Abstract
Sarcolipin (SLN) inhibits the cardiac sarco(endo)plasmic reticulum Ca2+ ATPase (SERCA2a) by direct binding and is superinhibitory if it binds as a binary complex with phospholamban (PLN). To demonstrate whether overexpression of SLN in the heart might impair cardiac function directly, transgenic (TG) mice with cardiac-specific overexpression of NF-SLN (SLN tagged at its N terminus with the FLAG epitope) were generated on a phospholamban (PLN) null (PLN KO) background. In NF-SLN TG/PLN KO cardiac microsomes, the apparent affinity of SERCA2a for Ca2+ was decreased compared with non-TG littermate PLN KO hearts. Analyses of isolated NF-SLN/PLN KO cardiomyocytes revealed impaired cardiac contractility, reduced calcium transient peak amplitude, and slower decay kinetics compared to PLN KO animals. In these cardiomyocytes, isoproterenol restored calcium dynamics to the levels seen in PLN KO. Invasive hemodynamic and echocardiographic analyses of NF-SLN/PLN KO mouse cardiac muscle in vivo showed no direct effects of NF-SLN overexpression when compared to PLN KO mice. A possible mechanism for the lack of effects in the whole heart may be a responsiveness to phosphorylation because we determined that NF-SLN can be phosphorylated in cardiomyocytes in response to isoproterenol, and we provide evidence that serine/threonine kinase 16 is a kinase that can phosphorylate NF-SLN. Site-directed mutagenesis showed that SLN Thr-5 is the target site for this kinase. These data show that overexpression of NF-SLN can inhibit SERCA2a in the absence of PLN and that the inhibition of SERCA2a is correlated with impairment of contractility and calcium cycling in cardiomyocytes.
Keywords: sarco(endo)plasmic reticulum ATPases
Sarco(endo)plasmic reticulum ATPases (SERCA) are ubiquitously expressed, integral membrane proteins that transport Ca2+ ions from the cytosol to the lumen of the sarco(endo)plasmic reticulum. In cardiac muscle, SERCA2a can associate with a 52-aa transmembrane protein, phospholamban (PLN) to form a PLN–SERCA2a complex that has a lower apparent affinity for Ca2+ (1). The inhibited complex can be disrupted by phosphorylation of PLN or elevation of cytosolic Ca2+, leading to the reversal of SERCA2a inhibition (1). Sarcolipin (SLN), a 31-aa protein, shows significant sequence identity and similarity in gene structure to PLN (2, 3) and copurifies with SERCA1a in fast-twitch skeletal muscle (4). Like PLN, SLN is an effective inhibitor of SERCA molecules (5–7).
Recently, we showed that when both SLN and PLN are coexpressed with SERCA1a or SERCA2a, superinhibition resulted in the apparent Ca2+ affinity being reduced by nearly 1 pCa unit, in contrast to reductions in the order of 0.2–0.3 pCa units for SLN or PLN alone (5, 6, 8). Biochemical analyses have shown that PLN and SLN form a very stable PLN–SLN binary complex when expressed at equal levels (6, 7). This complex interacts with SERCA molecules, and the additional binding sites supplied by the binary complex make the inhibited ternary complex more stable than either PLN–SERCA or SLN–SERCA binary complexes (7, 9).
What remains unknown is whether SLN alone is an effective inhibitor of SERCA2 in the heart. In this study, we investigated the biochemical and physiological consequences of cardiac-specific, transgenic (TG) overexpression of NF-SLN (SLN tagged at its N terminus with the FLAG epitope) in mouse hearts on a PLN KO background.
Results
NF-SLN Transgenic Animals on a PLN Null Background.
To determine whether sarcolipin can inhibit SERCA effectively, in vivo, we mated NF-SLN Tg mice against PLN knockout mice to obtain NF-SLN transgenic animals on a PLN null background and analyzed wild-type (WT) mice, PLN KO, NF-SLN Tg on a WT background, and NF-SLN Tg on the PLN KO background (see Fig. 1A). Immunoblot analyses of cardiac ventricular lysates from these animals show NF-SLN expression in both the NF-SLN Tg mice, together with the absence of PLN in the PLN KO backgrounds (Fig. 1B). Previously we showed that the presence of the flag does not appear to affect SLN function (5, 10). In these experiments, however, the actual expression levels of NF-SLN and endogenous SLN are unknown.
Fig. 1.
Characterization of NF-SLN transgenic mice. (A) PCR genotyping for the NF-SLN targeting construct, a WT PLN allele and the PLN knockout cassette. (B) Immunoblot analyses of cardiac lysates to detect NF-SLN (with Flag antibody) and PLN. (C) Immunoblot analyses of cardiac lysates. NCX1, sodium calcium exchanger 1; DHPR α1, dihydropyridine receptor α1 subunit; RyR2, ryanodine receptor 2. Numbers represent mean ± SEM. ∗, P < 0.05 versus WT.
Quantification of Sarcoplasmic Reticulum Proteins by Immunoblot Analysis.
To rule out compensatory changes in the expression of other Ca2+-signaling proteins, levels of the major Ca2+-signaling proteins were measured by semiquantitative Western blotting with appropriate antibodies. The expression of SERCA2a and dihydropyridine receptor (DHPR) α1 was not significantly different among any of the mice. We did observe a decrease in RyR2 protein levels in mice on the PLN KO background (11), although there was no significant difference between PLN KO and NF-SLN TG/PLN KO. We also observed a significant decrease in NCX1 levels. A decrease in NCX1 in the PLN KO has not been documented previously, but these results are consistent with a report indicating that PLN KO animals have depressed Na/Ca exchange activity (12). Differences in results may also have arisen from the different mouse backgrounds and different NCX1 antibodies.
