Abstract
The SWIRM domain is a module found in the Swi3 and Rsc8 subunits of SWI/SNF-family chromatin remodeling complexes, and the Ada2 and BHC110/LSD1 subunits of chromatin modification complexes. Here we report the high-resolution crystal structure of the SWIRM domain from Swi3 and characterize the in vitro and in vivo function of the SWIRM domains from Saccharomyces cerevisiae Swi3 and Rsc8. The Swi3 SWIRM forms a four-helix bundle containing a pseudo 2-fold axis and a helix–turn–helix motif commonly found in DNA-binding proteins. We show that the Swi3 SWIRM binds free DNA and mononucleosomes with high and comparable affinity and that a subset of Swi3 substitution mutants that display growth defects in vivo also show impaired DNA-binding activity in vitro, consistent with a nucleosome targeting function of this domain. Genetic and biochemical studies also reveal that the Rsc8 and Swi3 SWIRM domains are essential for the proper assembly and in vivo functions of their respective complexes. Together, these studies identify the SWIRM domain as an essential multifunctional module for the regulation of gene expression.
Keywords: chromatin regulation, DNA binding, RSC, SWI/SNF
DNA within the nucleus of the cell is wrapped around octamers of histone proteins, called nucleosomes that allow for compaction of DNA and assist in gene regulation. Each nucleosome core particle contains two copies of the four histone proteins H2A, H2B, H3, and H4 with almost two turns of DNA encircling its perimeter (1). Nucleosomes form arrays that fold into higher-order structures called chromatin. Chromatin structure is remarkably dynamic and its state of compaction is correlated with the state of gene expression; more compact chromatin is associated with gene silencing and less compact chromatin is associated with gene expression (2, 3). Two distinct classes of enzymes mediate the regulation of chromatin structure and in turn gene expression. The first class of enzymes utilizes the energy of ATP hydrolysis to reorganize or remodel chromatin (4). These enzymes are complexed with other proteins, although the specific mechanistic contribution of these associated subunits is not well understood. The second class of enzymes that regulate chromatin mediate the posttranslational modification of the histone proteins, predominantly on their N-terminal tails (5). These enzymes mediate the transfer or hydrolysis of acetyl, phosphate, methyl, or ubiquitin groups to specific histone residues; and are also often present in complexes with additional subunits that may serve to regulate enzymatic activity or complex targeting (6).
Remodeling and modifying complexes bear subunits/domains that mediate specific protein–protein and protein–nucleic acid interactions critical for their appropriate regulation of chromatin. Examples of such modules are bromodomains, chromodomains, and SANT domains (7–11). Bromodomains and chromodomains are found within the catalytic proteins of a subset of histone acetyltransferases, remodeling ATPases, and other transcription factors. Bromodomains specifically target acetyl-lysine residues (7), whereas chromodomains target methyl-lysine residues (10) or nucleic acids (12). Structures of bromodomains and chromodomains bound to their cognate protein targets have informed the mechanism of substrate recognition, although the functional consequence of these interactions is still not well established.
The SWIRM domain has recently been identified as an additional conserved module present in multiple remodeling and modifying complexes, but of entirely unknown function (13). This domain is named for its presence in the proteins Swi3, Rsc8, and Moira, which are homologous members in the SWI/SNF-family of ATP-dependent chromatin remodeling complexes SWI/SNF, RSC, and BRM, respectively. However, the SWIRM domain is also present in many other chromatin regulatory proteins such as the Gcn5 histone acetyltransferase subunit Ada2, and the recently identified histone demethylase BHC110/LSD1 (14, 15). Additionally, this domain is found in other proteins associated with ubiquitin-mediated protein degradation (13). Secondary structure predictions, based on multiple alignments indicate the presence of four distinct α-helices, suggesting that the domain may form a tetrahelical bundle. However, this domain shows no sequence conservation patterns with other α-helical bundles associated with transcriptional regulators such as the bromodomain or the SANT domain.
