorylation in senescent C3 cells, apparently in a p53- and p16INK4a-independent manner (Fig. 2B). Cyclin E and A levels were also decreased, but p21cip1 levels were elevated in both presenescent and senescent C3 cells (Fig. 5A, which is published as supporting information on the PNAS web site). Cyclin D1, CDK4, and CDK2 protein levels (Fig. 5A) and p14ARF transcript levels (Fig. 5B) did not change.
Cancer cell senescence that we characterized here shared many features with normal cell replicative senescence (3), except that it was not accompanied with wild-type p53 or p16INK4a induction. However, in vivo relevance of the replicative senescence observed in cell culture is debated (6). Therefore, we compared in vivo replicative potentials of C3 (PD 59) and C1 (PD 119) cells in CD-1 nude mice. C3 cells did not form visible tumors, whereas C1 cells were fully tumorigenic in the same set of animals (Fig. 2C), like parental Huh7 cells (data not shown; ref. 24). C1 tumors collected at day 35 displayed scattered but low-rate SABG-positive staining, but remnant C3 cell masses collected from their injection sites were fully SABG-positive (Fig. 2D Upper). For confirmation, these remnants were removed from two different animals, passaged twice in cell culture for selection, and examined. Nearly all cells displayed senescence features including enlarged size, flattened shape, and highly positive SABG staining (Fig. 2D Lower). We concluded that loss of C3 tumorigenicity was due to replicative senescence in vivo.
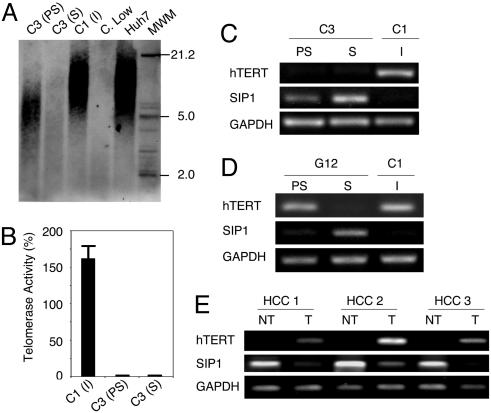
Replicative senescence, also called telomere-dependent senescence is associated with progressive telomere shortening due to inefficient telomerase activity (3). When compared to parental Huh7 cells, presenescent C3 cells at PD 57 had telomeres that have already been shortened to ≈7 kbp from ≈12 kbp. These cells eroded their telomeres to <5 kbp at the onset of senescence. In contrast, immortal C1 clone (PD 179) telomeres did not shorten (Fig. 3A). These observations showed a perfect correlation with telomerase activity and hTERT expression. Immortal C1 cells displayed robust telomerase activity, whereas both presenescent and senescent C3 cells had no detectable telomerase activity (Fig. 3B). Accordingly, the expression of hTERT gene was high in C1, but barely detectable in C3 cells (Fig. 3C). Thus, senescence observed with C3 cells was characterized with the loss of hTERT expression and telomerase activity, associated with telomere shortening.
Fig. 3.
C3 clonal cells undergo telomere-dependent replicative senescence associated with SIP1 expression and hTERT repression. SIP1 expression is lost, whereas hTERT is induced in primary HCC tumors. (A) Genomic DNAs from parental Huh7 and immortal C1 cells display long telomeres, whereas C3 telomeres are progressively shortened in presenescent and senescent stages, respectively. Equal amounts of genomic DNAs were blotted with a telomere repeat probe. C. Low, short telomere control DNA. (B) Presenescent and senescent C3 cells have lost telomerase activity, as measured by TRAP assay. Telomerase activity was shown as % value of test samples (± SD) compared to “high positive” control sample. (C) hTERT expression as tested by RT-PCR was high in immortal C1, but decreased to weakly detectable levels in C3 cells. Inversely, SIP1 expression tested by RT-PCR was undetectable in C1 cells, but showed a progressive increase in presenescent and senescent C3 cells. (D) Inverse relationship between SIP1 and hTERT expression was confirmed with another senescence-programmed Huh7 clone named G12 (for SABG and BrdUrd assays, see Fig. 7). hTERT expression in G12 showed a slight decrease in presenescent stage, followed by a loss at the onset of senescence. Inversely, the expression of SIP1 gene was weakly positive in presenescent G12, but highly positive in senescent G12 cells. C1 was used as control. PS, presenescent; S, senescent; I, immortal. (E) Negative correlation between hTERT and SIP1 expression in primary tumors (T) and nontumor liver tissues (NT).