NF-SLN Can Inhibit SERCA2 as Effectively as PLN.
We compared the effect of NF-SLN overexpression on the Ca2+ affinity of SERCA2a in an assay of the Ca2+ dependence of Ca2+ transport in ventricular lysates (Fig. 2). Samples from wild-type mice have a Ca2+-dependent Ca2+ transport activity with a KCa of 6.74, whereas PLN KO mice have an apparent KCa of 6.92 pCa units (P < 0.05) (Fig. 2 A and C). Interestingly, transport activities of NF-SLN Tg on the PLN KO background are nearly identical to wild type where inhibition occurs only through PLN, with a KCa of 6.68 (Fig. 2 A and C). Overexpression of NF-SLN on a WT background (i.e., in the presence of PLN) results in a supershift to a KCa of 6.43 (10). Analysis of mice overexpressing a superinhibitory PLN (I40A) or WT-PLN (PLN2X) have a transport activity with KCa of 6.13 and 6.11, respectively (Fig. 2 B and C). Crossing the NF-SLN mouse to these mice to generate NF-SLN/I40A and NF-SLN/PLN2X lines did not result in a greater superinhibition of Ca2+ uptake (KCa values of 6.11 and 6.14, respectively) (Fig. 2C).
Fig. 2.
Ca2+ dependence of Ca2+ transport activity. Calcium uptake activity was assessed in cardiac lysates. (A) Results obtained from WT, PLN KO, NF-SLN transgenic, and NF-SLN transgenic on a PLN KO background. (B) Calcium uptake data in PLN2x overexpressing transgenic lines, NF-SLN transgenics on the PLN2x background, I40A superinhibitory PLN transgenics, and NF-SLN transgenic mice crossed to the I40A PLN transgenics. (C) KCa (pCa) is the negative logarithm of the Ca2+ concentration required to attain the half-maximal Ca2+ uptake rate. Data are mean ± SEM. Symbols: ∗, P < 0.05 versus WT (+/+); ϕ, P < 0.05 versus PLN KO.
Measurement of Contractility in Isolated Ventricular Cardiomyocytes.
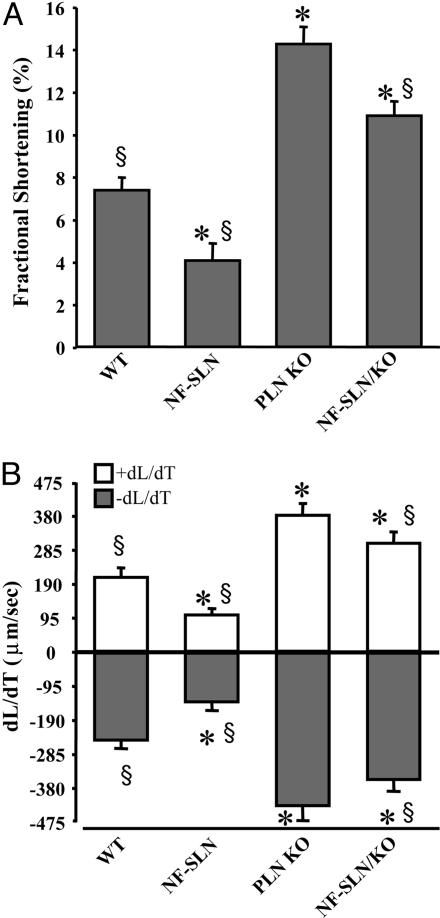
Contractile measurements in isolated cardiac myocytes showed significant differences between the groups in fractional cell shortening (FCS) (Fig. 3A) and in the rate of contraction and relaxation (+dL/dT and −dL/dT) (Fig. 3B). In particular, a significant decrease in FCS was seen in the NF-SLN TG compared to wild type, whereas a significant increase was found in the PLN KO compared to the wild type (Fig. 3A). The NF-SLN TG on the PLN KO background showed enhanced FCS compared to the wild type, but this shortening was significantly less than that observed in PLN KO. Similarly, dL/dTs values were smaller than the wild type in NF-SLN TG, higher than the wild type in PLN KO cells, and significantly different from the wild type and PLN KO in myocytes from NF-SLN TG/PLN KO (Fig. 3).
Fig. 3.
Contractile characteristics of isolated adult cardiomyocytes. Fractional cell shortening (A) and rates of shortening (+dL/dt) and relaxation (-dL/dt) (B) in single cells isolated from WT, PLN KO, NF-SLN transgenic, and NF-SLN transgenic on a PLN KO background. ∗, significantly different from WT, P < 0.05; §, significantly different from PLN KO, P < 0.05.
Measurement of Ca2+ Transients in Isolated Ventricular Cardiomyocytes.
Intracellular Ca2+ transients amplitudes (ΔCa2+) and kinetics were examined in Indo-loaded left ventricular myocytes at a stimulation frequency of 1 Hz (Fig. 4). Under baseline conditions (solid lines in Fig. 4A, black bars in Fig. 4B), the calcium transient amplitude was lower in NF-SLN TG and enhanced in the PLN KO, compared to the wild type. The NF-SLN TG/PLN KO calcium transient amplitudes were not different from the wild type but were different compared to PLN KO. In the presence of isoproterenol (ISO) (dashed lines in Fig. 4A and gray bars in Fig. 4B), myocytes from the wild type and NF-SLN TG showed an increase in ΔCa2+, whereas the PLN KO did not. Similarly, the peak calcium amplitudes in NF-SLN TG/KO show a significant increase in response to ISO treatment compared to the baseline.