In this study, we report the crystal structure of the SWIRM domain from yeast Swi3 and use the structure as a scaffold to carry out a functional analysis of the SWIRM domain from yeast Swi3 and Rsc8. The Swi3 SWIRM structure reveals a globular helical domain containing an embedded helix–turn–helix motif, often associated with DNA-binding proteins. Functional analysis of the Swi3 and Rsc8 SWIRM domains and selected surface mutations in vivo and in vitro is consistent with a role for the SWIRM domain in protein and complex stability and nucleosome targeting through association with DNA.
Results
Overall Structure of the Swi3 SWIRM Domain and Functional Implications.
We prepared a recombinant protein encoding the SWIRM domain of Swi3 (residues 309–398) by overexpression and purification from Escherichia coli as an N-terminal 6× His fusion. The 6× His-tagged fusion protein was crystallized in space group P212121 containing one molecule per asymmetric unit cell, and the structure was determined by multiwavelength anomalous dispersion using selenomethionine (SeMet)-derivatized protein. The final native structure was refined to 1.4-Å resolution with excellent refinement statistics and geometrical parameters (Table 1).
Table 1.
Data collection, phasing, and refinement statistics for Swi3 SWIRM domain
| Data set | SeMet MAD |
Native | ||
|---|---|---|---|---|
| Peak | Edge | Remote | ||
| Space group | PB212121 | P212121 | ||
| Cell parameters, Å | a = 44.226 | a = 44.086 | ||
| b = 44.309 | b = 44.409 | |||
| c = 48.792 | c = 48.687 | |||
| Wavelength, Å | 0.9788 | 0.9791 | 0.9562 | 1.1 |
| Resolution, Å | 50–1.75 | 50–1.75 | 50–1.75 | 50–1.40 |
| Unique reflections | 10,127 | 9,903 | 10,126 | 19,520 |
| Completeness, % | 99.7 (97.6) | 97.5 (78.5) | 99.5 (95.6) | 99.8 (99.8) |
| Multiplicity | 11.2 (8.7) | 10.8 (5.7) | 11.2 (8.3) | 13.1 (5.5) |
| I/σ | 25.9 (7.3) | 27.4 (3.5) | 31.9 (6.1) | 30.1 (5.6) |
| Rmerge, %* | 6.3 (27.9) | 6.6 (38.8) | 5.7 (30.7) | 6.2 (37.2) |
| Phasing | ||||
| Number of sites | 1 | |||
| Figure of merit (SOLVE) | 0.73 | |||
| Refinement | ||||
| Resolution | 50–1.4 | |||
| Rfree, %† | 21.3 | |||
| Rworking, %‡ | 20.7 | |||
| Number of atoms | ||||
| Protein | 722 | |||
| Water | 123 | |||
| B-factor (Å2) | 24.4 | |||
| rms deviations | ||||
| Bond length, Å | 0.0047 | |||
| Bond angle, ° | 1.04 | |||
MAD, multiwavelength anomalous dispersion.
*Rmerge = Σ|I − 〈I〉|/Σ〈I〉.
†Rfree = ΣT||Fo| − |Fc||/ΣTb|Fo| (where T is a test data set of 10% of the total reflections randomly chose and set aside in prior to refinement).
‡Rworking = Σ||Fo| − |Fc||/Σ|Fo|.
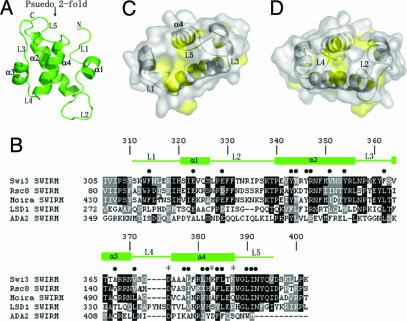
Residues 311–395 of the Swi3 SWIRM domain are well resolved in the final electron density map, whereas the N-terminal residues 309–310 and the 6× His tag and C-terminal residues 396–398 are not visible and are presumably disordered. The structure forms a globular antiparallel four helical bundle with the N and C termini on the same side of the domain (Fig. 1A). Helices 1 and 3 are relatively short, one and a half and two turns long, and helices 2 and 4 are longer with a length between three and five turns. There is a pseudo 2-fold axis that relates helices 1 and 2 to helices 3 and 4, although the loops that connect the helices do not obey this pseudosymmetry. Surprisingly, the loops connecting the helices are all well ordered and comprise ≈50% of the structure. The 13-residue L2 loop connecting helices 1 and 2 is at one end of the molecule and the N- and C-terminal loops that are 10 and 8 residues in length, respectively, are at the opposite end of the molecule.