Mechanisms of hTERT expression are presently unclear, but several genes including SIP1, hSIR2, c-myc, Mad1, Menin, Rak, and Brit1 have been implicated (14, 25). Therefore, we analyzed their expression in C1 and C3 clones. All tested genes, except SIP1, were expressed at similar levels in both C1 and C3 clones, independent of hTERT expression (Fig. 6, which is published as supporting information on the PNAS web site). SIP1 transcripts were undetectable in C1 cells, but elevated in C3 cells, moderately in presenescent, but strongly in senescent stages (Fig. 3C). We verified these findings with another Huh7-derived clone (G12) that displayed replicative senescence resulting in permanent cell proliferation arrest. Like C3, presenescent G12 cells that displayed low SABG staining with high BrdUrd index (98 ± 1%), became fully positive for SABG, and nearly negative for BrdUrd (3 ± 2%) at the onset of senescence (Fig. 7, which is published as supporting information on the PNAS web site). Presenescent G12 cells displayed only a weak hTERT repression associated with a slight increase in SIP1 expression, whereas SIP1 was strongly elevated in hTERT-negative senescent cells (Fig. 3D). Thus, there was a close correlation between SIP1 expression and hTERT repression in all Huh7 clones tested. The analysis of SIP1 and hTERT expression in primary HCCs and their corresponding nontumor liver tissues confirmed this relationship. SIP1 transcript levels were high, but hTERT expression was low in nontumor liver tissues, whereas respective HCC tumors displayed diminished SIP1 expression associated with up-regulated hTERT expression (Fig. 3E).
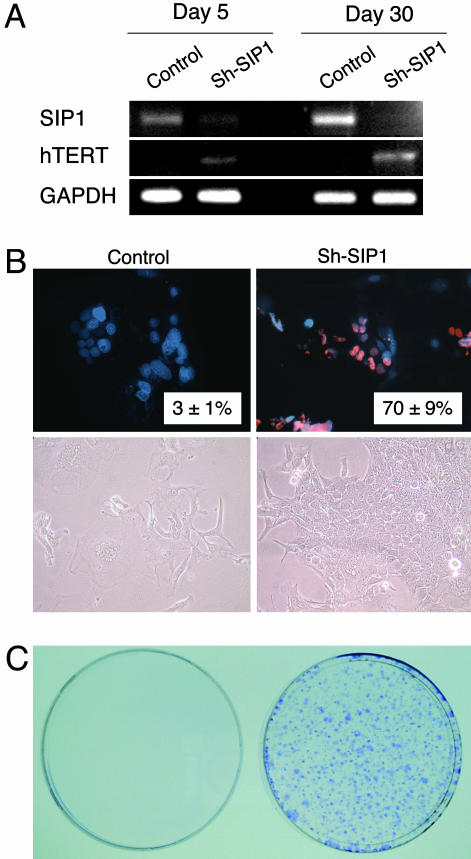
The SIP1 gene (Zinc finger homeobox 1B; ZFHX1B) encodes a transcriptional repressor protein that interacts with SMAD proteins of the TGF-β signaling pathway and CtBP corepressor (12, 13). This gene has recently been implicated in TGF-β-dependent regulation of hTERT expression in breast cancer cells (14). Our observations implicated SIP1 gene as a candidate regulator of replicative senescence in HCC cells. To investigate whether SIP1 expression constitutes a protective barrier against hTERT expression and senescence bypass, we constructed SIP1 short hairpin RNA (shRNA)-expressing plasmids, based on a reported effective SIP1 siRNA sequence (14). SIP1 shRNA was expressed by using either G-418-resistance plasmid pSuper.retro.neo+GFP or puromycin-resistance plasmid pSUPER.puro (see shRNA in Methods). Presenescent C3 cells at PD 75 were used for transfections, 3–4 weeks before expected senescence arrest stage. pSuper.retro.neo+GFP-based SIP1 shRNA suppressed the accumulation in SIP1 when expressed transiently (Fig. 4A, day 5). This resulted in a weak increase in hTERT expression. Transfected cells were maintained in culture in the presence of 500 μg/ml G-418 and observed for 30 days. At this period, C3 cells transfected with a control plasmid reached senescence-arrested stage with further up-regulation of SIP1 expression (Fig. 4A, day 30) and resistance to BrdUrd incorporation after mitogenic stimuli (BrdUrd index = 3 ± 1%; Fig. 4B Upper Left). In sharp contrast, SIP1 shRNA-transfected cells lost SIP1 expression and up-regulated hTERT transcripts (Fig. 4A, day 30). Furthermore, SIP1-inactivated cells escaped senescence, as evidenced with 70 ± 9% BrdUrd index (Fig. 4B Upper Right). Morphologically, SIP1 shRNA-transfected cells formed proliferating clusters, whereas cells transfected with control plasmid displayed hallmarks of senescence such as scattering, enlargement, and multiple nuclei (Fig. 4B Lower). Twelve independent clones were selected from SIP1 shRNA-transfected C3 cells. All but one of these clones have performed so far >15 PD beyond the expected senescence barrier (data not shown). As an additional confirmatory assay, C3 cells were transfected with the puromycin-selectable pSUPER.puro-based SIP1 shRNA vector and subjected to puromycin selection. SIP1 shRNA-transfected cells survived and formed large number of colonies after 30 days of puromycin selection. In contrast, no surviving colony was obtained from cells transfected with the control plasmid, as expected (Fig. 4C).