Fig. 4.
Calcium transients in isolated adult cardiomyocytes. (A) Representative calcium transients obtained in field stimulated cardiomyocytes in the absence and presence of 1 μM ISO. (B) Quantification of calcium transient amplitude (Upper) and decay time (Lower). ∗, significant difference, P < 0.05.
Comparable results were seen with the calcium decay kinetics. In control conditions, calcium transient decays were slower in NF-SLN TG than controls, whereas PLN KO showed a faster decay (Fig. 4B). In NF-SLN TG/PLN KO, the decay of the transients was faster than controls. In the presence of ISO, NF-SLN TG/PLN KO myocytes responded with a significant acceleration of the decay, whereas the PLN KOs did not. Both the wild type and NF-SLN TG showed substantial reductions in decay times with ISO treatment.
Prediction of SLN Phosphorylation.
Computational prediction of putative SLN phosphorylation sites was performed by using the netphos2.0 database (www.cbs.dtu.dk/services/NetPhos). The predicted phosphorylation sites for mouse and rat (MERSTQE), human (MGINTRE), rabbit, cow, and pig (MERSTRE) SLN sequences are shown in Fig. 7, which is published as supporting information on the PNAS web site. The mouse, rat, and human sequence have a predicted site at Thr-5, whereas the rabbit/cow/pig sequences all show predicted phosphorylation of Ser-4.
Identification of NF-SLN Phosphorylation and Putative Kinase.
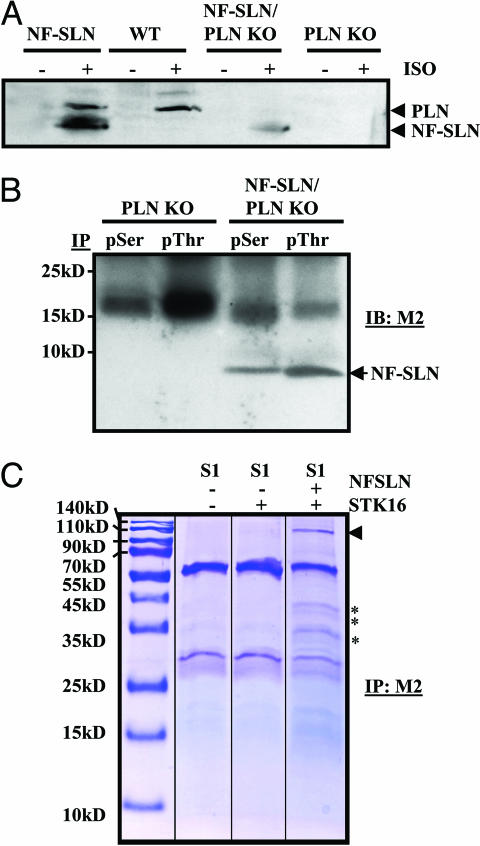
We investigated the mechanism by which ISO results in altered contractility and calcium transients in the NF-SLN Tg/KO mouse. Postnuclear ventricular lysates from animals injected with systemic ISO (2 μg/g) and collected 5 min later were immunoblotted by using Ab285, a phospho-serine-specific antibody raised against pSer16 of PLN. In the NF-SLN transgenic mouse, pSer bands were observed at ≈10 kDa and ≈6 kDa, representing PLN and SLN, respectively, which were more intense after ISO treatment (Fig. 5A). Although the upper PLN band was not seen in untreated WT mice, it was clearly evident in WT ISO-treated samples. The lower band was not present in untreated NF-SLN Tg/PLN KO hearts, but was seen in ISO-treated NF-SLN Tg/PLN KO hearts. No reactivity was seen in the PLN KO.
Fig. 5.
Phosphorylation of NF-SLN. (A) Representative immunoblot of cardiac lysates blotted with the PLN phosphorylation-specific antibody (A285) under control conditions or in response to systemic ISO infusion. (B) IP assays using cardiac lysates and pSerine- (pSer) and pThreonine- (pThr) specific antibodies, followed by detection of FLAG by using the M2 antibody. (C) Identification of a NF–SLN interacting kinase. Cotransfection of SERCA1 (S1), NF-SLN, and serine-threonine kinase 16 (STK16) into HEK293 cells was followed by M2 immunoprecipitation. Eluates were separated by SDS/PAGE and stained with Coomassie blue. arrowhead, SERCA1; ∗, specific bands corresponding to the molecular weight of STK16.
We performed coimmunoprecipitation assays by using commercially available pSer and pThr antibodies, followed by anti-flag (M2) immunoblots (Fig. 5B). We did not detect any reactivity in the PLN KO hearts, but clear bands at ≈6 kDa were seen in the NF-SLN Tg/PLN KO samples for both pSer and pThr, indicating either cross reactivity of the antibodies or possible phosphorylation of both serine and threonine phosphorylation of NF-SLN. Control experiments with 1D11 as the immunoprecipitating antibody, followed by pSer and pThr immunoblots, failed to show any specific bands (results not shown).