Fig. 1.
Structure of the Swi3 SWIRM domain and sequence alignment of SWIRM proteins. (A) Overall structure of the Swi3 SWIRM domain. Helices 1–4 and loops 1–5 are denoted according to their order of appearance from the N to C termini, and the psuedo 2-fold axis of the domain is indicated. (B) Sequence alignment of a subset of SWIRM domains. The closest Swi3 SWIRM homologues, Rsc8 and Moira are shown as well as the SWIRM domains from the Ada2 and BHC110/LSD1 proteins. Indicated above the sequence alignment are the secondary structure elements from the Swi3 SWIM domain, the Swi3 residues that mediate intra-atomic interactions (●), and residues proposed to mediate DNA binding (*). (C) Strictly or highly conserved residues within the SWIRM domain are highlighted in yellow on a surface representation of the SWIRM domain. The L1, L3, and L5 loops that contribute to this region of conservation are indicated. Tryptophan 313 of the L1 loop is also conserved but is removed along with residues 311 and 312 from this view to highlight several conserved residues that are located behind it. The α4 helix is also indicated. (D) Opposite view from C highlighting a second region of conservation that maps to the L2 and L4 that are indicated.
A correlation of the structure to sequence conservation within the SWIRM domains reveals that nearly all of the strictly or highly conserved hydrophobic residues are involved in stabilizing the protein tertiary structure strongly, suggesting that each of the SWIRM domains from different proteins have a common tertiary fold (Fig. 1B). Interestingly, many of the other conserved polar residues map to solvent exposed residues in or near the loop regions resulting in two solvent exposed patches on opposite ends of the molecule, one that is flanked by loops L1, L3, and L5 and the other that is flanked by loops L2 and L4 (Fig. 1 C and D). These observations suggest that these two ends of the SWIRM structure may mediate functionally important small molecule and/or macromolecule interactions.
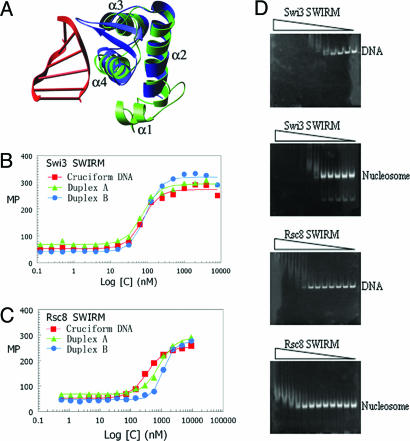
To derive possible functional insights from the SWIRM structure, we queried the Protein Data Bank using the program dali (16) for structural homologues of the SWIRM structure with known function. The highest similarities were observed with the Z-DNA binding domain from ADAR1 (Z score = 5.3, rms deviation = 1.6) (17) and the restriction endonuclease FokI (Z score = 4.7, rms deviation = 3.0) (18). Both structures revealed a high degree of superposition with only the last three α-helices of the Swi3 SWIRM domain structure comprising a helix–turn–helix motif (Fig. 2A). The DNA binding properties of the structural homologues of the Swi3 SWIRM domain led us to test the hypothesis that the Swi3 SWIRM domain may function in nucleic acid binding.
Fig. 2.