Fig. 4.
ShRNA-mediated down-regulation of endogeneous SIP1 transcripts releases hTERT repression and rescues C3 cells from senescence arrest. (A) At day 5 after transfection, SIP1 shRNA-transfected cells (Sh-SIP1) show decreased expression of SIP1 and weak up-regulation of hTERT expression. At day 30, the expression of SIP1 is lost completely, and hTERT expression is stronger. (B) Cells transfected with empty vector (Control) are senescence-arrested as evidenced by resistance to BrdUrd incorporation (Upper Left) and morphological changes (Lower Left), but cells transfected with SIP1 shRNA vector (Sh-SIP1) escaped senescence arrest as indicated by high BrdUrd index (Upper Right) and proliferating cell clusters (Lower Right). (C) Colony-forming assay shows that C3 cells formed large number of colonies following puromycin selection after transfection with a puromycin-resistant SIP1-shRNA-expressing plasmid (Right), whereas cells transfected with empty vector did not survive (Left). SIP1 shRNA was expressed by using either G-418-resistance plasmid pSuper.retro.neo+GFP (A and B) or puromycin-resistance plasmid pSUPER.puro (C). Presenescent C3 cells at PD 75 were transfected with either SIP1 shRNA-expressing or empty plasmid vectors, maintained in culture in the presence of appropriate selection media and tested at days 5 (A) and 30 (A–C).
Discussion
Our observations provide experimental evidence for the generation of senescence-arrested clones from immortal HCC and breast cancer cell lines. Detailed analysis of clones from HCC-derived Huh7 cell line further indicates that what we observe is a replicative senescence, but not a stress-induced premature senescence-like arrest. Clonal C3 cells displayed telomerase repression, progressive telomere shortening, and permanent growth arrest after ≈80 PD with senescence-associated morphological changes and positive SABG staining. Similar changes have also been observed with G12, another independently derived clone. Thus, we demonstrate that immortal cancer cells have the intrinsic ability to reprogram the replicative senescence. As expected, this shift in cell fate results in a complete loss of tumorigenicity. The replicative senescence arrest that we identified with clonal C3 cells was not accompanied with the induction of the p53, p16INK4a, p14ARF, or p21Cip1 gene. The nonparticipation of p53 and p16INK4a to the senescence arrest described here was expected, in the light of published observations showing that Huh7 cells express a mutant p53 protein (20–22) and they are deficient in p16INK4a expression (23). Although the levels of p21Cip1 protein displayed a slight increase in C3 cells, this was not related to senescence arrest, as early passage proliferating C3 cells also displayed this slight increase (Fig. 5). The early loss of hTERT expression in this clone could contribute to early p21Cip1 up-regulation, because hTERT is known to down-regulate p21Cip1 promoter activity (26). p53, p16INK4a, p14ARF, and p21Cip1 form a group of replicative senescence-related cell cycle checkpoint genes. The lack of induction of these genes in senescence-arrested C3 cells clearly indicates that there are additional genes involved in senescence arrest in these tumor-derived cells.