In a previous study in which we used gel-free mass spectrometry and mudpit technology, we identified proteins in mouse cardiac lysates that showed specific interactions with NF-SLN (13). One of these proteins was serine/threonine protein kinase 16 (STK16) (Table 2, which is published as supporting information on the PNAS web site). In this study, we overexpressed NF-SLN and STK16 transiently in HEK-293 cells to verify whether these proteins interact. We tried to use the commercial STK16 antibody (Abgent, San Diego) in immunoprecipitation (IP) assays, either as the precipitating antibody or the immunoblotting antibody, but were unable to visualize STK16 in positive control experiments and, therefore, could not perform these assays. As an alternative, we precipitated lysates by using anti-flag antibody and subjected the IP eluates to SDS/PAGE and Coomassie blue staining (Fig. 5C). In the SERCA1, NF-SLN, and STK16 cotransfection experiments, specific bands at ≈110 kDa, ≈42 kDa, 40 kDa, and 35 kDa were evident. SERCA2 is a 110-kDa protein that interacts with SLN, and STK16 is a 35-kDa protein that migrates on SDS/PAGE gels between 35 kDa and 42 kDa. Similar size bands were not evident in the M2 immunoprecipitations from SERCA1, or SERCA1 plus STK16 lysates, nor from the lysates with SERCA1 coexpressed with NF-SLN (13).
Functional consequences of STK16 on SLN and SERCA1 were studied by Ca2+ uptake assays by using an assay of the Ca2+ dependence of Ca2+ transport in isolated microsomes from HEK-293 cells transfected with SERCA1 (S1) alone, S1 and NF-SLN, and S1 coexpressed with NFSLN and STK16. SERCA1 alone had an apparent KCa of 6.50 ± 0.07 pCa units but with NF-SLN, it had a significantly lower (P < 0.05) apparent KCa of 6.13 ± 0.09 pCa units (Fig. 6A and C). The coexpression of both NF-SLN and STK16 with SERCA1 led to reversal of the inhibitory properties of NF-SLN so that the affinity of SERCA1 for Ca2+ was nearly identical to that observed for SERCA1 alone (KCa of 6.44 ± 0.08). We next identified which of the potential sites on SLN STK16 might phosphorylate according to the netphos2.0 computational algorithm. Serine-4 and/or threonine-5 were identified to be candidate phosphorylation sites (Fig. 7). The sites in our rabbit SLN cDNA were mutated to generate Ser4Ala and Thr5Ala mutants, each of which was cotransfected with SERCA1 and/or STK16 (Fig. 6C) into HEK cells. The S4A mutant inhibited SERCA1 (KCa of 6.40 ± 0.09), although it was not as effective as NF-SLN. The inhibitory activity of S4A was relieved by coexpression with STK16 (KCa of 6.58 ± 0.02; Fig. 6C). T5A did not inhibit SERCA1 (KCa of 6.58 ± 0.05), and no differences were observed when STK16 was coexpressed (KCa of 6.61 ± 0.05; Fig. 6C). Immunoblots were performed to verify expression of these constructs and are shown in Fig. 6D.
Fig. 6.
STK16 regulates SLN inhibition of SERCA. (A) Ca2+ uptake assays were performed in the presence or absence of cotransfection with STK16 and NF-SLN. (B) Putative phosphorylation sites in SLN were identified to be serine-4 and threonine-5 (Fig. 7). These amino acids were subjected to site-directed alanine mutagenesis to generate Ser4Ala (S4A) and Thr5Ala (T5A) mutants. Ca2+ uptake assays for these mutants were performed in the presence or absence of STK16. Analyses of NF-SLN mutants, S4A and T5A, in the presence or absence of STK16 cotransfection. (C) KCa (pCa) is the negative logarithm of the Ca2+ concentration required to attain the half-maximal Ca2+ uptake rate. (D) S4A and T5A mutants detected by M2 immunoblots. Data are mean ± SEM. ∗, P < 0.05 versus SERCA1 alone.
Assessment of Cardiac Function.
To examine the effect of NF-SLN overexpression on cardiac function, echocardiography and hemodynamic measurements were performed in 10- to 12-week-old mice (Table 1). NF-SLN Tg mice have mild hypertrophy, although there were no functional differences between the PLN KO and NF-SLN/PLN KO hearts. Similar results were obtained from 8-month-old animals (results not shown). Based on the lack of differences between these groups, echocardiographic and hemodynamic measurements after ISO treatment were not performed.
Table 1.