The DNA and nucleosome binding properties of the SWIRM domain from Swi3 and Rsc8. (A) The Swi3 SWIRM domain structure (green) is superposed with the ADAR1/DNA complex structure (with ADAR1 blue and the associated Z DNA in red). (B) Results of Swi3 SWIRM domain DNA-binding assays using fluorescence polarization. Recombinant Swi3 protein is titrated against fluorescein-labeled DNA substrates. The DNA substrate conformers used were cruciform and duplex and were prepared as described by Bianchi et al. (18) and summarized in Materials and Methods. The Swi3 SWIRM domain is titrated with a 2-fold dilution series of 7.8 nM to 16 μM. The dissociation constants calculated from the binding data are as follows. Swi3 SWIRM: for cruciform DNA, 75.10 ± 6.26 nM; for duplex A, 72.67 ± 4.83 nM; for duplex B, 90.23 ± 6.83 nM. Rsc8 SWIRM: for cruciform DNA, 340.89 ± 18.73 nM; for duplex A, 829.60 ± 71.57 nM; for duplex B, 1130.37 ± 49.21 nM. (C) Same as in A but using the recombinant Rsc8 SWIRM domain with a protein titration range of 9.0 nM to 18.4 μM. (D) Results of the mobility shift assay using reconstituted mononucleosomes containing a 172-bp DNA duplex or the isolated DNA duplex with increasing concentrations of recombinant SWIRM domain from either Swi3 or Rsc8 with 2-fold serial dilutions ranging from 1.6 μM to 7.8 nM and 1.8 μM to 9.0 nM for the SWIRM domains from Swi3 and Rsc8, respectively.
DNA-Binding Properties of the SWIRM Domain.
To directly test whether the Swi3 SWIRM domain bound to nucleic acid, we initially used a florescence polarization assay in which fluorescein-labeled nucleic acid were used for the binding assay. The nucleic acid substrates included B-form and cruciform DNA conformers (19). As shown in Fig. 2B, the Swi3 SWIRM domain bound tightly to each of these nucleic acid substrates with an apparent dissociation constant ranging from 72 to 90 nM and with no apparent specificity for the type of nucleic acid conformer.
For comparison, we prepared a recombinant version of the SWIRM domain from Rsc8 (residues 84–173) and carried out similar DNA binding studies. As shown in Fig. 2C, the Rsc8 SWIRM domain also has appreciable affinity for nucleic acid, although its apparent affinity is significantly reduced (by ≈4- to 10-fold) relative to the SWIRM domain from Swi3, despite the nearly identical PI values of these SWIRM domains (9.85 for the Swi3 SWIRM domain and 9.52 for the Rsc8 SWIRM domain). These results suggest that one of the functions of the Swi3 SWIRM domain, and to a lesser extent the Rsc8 SWIRM domain, may be to bind nucleic acid with no apparent preference for DNA conformation.
To carry this observation further, we asked whether the recombinant SWIRM domains from Swi3 and Rsc8 could bind to DNA in the context of a nucleosome core particle. For this analysis, we used recombinant yeast histone proteins and a 172-bp DNA duplex to prepare recombinant mononucleosomes. These nucleosomes were used in gel shift assays employing nanomolar concentrations of nucleosomes and titrations between micromolar and nanomolar concentrations of recombinant SWIRM domain from Swi3 and Rsc8. To directly establish the effect of the histone proteins on nucleosome binding by the SWIRM domain, similar gel-shift assays were also carried out by using only the 172-bp DNA duplex used for assembling the recombinant mononucleosome. As can be seen in Fig. 2D, the Swi3 SWIRM domain shifts the mononuclesomes at a concentration of ≈62 nM. With increasing concentrations of protein, the shifted species appears as a heterogeneous smear rather than as a distinct band, suggesting that the binding can occur at many locations along nucleosomal DNA and/or that more than one SWIRM domain can bind to the same nucleosome. Interestingly, the band shift with the isolated 172-bp DNA shows a similar type of band shift at a comparable protein concentration, suggesting that binding is mediated through the direct association with DNA with no significant contribution from the histone proteins. This finding is consistent with isothermal titration calorimetry studies in which binding of the Swi3 SWIRM domains to the N-terminal tail regions from histones H3 and H4 was not detectable (data not shown). Also notable from the band shift assays is that the histone proteins do not present a significant barrier for binding of the Swi3 SWIRM domain. Correlating well with these studies, the apparent dissociation constant calculated from the band shift studies employing the 172-bp DNA is comparable to the dissociation constant calculated by using the fluorescence polarization experiment and employing a 36-bp DNA fragment where a dissociation constant in the range of 72 to 90 nM is calculated.