The loss of hTERT expression in senescence programmed clones prompted us to analyze the expression of genes that have been implicated in hTERT regulation. Among seven candidate genes studied, only one, the SIP1 gene, displayed a differential expression between immortal and senescence-programmed clones. This gene has been identified as a mediator of TGF-β-regulated repression of hTERT expression in a breast cancer cell line, although it was not effective in an osteosarcoma cell line (14). In our studies, SIP1 was not expressed in immortal hTERT-expressing C1 clone, but expressed in senescence-programmed hTERT-repressed C3 and G12 clones (Fig. 3 B and C). Furthermore, experimental depletion of SIP1 transcripts resulted in hTERT up-regulation in C3 clonal cells (Fig. 4A). This effect has been confirmed by using SKHep1, another HCC cell line (data not shown). Thus, we demonstrate that the SIP1 gene acts as an hTERT repressor in HCC cells. More importantly, we also showed the bypass of senescence arrest after functional inactivation of SIP expression by shRNA in senescence-programmed C3 clonal cells. In contrast to C3 cells transfected with a control plasmid, SIP1 shRNA-treated cells displayed continued proliferation beyond PD ≈80 as evidenced by 70% BrdUrd incorporation index, and formation of large number of colonies. Selected shRNA-transfected clones from these experiments have already performed >15 PD beyond the senescence barrier. Thus, our findings indicate that the functional inactivation of SIP1 in senescence-programmed cancer cells is sufficient to bypass senescent arrest.
SIP1 is a zinc finger and homeodomain containing transcription factor that exerts a repressive activity by binding to CACCT sequences in regulatory elements of target genes (12, 27). The SIP1 gene is expressed at high levels in almost all human somatic tissues tested, including liver (28). Therefore, we also performed comparative analysis of hTERT and SIP1 expression in nontumor liver and primary HCC tissues. SIP1 was strongly positive in nontumor liver samples, but its expression was significantly decreased in corresponding HCC samples. Inversely, hTERT expression was negative or low in nontumor liver samples, but highly positive in HCC tumors (Fig. 3E). We also detected complete loss of SIP1 expression in 5 of 14 (36%) of HCC cell lines (data not shown). Taken together with in vitro studies, these observations strongly suggest that SIP1 acts as a tumor suppressor gene in HCC. Although SIP1, as a repressor of E-cadherin promoter, has been suggested to be a promoter of invasion in malignant epithelial tumors (29), a tumor suppressive activity by the repression of hTERT and inhibition of senescence arrest is not precluded.
Hepatocellular carcinoma is one of the most common cancers worldwide. Liver cirrhosis is the major etiology of this tumor with limited therapeutic options (30, 31). Telomere shortening and senescence play a major role in liver cirrhosis, from which the neoplastic HCC cells emerge with high rates of telomerase reactivation (32). Furthermore, p53 and p16INK4a are the most frequently inactivated genes in these tumors. This fact enhances the importance of our findings for potential therapeutic applications of replicative senescence programming in HCC.
Methods
Tissues, Cells, and Clones.