Echocardiographic and hemodynamic parameters in SLN overexpressing transgenic mice on wild-type and PLN KO background
| Parameter | WT | NF-SLN | PLN KO | NF-SLN/PLNKO |
|---|---|---|---|---|
| Age, wk | 10–12 | 10–12 | 10–12 | 10–12 |
| n | 8 | 8 | 8 | 8 |
| Gender | M | M | M | M |
| H/BW, mg/g | 4.1 ± 0.1 | 4.6 ± 0.1* | 4.2 ± 0.1 | 4.3 ± 0.1 |
| H/TL, mg/mm | 6.1 ± 0.2 | 7.4 ± 0.2* | 6.2 ± 0.2 | 6.2 ± 0.2 |
| HR, bpm | 558 ± 9 | 561 ± 6 | 563 ± 9 | 565 ± 9 |
| AW, mm | 0.70 ± 0.01 | 0.71 ± 0.01 | 0.69 ± 0.01 | 0.69 ± 0.01 |
| PW, mm | 0.70 ± 0.01 | 0.71 ± 0.01 | 0.69 ± 0.01 | 0.69 ± 0.01 |
| LVEDD, mm | 4.06 ± 0.05 | 4.09 ± 0.06 | 4.06 ± 0.04 | 4.08 ± 0.03 |
| LVESD, mm | 1.92 ± 0.03 | 2.33 ± 0.05* | 1.88 ± 0.02 | 1.93 ± 0.03 |
| FS, % | 53.4 ± 0.8 | 43.1 ± 1.2* | 54.1 ± 1.1 | 52.9 ± 0.9 |
| VCFc, circ/s | 10.8 ± 0.2 | 8.4 ± 0.23* | 13.1 ± 0.24# | 12.3 ± 0.25# |
| PAVc, cm/s | 106.9 ± 1.5 | 93.6 ± 1.5* | 123.7 ± 2.2# | 121.5 ± 1.9# |
| AVA, m/s2 | 62.7 ± 1.0 | 54.2 ± 0.9* | 90.4 ± 2.8# | 86.2 ± 2.9# |
| E wave, cm/s | 83.6 ± 1.4 | 73.8 ± 1.2* | 92.2 ± 1.7# | 91.6 ± 1.1# |
| LVESP, mmHg | 121.2 ± 7.1 | 116.9 ± 3.9 | 119.5 ± 4.1 | 117.1 ± 3.5 |
| LVEDP, mmHg | 2.7 ± 1.4 | 6.0 ± 1.9 | 2.5 ± 1.8 | 6.5 ± 2.1 |
Values are mean ± SEM. HR, heart rate; H/BW, heart/body weight ratio; H/TL, heart/tibial length ratio; AW, anterior wall thickness; PW, posterior wall thickness (left ventricle); LVEDD, left ventricular end diastolic dimension LVESD, left ventricular end systolic dimension, respectively; FS, fractional shortening (LVEDD − LVESD)/LVEDD × 100%; ETC, ejection time corrected for HR; VCFc, velocity of circumferential shortening corrected for HR (FS/ETC); PAVC, peak aortic velocity corrected for HR; AVA, aortic velocity acceleration (PAVc/acceleration time); E wave, early-filling transmitral diastolic wave; LVESP, left ventricular end systolic pressure; LVEDP, left ventricular end diastolic pressure. ∗, P < 0.05 compared with all other groups; #, P < 0.05 compared with WT.
Discussion
In this study, we show that: (i) SERCA2a is inhibited by NF-SLN in vivo as effectively as by PLN; (ii) NF-SLN inhibition of SERCA2a can be regulated by phosphorylation; (iii) the serine/threonine kinase STK16 regulates NF-SLN phosphorylation; and (iv) Thr-5 of NF-SLN appears to be the critical amino acid in STK16-dependent phosphorylation.
Interest in SLN in the heart has centered largely on the fact that PLN and SLN form a superinhibitory binary complex that supershifts calcium affinity for SERCA2a (6, 7) and could have serious consequences for cardiac contractility and progression to failure. However, because most relevant experiments were performed in a heterologous experimental system, i.e., HEK293 cells, it was not clear what the effects would be in cardiac muscle. In a previous study, we generated TG mice with the cardiac-specific overexpression of NF-SLN as a first step in the investigation of a potential role for SLN in cardiac function. We observed a phenotype in the NF-SLN transgenic consistent with the cell culture work: a decreased affinity of SERCA2a for Ca2+; impaired contractility as determined by hemodynamic and echocardiographic assessments, reduced amplitude of Ca2+ transients, and longer decay time for the Ca2+ transient (10). These experiments confirmed that the coexpression of SLN and PLN are superinhibitory and affect Ca2+ cycling in cardiac muscle. The impact of SLN alone on cardiac muscle remained unknown. As a result, we mated the Tg mice onto the PLN KO line to obtain cardiac specific NF-SLN expression on a PLN null background.
The Ca2+ uptake assays of isolated microsomes from ventricular tissues of WT, NF-SLN Tg/PLN KO, and PLN KO mice showed that NF-SLN inhibits SERCA2a as effectively as PLN, i.e., the KCa of NF-SLN Tg/PLN KO (expression of only NF-SLN in the ventricle) is identical to WT mice (expression of only PLN in the ventricle) (Fig. 2A). Consistent with these findings, NFSLN-Tg/PLN KO mice display reduced contractility in isolated cardiomyocytes and decreased amplitude and longer decay times of the Ca2+ transient.
The coexpression of NF-SLN with 2× PLN overexpressing or PLN I40A transgenic mice failed to induce further inhibition of SERCA2a. These results indicate that further inhibition of Ca2+ uptake via the ternary SLN/PLN/SERCA complex does not occur. This threshold of SERCA inhibition most likely results from saturation of binding kinetics and on conformational constraints of SERCA2a (6, 9). We did not observe progression to overt heart failure in any of the mouse lines. These results can be evaluated in relation to results obtained by the overexpression of superinhibitory and other mutant forms of PLN in Tg mouse hearts. The 2-fold overexpression of WT PLN reduced cardiac contractility but did not progress to cardiac hypertrophy (14). Overexpression of the superinhibitory mutants PLN L37A and PLN I40A impaired contractility and induced minimal hypertrophy but did not lead to heart failure (15). Overexpression of the PLN N27A superinhibitory mutant impaired contractility more severely, and function was not fully restored by ISO (16). By contrast, the PLN R9C mutant (17) was not superinhibitory, but its failure to be phosphorylated resulted in a depressed response to β-adrenergic agonists. This mutation led to dilated cardiomyopathy by 24 weeks. In summary, these results suggest that superinhibition of SERCA2a will induce cardiac hypertrophy but will not progress to heart failure if β-adrenergic stimulation of the heart can reverse SERCA2a inhibition.