Consistent with the results of the fluorescence polarization experiments, the SWIRM domain from Rsc8 is also able to band shift nucleosomes and free DNA, albeit ≈10-fold more weakly than the SWIRM domain of Swi3 (Fig. 2D). Taking these in vitro studies together, we suggest that the Swi3 SWIRM domain, and to a lesser extent the Rsc8 SWIRM domain, may function in vivo as a nucleosome targeting module.
In Vivo Function of the SWIRM Domains of Swi3 and Rsc8.
Swi3 and Rsc8 are paralogs present in either the SWI/SNF or RSC remodeling complexes, respectively. These two complexes are highly similar in composition and activities but have separate functions in vivo, as exemplified by their distinct mutant phenotypes and different remodeling targets. Loss of Swi3 or Rsc8 confers a full loss of SWI/SNF or RSC function, respectively: swi3Δ confers all swi/snf phenotypes (detailed below), whereas rsc8Δ confers lethality, due to the essential nature of RSC (20). To determine the importance of the SWIRM domain for Swi3 and Rsc8 function, we initially prepared a Swi3 derivative lacking the SWIRM domain (swi3ΔS) and tested for complementation of swi3Δ phenotypes. We found that swi3ΔS failed to complement any swi/snf phenotypes, consistent with a requirement for the SWIRM domain for Swi3 function (Fig. 3A). For example, swi3ΔS mutants failed to grow on a medium containing raffinose, as SWI/SNF remodeling of SUC2 (which encodes invertase) is required for raffinose utilization (21). Further tests of swi3ΔS were consistent with a complete loss of SWI/SNF function (Table 2 and Table 3, which is published as supporting information on the PNAS web site).
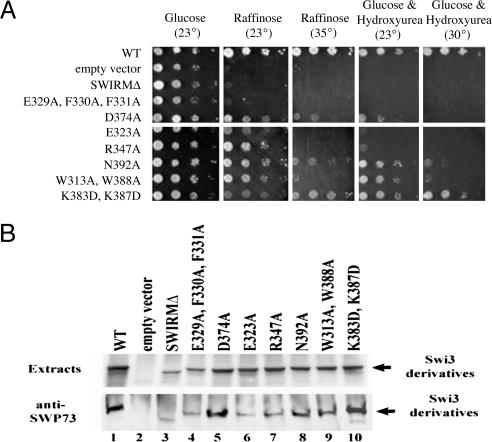
Fig. 3.
Growth phenotypes and stability/assembly properties of swi3 SWIRM mutants. (A) Growth phenotypes of SWIRM domain mutants. A swi3δ strain (YBC2051) was transformed with LEU2-bearing plasmids containing either SWI3 or swi3S derivatives, and tested for growth ability on various media and conditions (as indicated). (B) Stability and assembly properties of SWIRM domain mutants. Swi3 derivative expression (Upper) and Swi3 derivative assembly into SWI/SNF (Lower) are shown.
To determine whether swi3ΔS encoded a stable protein, we performed immunoblot analyses and observed a significant decrease in Swi3ΔS protein levels (Fig. 3B). Furthermore, instability was also observed with rsc8 SWIRM mutants (data not shown), suggesting that the SWIRM domain is important for Swi3/Rsc8 structural stability. Thus, further study of the SWIRM domain required the preparation of mutations that impair function but retain stability. To this end, we prepared single (or multiple) amino acid substitutions in the SWIRM domains of Swi3 (swi3S) or Rsc8 (rsc8S). Here, we prepared a large series of site directed mutations, with residue substitutions guided by the Swi3 structure and SWIRM domain alignments. For the Swi3 SWIRM, we prepared 28 different swi3S alleles (encoding 22 single or six multiple substitutions). The eight SWIRM domain mutants that showed strong to moderate phenotypes are presented in Fig. 3A, and the remainder are summarized in Table 3. Importantly, many of the swi3S mutants displayed the full spectrum of swi/snf mutant phenotypes, with varying expressivity. Analogous rsc8S mutations (SSW# for Swi3 SWIRM correspond to RSW# for Rsc8 SWIRM; Table 4, which is published as supporting information on the PNAS web site) conferred phenotypes ranging from slow growth to inviability, with some conferring temperature dependence. Importantly, residue replacements at equivalent positions in the Swi3 and Rsc8 SWIRM domains confer similar levels of impairment, suggesting structural and functional conservation between Swi3 and Rsc8 SWIRM domains.