Snap-frozen HCC and nontumor liver tissues were used. HCC and breast cancer cell lines T-47D (ATCC) and BT-474 (ATCC) were cultivated as described (33). hTERT-HME cells (Clontech) were cultivated in DMEM/Ham’s F-12 (Biochrom) containing insulin (3.5 μg/ml), EGF (0.1 ng/ml), hydrocortison (0.5 μg/ml), and 10% FBS (Biochrom). Huh7- and Hep3B-derived isogenic clones were obtained by either G-418 selection after transfection with neomycin-resistance pcDNA3.1 (Invitrogen) or pEGFP-N2 (Clontech) plasmids, or by low-density cloning. Huh7-derived isogenic clones C1 and C3 were obtained with pCDNA3.1, and G12 with pEGFP-N2. Huh7-derived C11, and Hep3B-derived 3B-C6, 3B-C11 and 3B-C13 were obtained by low-density cloning. Cells transfected with calcium phosphate/DNA-precipitation method were cultivated in the presence of geneticin G-418 sulfate (500 μg/ml; GIBCO), and isolated single cell-derived colonies were picked up by using cloning cylinders and expanded in the presence of 200 μg/ml geneticin G-418 sulfate. For low-density cloning, cells were plated at 30 cells per cm2 and single-cell derived colonies were expanded. Initial cell stocks were prepared when total number of cells became 1–3 × 107, and the number of accumulated population doubling (PD) at this stage was estimated to be 24, assuming that the progeny of the initial colony-forming cells performed at least 24 successive cell divisions until that step. Subsequent passages were performed every 4–7 days, and the number of additional PD was determined by using a described protocol (34).
Low-Density Clonogenic Assay.
Cells (30–50 per cm2) were plated in six-well plates and grown 1–3 weeks to obtain isolated colonies formed with 100–1,000 cells. The medium was changed every 4 days, and colonies were subjected to SABG staining (see below).
In Vivo Studies.
Cells were injected s.c. into CD-1 nude mice (Charles River Breeding Laboratory). Tumors and nontumorigenic cells at the injection sites were collected at day 35 and analyzed directly or after in vitro culture by SABG assay (see below). These experiments have been approved by the Bilkent University Animal Ethics Committee.
SABG Assay.
SABG activity was detected by using a described protocol (15). After DAPI or eosin counterstaining, SABG-positive and negative cells were identified and counted.
BrdUrd Incorporation Assay.
Subconfluent cells were labeled with BrdUrd for 24 h in freshly added culture medium and tested as described (33), using anti-BrdUrd antibody (Dako) followed by tetramethylrhodamine B isothiocyanate-labeled secondary antibody (Sigma). DAPI (Sigma) was used for counterstaining.
Immunoblotting.
Antibodies against cyclin D1, CDK4, CDK2, p21Cip1, pRb (all from Santa Cruz Biotechnology), cyclin E (Transduction), cyclin A (Abcam), p16INK4a (Abcam), p53 (clone 6B10; ref. 35), and calnexin (Sigma) were used for immunoblotting as described (33).
RT-PCR.
RT-PCR expression analysis was performed as described (33), using primers listed in Table 1, which is published as supporting information on the PNAS web site.
TRAP and Telomere Length Assays.
Telomerase activity and telomere length assays were performed by using TeloTAGGG Telomerase PCR ELISAPLUS and TeloTAGGG Telomere Length Assay (Roche Diagnostics), following kit instructions.
shRNA.
SIP1-directed shRNA was designed according to a previously described effective siRNA sequence (14) using the pSUPER RNAi system instructions (Oligoengine) and cloned into pSuper.retro.neo+GFP and pSUPER.puro (Oligoengine), respectively. SIP1 shRNA-encoding sequence was inserted by using 5′-GATCCCCCTGCCATCTGATCCGCTCTTTCAAGAGAAGAGCGGATCAGATGGCAGTTTTTA-3′ (sense) and 5′-AGCTTAAAAACTGCCATCTGATCCGCTCTTCTCTTGAAAG AGCGGATCAG ATGGCAGGGG-3′ (antisense) oligonucleotides.
The integrity of the inserted shRNA-coding sequence has been confirmed by nucleic acid sequencing of recombinant plasmids. Clone C3 cells were transfected with calcium phosphate precipitation method, using either pSuper.retro.neo+GFP-based or pSUPER.puro-based SIP1 shRNA expression plasmid, and cells were maintained in the presence of 500 μg/ml geneticin G-418 sulfate and 2 μg/ml puromycin (Sigma), respectively. Empty vectors were used as control. Media changed every 3 days, and cells were tested at days 5 and 30.