Our data showing that NF-SLN Tg or the NF-SLN Tg/PLN KO mice do not develop cardiac hypertrophy may be explained by their unanticipated ability to respond to β-adrenergic stimulation. In fact, the significant inhibition of Ca2+ uptake and Ca2+ transients that are not translated into significant changes at the level of the whole heart would be consistent with this idea. For instance, cellular compensation may occur in the heart, likely as a result of phosphorylation-dependent relief of NF-SLN inhibition. This compensation may be explained by our data showing STK16 phosphorylation of NF-SLN and its effect on SERCA2a activity.
The physiological role of STK16 in the regulation of SLN remains to be addressed. Serine threonine kinase 16, also referred to as PKL12 (protein kinase expressed in day 12 fetal liver; kinase related to Saccharomyces cerevisiae and Arabidopsis thaliana, embryo-derived protein kinase, myristoylated and palmitoylated serine-threonine kinase-1, and TGF-β stimulated factor 1) is a ubiquitously expressed ≈35 kDa protein kinase that acts on both serine and threonine residues, although it appears to prefer threonine (18–21). In our previous studies, STK16 was shown to be a SLN-interacting protein, but the functional significance of the interaction was not clear at the time (13). The results of this study showing that SLN is responsive to ISO treatment suggested that SLN may be phosphorylated and suggested an involvement of STK16 in SLN function. In fact, ISO acting through cAMP signaling to regulate SLN activity would be entirely consistent with a previous study showing that STK16 is regulated by TGF-β activation and cAMP signaling (22).
In the normal adult heart, SLN is expressed in the atria preferentially, with little expression at the mRNA or protein level in the ventricle (23, 24), and this pattern of SLN expression is not affected in the PLN KO mouse (24). The lack of stimulation by ISO, which should lead to phosphorylation and loss of the inhibitory effects of any endogenous SLN expressed in PLN KO hearts, is consistent with this pattern of expression because the PLN KO mice did not show any potential phosphorylation products or any Ca2+-transient response to ISO treatment. These data indicate that, in the normal heart, there is little physiological role for SLN in the ventricle. It is possible that under certain developmental or disease conditions, SLN expression in the ventricle may be increased, resulting in the formation of a SLN/PLN/SERCA superinhibitory complex. However, such conditions have not been described to date. Alternatively, the importance of the SLN/SERCA and the SLN/PLN/SERCA complex may become manifest in tissues such as the atria, where the three proteins are known to exist, or skeletal muscle, where SLN may be expressed in higher concentrations (24).
The inability of the T5A mutant to show any inhibitory function on SERCA suggests that this amino acid is crucial for SLN function, perhaps by providing a stable binding interaction site with SERCA2a. It would be consistent with the view that phosphorylation of the threonine by STK16 would mimic the conditions seen with the T5A mutant alone, and the role of Thr-5 then becomes one of an interaction site with SERCA. The 100% conservation of T5 in SLN across six species supports the view that this amino acid has a key role in SLN function. Future studies examining cross-linking between SLN T5 and SERCA would provide a structural model for this interaction.
Materials and Methods
Immunoblots and IPs.
Monoclonal antibody against PLN (1D11) and phosphorylated PLN (A285) were kindly provided by R. Johnson (Merck, West Point, PA). mAB IID8F6 against SERCA2 was a gift from K. Campbell (University of Iowa, Iowa City). All other antibodies were obtained commercially: FLAG antibody, M2 (Sigma); sodium calcium exchanger polyclonal antibody (pAb), NCX1 (Abcam, Inc., Cambridge, MA); dihydropyridine receptor α 1 subunit mAb (Affinity BioReagents, Golden, CO); ryanodine receptor 2 mAb, α-actinin mAb (Sigma), and phospho-serine and phospho-threonine specific pAbs (Research Diagnostics, Concord, MA). IPs were performed as described in refs. 5, 8, and 25. Postnuclear homogenates (250 μl) from ventricular samples were subjected to IP by using 10 μg of M2 antibody conjugated to 50 μl of protein-G Sepharose and incubated overnight with 500 μl of Seize-X Protein G IP buffer (Pierce), 7.5 μl of Nonidet P-40, and 7.5 μl of activated sodium orthovanadate. Samples were washed with IP buffer and eluted with 190 μl Seize-X Elution buffer (Pierce), separated by SDS/PAGE gels and either transferred to nitrocellulose for immunoblotting or stained by using Coomassie brilliant blue.
Transgenic Mice.
NF-SLN transgenic mice were described in ref. 10. The PLN KO and 2× PLN overexpressing mice were obtained from Evangelia Kranias (University of Cincinnati, Cincinnati), and the I40A PLN transgenic was described in ref. 15. NF-SLN transgenic mice were mated onto PLN KO backgrounds (26), PLN I40A superinhibitory transgenics (15), and 2× PLN overexpressing transgenic mice (14) to obtain NF-SLN overexpression on a PLN null background, or in the presence of WT PLN overexpression or I40A superinhibitor overexpression.