Impact of SWIRM Domain Mutations in Vivo.
Many of the highly conserved residues of the SWIRM domain are buried within the hydrophobic core of the Swi3 SWIRM structure, and are therefore likely to be important for domain folding. Consistent with this, we find that replacements involving core residues conferred greater growth deficiencies than surface replacements (Fig. 1B and Table 2).
Table 2.
In vivo properties of Swi3 SWIRM domain mutants
| Mutation | Raf, °C | Ino, °C | Made and assembled into Swi/SNf |
|---|---|---|---|
| WT | None | None | Yes |
| Empty vector | 23 | 23 | No |
| SWIRM deletion | 23 | 23 | Yes |
| E329A, F330A, F331A | 23 | 23 | Yes |
| D374A | 23 | 30 | Yes |
| E317A | None | None | ND |
| E323A | 23 | 23 | Yes |
| Y346A | None | None | ND |
| R347A | 23 | 23 | Yes |
| N348A | 23 | None | ND |
| L378A | 33 | None | ND |
| L381A | 23 | None | ND |
| L385A | 23 | None | ND |
| W388A | 23 | None | ND |
| L390A | 23 | None | ND |
| W313A | 23 | 35 | Yes |
| E329A | 23 | 33 | Yes |
| N357A | None | None | Yes |
| Y361A | None | None | Yes |
| S363A | None | None | Yes |
| N392A | 23 | 33 | Yes |
| Y393A | None | None | Yes |
| Y310A | None | None | ND |
| N315A, K318D | None | None | Yes |
| N333A, R334E | 23 | 35 | Yes |
| S337A | 23 | None | Yes |
| N359A, E360K | 23 | None | Yes |
| K383D | None | None | Yes |
| K387D | None | None | Yes |
| W313A, W388A | 23 | 33 | Yes |
| K383D, K387D | 23 | 35 | Yes |
Media: Raf, YP + 2% raffinose; Ino, SC lacking inositol; HU, YP + 150 mM hydroxyurea (YP, yeast peptone media; SC, synthetic complete media). For phenotypes, the temperature listed is the lowest temperature at which slow growth was detected. None, no detectable phenotype at any temperature; ND, not determined.
To further examine the effect of Swi3 SWIRM domain surface replacements on protein stability and complex assembly, we examined Swi3S protein levels (by immunoblots) and their capacity to assemble into the SWI/SNF complex, through coprecipitation with the SWI/SNF member Swp73 (Fig. 3B). Notably, most Swi3S proteins were present at reduced levels and/or were defective in their association with the SWI/SNF complex (Fig. 3B and data not shown). These results further suggest that the SWIRM domain is important for Swi3 stability and SWI/SNF complex assembly. However, four swi3S mutants were isolated that conferred moderate phenotypes but retained wild-type or near wild-type stability and assembly properties: D374A, K383D/K387D, N392A, and W313A/W388A (Fig. 3). Because the residues replaced in these mutants are located on the surface of the SWIRM domain, these residues are unlikely to contribute to the stability of the SWIRM domain itself. Interestingly, residues 313, 388, and 392 map to the L1 and L5 loops flanking the most highly conserved surface patch of the SWIRM domains (Fig. 1C), and residues 374, 383, and 387 map to another region of SWIRM domain conservation, the α4 helix (Fig. 1C), a putative DNA recognition helix (Fig. 2A). Taken together, these findings suggest that the SWIRM domain has a functional role that extends beyond Swi3 stability and SWI/SNF complex stability, consistent with the DNA-binding studies described above.