Supplementary Material
Acknowledgments
We thank E. Galun, G. Hotamisligil, F. Saatcioglu, and A. Sancar for reading the manuscript and helpful suggestions. This work was supported by Grant SBAG-2774/104S045 from the Scientific and Technological Research Council of Turkey (TUBITAK) and funds from Bilkent University and Turkish Academy of Sciences (TUBA).
Glossary
Abbreviations:
- HCC
hepatocellular carcinoma
- PD
population doubling
- SABG
senescence-associated β-galactosidase
- shRNA
short hairpin RNA.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Nowell P. C. Science. 1976;194:23–28. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- 2.Vogelstein B., Kinzler K. W. Trends. Genet. 1993;9:138–141. doi: 10.1016/0168-9525(93)90209-z. [DOI] [PubMed] [Google Scholar]
- 3.Shay J. W., Wright W. E. Carcinogenesis. 2005;26:867–874. doi: 10.1093/carcin/bgh296. [DOI] [PubMed] [Google Scholar]
- 4.Campisi J. Cell. 2005;120:513–522. doi: 10.1016/j.cell.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 5.Dimri G. P. Cancer Cell. 2005;7:505–512. doi: 10.1016/j.ccr.2005.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ben-Porath I., Weinberg R. A. J. Clin. Invest. 2004;113:8–13. doi: 10.1172/JCI200420663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shay J. W., Bacchetti S. Eur. J. Cancer. 1997;33:787–791. doi: 10.1016/S0959-8049(97)00062-2. [DOI] [PubMed] [Google Scholar]
- 8.Sherr C. J., McCormick F. Cancer Cell. 2002;2:103–112. doi: 10.1016/s1535-6108(02)00102-2. [DOI] [PubMed] [Google Scholar]
- 9.Boehm J. S., Hahn W. C. Curr. Opin. Genet. Dev. 2005;15:13–17. doi: 10.1016/j.gde.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 10.Roninson I. B. Cancer Res. 2003;63:2705–2715. [PubMed] [Google Scholar]
- 11.Shay J. W., Roninson I. B. Oncogene. 2004;23:2919–2933. doi: 10.1038/sj.onc.1207518. [DOI] [PubMed] [Google Scholar]
- 12.Verschueren K., Remacle J. E., Collart C., Kraft H., Baker B. S., Tylzanowski P., Nelles L., Wuytens G., Su M. T., Bodmer R., et al. J. Biol. Chem. 1999;274:20489–20498. doi: 10.1074/jbc.274.29.20489. [DOI] [PubMed] [Google Scholar]
- 13.Postigo A. A., Depp J. L., Taylor J. J., Kroll K. L. EMBO J. 2003;22:2453–2462. doi: 10.1093/emboj/cdg226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin S. Y., Elledge S. J. Cell. 2003;113:881–889. doi: 10.1016/s0092-8674(03)00430-6. [DOI] [PubMed] [Google Scholar]
- 15.Dimri G. P., Lee X., Basile G., Acosta M., Scott G., Roskelley C., Medrano E. E., Linskens M., Rubelj I., Pereira-Smith O., et al. Proc. Natl. Acad. Sci. USA. 1995;92:9363–9367. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lasfargues E. Y., Coutinho W. G., Redfield E. S. J. Natl. Cancer Inst. 1978;61:967–978. [PubMed] [Google Scholar]
- 17.Keydar I., Chen L., Karby S., Weiss F. R., Delarea J., Radu M., Chaitcik S., Brenner H. J. Eur. J. Cancer. 1979;15:659–670. doi: 10.1016/0014-2964(79)90139-7. [DOI] [PubMed] [Google Scholar]
- 18.Nakabayashi H., Taketa K., Miyano K., Yamane T., Sato J. Cancer Res. 1982;42:3858–3863. [PubMed] [Google Scholar]