Cardiac Lysates.
Mouse ventricles were isolated, washed in PBS, then homogenized for 30 sec in 250 mM sucrose/50 mM Tris·HCl pH 7.4/1 mM PMSF/20 μg/ml aprotinin by using a Polytron homogenizer set to maximum. Lysates were isolated after a 15-min centrifugation at 8000 × g, and the supernatant was collected and used.
Ca2+ Transport.
ATP and oxalate-dependent Ca2+ uptake activity in cardiac lysates was measured by using a Millipore filtration method (10, 13). The Ca2+ concentration required for half-maximal velocity for Ca2+ uptake (EC50) was determined by nonlinear curve fitting.
In Vivo Echocardiographic and Hemodynamic Assessment of Cardiac Function.
M mode and Doppler echocardiography were performed for noninvasive assessment of left ventricular function and dimensions by using methods described in refs. 15 and 27–29.
Single Myocyte Studies.
Mouse ventricular myocytes were isolated and studied as described in refs. 10 and 30. For the ISO studies, ISO (1 μM) was added to the perfusate. For all experiments, at least 15 cells per group were analyzed.
cDNAs and Other Reagents.
The rabbit NF-SLN expression vectors were described in refs. 5, 6, and 8. Full-length human STK16 cDNA (no. TC108468) was obtained from OriGene Technologies, Rockville, MD. Mutagenesis of NF-SLN to generate NF-SLN S4A and T5A mutants was performed as described in ref. 13.
Statistics.
Data are presented as mean ± SEM. In all cases, results shown are from a minimum of three separate experiments. Comparisons were by Student’s t test or ANOVA followed by appropriate post hoc test. P values <0.05 were considered significant.
Supplementary Material
Acknowledgments
This work was supported by research grants from the Heart and Stroke Foundation Ontario (HSFO) (D.H.M. and P.H.B.) and the Canadian Institutes of Health Research (CIHR) (D.H.M. and P.H.B.). A.O.G. and M.G.T. were Research Fellows of Heart and Stroke Foundation of Canada. We also acknowledge fellowships from Tailored Advanced Collaborative Training in Cardiovascular Science (M.G.T. and G.Y.O.) and from the CIHR (G.Y.O.). P.H.B. is a Career Investigator of HSFO.
Glossary
Abbreviations:
- IP
immunoprecipitation
- ISO
isoproterenol
- NF-SLN
sarcolipin tagged N-terminally with a FLAG epitope
- PLN
phospholamban
- SERCA
sarco(endo)plasmic reticulum Ca2+-ATPase
- SLN
sarcolipin
- STK
serine/threonine protein kinase
- TG
transgenic.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Simmerman H. K., Jones L. R. Physiol. Rev. 1998;78:921–947. doi: 10.1152/physrev.1998.78.4.921. [DOI] [PubMed] [Google Scholar]
- 2.Wawrzynow A., Theibert J. L., Murphy C., Jona I., Martonosi A., Collins J. H. Arch. Biochem. Biophys. 1992;298:620–623. doi: 10.1016/0003-9861(92)90457-8. [DOI] [PubMed] [Google Scholar]
- 3.Odermatt A., Taschner P. E., Scherer S. W., Beatty B., Khanna V. K., Cornblath D. R., Chaudhry V., Yee W. C., Schrank B., Karpati G., et al. Genomics. 1997;45:541–553. doi: 10.1006/geno.1997.4967. [DOI] [PubMed] [Google Scholar]
- 4.MacLennan D. H., Yip C. C., Iles G. H., Seeman P. The Mechanism of Muscle Contraction. Vol. 37. Plainview, NY: Cold Spring Harbor Lab. Press; 1972. pp. 469–478. [Google Scholar]
- 5.Odermatt A., Becker S., Khanna V. K., Kurzydlowski K., Leisner E., Pette D., MacLennan D. H. J. Biol. Chem. 1998;273:12360–12369. doi: 10.1074/jbc.273.20.12360. [DOI] [PubMed] [Google Scholar]
- 6.Asahi M., Kurzydlowski K., Tada M., MacLennan D. H. J. Biol. Chem. 2002;277:26725–26728. doi: 10.1074/jbc.C200269200. [DOI] [PubMed] [Google Scholar]
- 7.Asahi M., Sugita Y., Kurzydlowski K., De Leon S., Tada M., Toyoshima C., MacLennan D. H. Proc. Natl. Acad. Sci. USA. 2003;100:5040–5045. doi: 10.1073/pnas.0330962100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kimura Y., Kurzydlowski K., Tada M., MacLennan D. H. J. Biol. Chem. 1997;272:15061–15064. doi: 10.1074/jbc.272.24.15061. [DOI] [PubMed] [Google Scholar]