- 19.Wei W., Sedivy J. M. Exp. Cell Res. 1999;253:519–522. doi: 10.1006/excr.1999.4665. [DOI] [PubMed] [Google Scholar]
- 20.Bressac B., Galvin K. M., Liang T. J., Isselbacher K. J., Wands J. R., Ozturk M. Proc. Natl. Acad. Sci. USA. 1990;87:1973–1977. doi: 10.1073/pnas.87.5.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Volkmann M., Hofmann W. J., Muller M., Rath U., Otto G., Zentgraf H., Galle P. R. Oncogene. 1994;9:195–204. [PubMed] [Google Scholar]
- 22.Kubica S., Trauwein C., Niehof M., Manns M. Hepatology. 1997;25:867–873. doi: 10.1002/hep.510250414. [DOI] [PubMed] [Google Scholar]
- 23.Roncalli M., Bianchi P., Bruni B., Laghi L., Destro A., Di Gioia S., Gennari L., Tommasini M., Malesci A., Coggi G. Hepatology. 2002;36:427–432. doi: 10.1053/jhep.2002.34852. [DOI] [PubMed] [Google Scholar]
- 24.Kaneko S., Hallenbeck P., Kotani T., Nakabayashi H., McGarrity G., Tamaoki T., Anderson W. F., Chiang Y. L. Cancer Res. 1995;55:5283–5287. [PubMed] [Google Scholar]
- 25.Wang J., Xie L. Y., Allan S., Beach D., Hannon G. J. Genes Dev. 1998;12:1769–1774. doi: 10.1101/gad.12.12.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Young J. I., Sedivy J. M., Smith J. R. J. Biol. Chem. 2003;278:19904–19908. doi: 10.1074/jbc.M301685200. [DOI] [PubMed] [Google Scholar]
- 27.Remacle J. E., Kraft H., Lerchner W., Wuytens G., Collart C., Verschueren K., Smith J. C., Huylebroeck D. EMBO J. 1999;18:5073–5084. doi: 10.1093/emboj/18.18.5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cacheux V., Dastot-Le Moal F., Kaariainen H., Bondurand N., Rintala R., Boissier B., Wilson M., Mowat D., Goossens M. Hum. Mol. Genet. 2001;10:1503–1510. doi: 10.1093/hmg/10.14.1503. [DOI] [PubMed] [Google Scholar]
- 29.Comijn J., Berx G., Vermassen P., Verschueren K., van Grunsven L., Bruyneel E., Mareel M., Huylebroeck D., van Roy F. Mol. Cell. 2001;7:1267–1278. doi: 10.1016/s1097-2765(01)00260-x. [DOI] [PubMed] [Google Scholar]
- 30.Thorgeirsson S. S., Grisham J. W. Nat. Genet. 2002;31:339–346. doi: 10.1038/ng0802-339. [DOI] [PubMed] [Google Scholar]
- 31.Bruix J., Boix L., Sala M., Llovet J. M. Cancer Cell. 2004;5:215–219. doi: 10.1016/s1535-6108(04)00058-3. [DOI] [PubMed] [Google Scholar]
- 32.Satyanarayana A., Manns M. P., Rudolph K. L. Hepatology. 2004;40:276–283. doi: 10.1002/hep.20308. [DOI] [PubMed] [Google Scholar]
- 33.Erdal E., Ozturk N., Cagatay T., Eksioglu-Demiralp E., Ozturk M. Int. J. Cancer. 2005;115:903–910. doi: 10.1002/ijc.20972. [DOI] [PubMed] [Google Scholar]
- 34.Masutomi K., Yu E. Y., Khurts S., Ben-Porath I., Currier J. L., Metz G. B., Brooks M. W., Kaneko S., Murakami S., DeCaprio J. A., et al. Cell. 2003;114:241–253. doi: 10.1016/s0092-8674(03)00550-6. [DOI] [PubMed] [Google Scholar]
- 35.Yolcu E., Sayan B. S., Yagci T., Cetin-Atalay R., Soussi T., Yurdusev N., Ozturk M. Oncogene. 2001;15:1398–1401. doi: 10.1038/sj.onc.1204240. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.