- 9.Toyoshima C., Asahi M., Sugita Y., Khanna R., Tsuda T., MacLennan D. H. Proc. Natl. Acad. Sci. USA. 2003;100:467–472. doi: 10.1073/pnas.0237326100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Asahi M., Otsu K., Nakayama H., Hikoso S., Takeda T., Gramolini A. O., Trivieri M. G., Oudit G. Y., Morita T., Kusakari Y., et al. Proc. Natl. Acad. Sci. USA. 2004;101:9199–9204. doi: 10.1073/pnas.0402596101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chu G., Ferguson D. G., Edes I., Kiss E., Sato Y., Kranias E. G. Ann. N.Y. Acad. Sci. 1998;853:49–62. doi: 10.1111/j.1749-6632.1998.tb08256.x. [DOI] [PubMed] [Google Scholar]
- 12.Li L., Chu G., Kranias E. G., Bers D. M. Am. J. Physiol. 1998;274:H1335–H1347. doi: 10.1152/ajpheart.1998.274.4.H1335. [DOI] [PubMed] [Google Scholar]
- 13.Gramolini A. O., Kislinger T., Asahi M., Li W., Emili A., MacLennan D. H. Proc. Natl. Acad. Sci. USA. 2004;101:16807–16812. doi: 10.1073/pnas.0407815101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kadambi V. J., Ponniah S., Harrer J. M., Hoit B. D., Dorn G. W., 2nd, Walsh R. A., Kranias E. G. J. Clin. Invest. 1996;97:533–539. doi: 10.1172/JCI118446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zvaritch E., Backx P. H., Jirik F., Kimura Y., de Leon S., Schmidt A. G., Hoit B. D., Lester J. W., Kranias E. G., MacLennan D. H. J. Biol. Chem. 2000;275:14985–14991. doi: 10.1074/jbc.275.20.14985. [DOI] [PubMed] [Google Scholar]
- 16.Zhai J., Schmidt A. G., Hoit B. D., Kimura Y., MacLennan D. H., Kranias E. G. J. Biol. Chem. 2000;275:10538–10544. doi: 10.1074/jbc.275.14.10538. [DOI] [PubMed] [Google Scholar]
- 17.Schmitt J. P., Kamisago M., Asahi M., Li G. H., Ahmad F., Mende U., Kranias E. G., MacLennan D. H., Seidman J. G., Seidman C. E. Science. 2003;299:1410–1413. doi: 10.1126/science.1081578. [DOI] [PubMed] [Google Scholar]
- 18.Berson A. E., Young C., Morrison S. L., Fujii G. H., Sheung J., Wu B., Bolen J. B., Burkhardt A. L. Biochem. Biophys. Res. Commun. 1999;259:533–538. doi: 10.1006/bbrc.1999.0811. [DOI] [PubMed] [Google Scholar]
- 19.Ligos J. M., Gerwin N., Fernandez P., Gutierrez-Ramos J. C., Bernad A. Biochem. Biophys. Res. Commun. 1998;249:380–384. doi: 10.1006/bbrc.1998.9163. [DOI] [PubMed] [Google Scholar]
- 20.Stairs D. B., Perry Gardner H., Ha S. I., Copeland N. G., Gilbert D. J., Jenkins N. A., Chodosh L. A. Hum. Mol. Genet. 1998;7:2157–2166. doi: 10.1093/hmg/7.13.2157. [DOI] [PubMed] [Google Scholar]
- 21.Kurioka K., Nakagawa K., Denda K., Miyazawa K., Kitamura N. Biochim. Biophys. Acta. 1998;1443:275–284. doi: 10.1016/s0167-4781(98)00224-3. [DOI] [PubMed] [Google Scholar]
- 22.Ohta S., Takeuchi M., Deguchi M., Tsuji T., Gahara Y., Nagata K. Biochem. J. 2000;350:395–404. [PMC free article] [PubMed] [Google Scholar]
- 23.Minamisawa S., Wang Y., Chen J., Ishikawa Y., Chien K. R., Matsuoka R. J. Biol. Chem. 2003;278:9570–9575. doi: 10.1074/jbc.m213132200. [DOI] [PubMed] [Google Scholar]
- 24.Vangheluwe P., Schuermans M., Zador E., Waelkens E., Raeymaekers L., Wuytack F. Biochem. J. 2005;389:151–159. doi: 10.1042/BJ20050068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Asahi M., Kimura Y., Kurzydlowski K., Tada M., MacLennan D. H. J. Biol. Chem. 1999;274:32855–32862. doi: 10.1074/jbc.274.46.32855. [DOI] [PubMed] [Google Scholar]
- 26.Luo W., Grupp I. L., Harrer J., Ponniah S., Grupp G., Duffy J. J., Doetschman T., Kranias E. G. Circ. Res. 1994;75:401–409. doi: 10.1161/01.res.75.3.401. [DOI] [PubMed] [Google Scholar]
- 27.Crackower M. A., Oudit G. Y., Kozieradzki I., Sarao R., Sun H., Sasaki T., Hirsch E., Suzuki A., Shioi T., Irie-Sasaki J., et al. Cell. 2002;110:737–749. doi: 10.1016/s0092-8674(02)00969-8. [DOI] [PubMed] [Google Scholar]
- 28.Crackower M. A., Sarao R., Oudit G. Y., Yagil C., Kozieradzki I., Scanga S. E., Oliveira-dos-Santos A. J., da Costa J., Zhang L., Pei Y., et al. Nature. 2002;417:822–828. doi: 10.1038/nature00786. [DOI] [PubMed] [Google Scholar]
- 29.Oudit G. Y., Sun H., Kerfant B. G., Crackower M. A., Penninger J. M., Backx P. H. J. Mol. Cell Cardiol. 2004;37:449–471. doi: 10.1016/j.yjmcc.2004.05.015. [DOI] [PubMed] [Google Scholar]
- 30.Wickenden A. D., Kaprielian R., Parker T. G., Jones O. T., Backx P. H. J. Physiol. 1997;504:271–286. doi: 10.1111/j.1469-7793.1997.271be.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.