In Vitro and in Vivo Analysis of Putative Swi3 SWIRM Domain DNA-Binding Mutants.
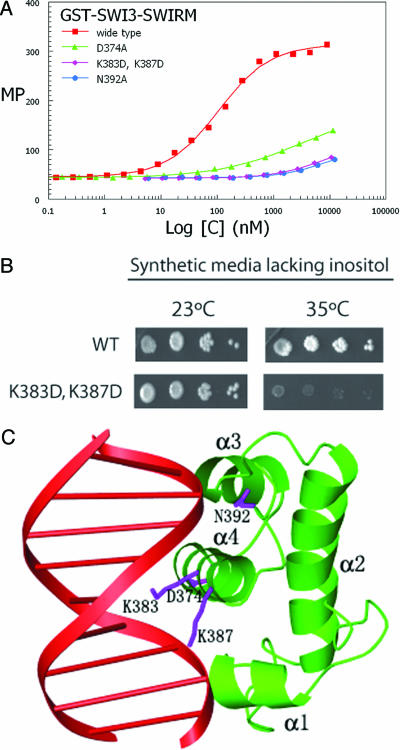
The in vivo studies described above, coupled with our structural analysis and in vitro DNA-binding activity of the Swi3 SWIRM domain suggests that residues 374, 383, 387 and 392 might be used for DNA binding by the Swi3 SWIRM domain. To directly test this hypothesis, we prepared recombinant Swi3 SWIRM domain harboring the three substitution mutations that were characterized in vivo and that map to the putative DNA binding region of the SWIRM domain, D374, K383D/K387D, and N392A, and tested them for DNA binding activity in vitro. For these studies, we prepared the substitution mutants, as well as the wild-type Swi3 SWIRM domain as N-terminal GST fusion proteins. As can be seen in Fig. 4A, each of these substitution mutants show a dramatic reduction in DNA binding activity, consistent with their importance in mediating DNA-binding activity by the Swi3 SWIRM domain.
Fig. 4.
In vitro and in vivo characterization of putative Swi3 SWIRM domain DNA-binding mutants. (A) Results of DNA-binding assays using recombinant GST-Swi3 SWIRM domain and substitution mutants. The assay was carried out as described in Fig. 2B, and the substitution mutants of the Swi3-SWIRM domain used in the assay are indicated. The dissociation constant calculated from the binding data for GST-Swi3 SWIRM: cruciform DNA is 105.36 ± 7.65 nM. CD spectra of native and GST-SWIRM domain mutants are essentially superimposable, suggesting that the SWIRM domain mutants are properly folded (data not shown). (B) The Swi3 SWIRM double mutation K383D K387D fails to grow on media lacking inositol at elevated temperatures. A swi3δ strain (YBC2051) was transformed with LEU2-bearing plasmids containing the K derivatives, grown on synthetic medium lacking leucine, and then spotted in 10-fold serial dilutions on a medium lacking inositol, and grown at either 23°C or 35°C for 4 days. (C) Model for a Swi3 SWIRM domain/DNA complex. The model for the protein/DNA complex was created by superimposing the helix–turn-helix of the Swi3 SWIRM domain (green) with the c-Myb/DNA complex. The DNA from C-Myb is shown in red, and the c-Myb protein is removed for clarity. Residues of the Swi3 SWIRM implicated for DNA binding from the in vivo and in vitro studies (D374, K383, K387, and N392A) are highlighted as blue side chains.
We further characterized the K383D/K387D Swi3 SWIRM domain substitution mutant in vivo. Interestingly, this mutant conferred particular conditional phenotypes; a moderate loss of growth ability at low temperatures on media containing raffinose (Fig. 3A) and a pronounced loss of growth ability at higher temperatures on media lacking inositol (Fig. 4B). These results further suggest that the residues in the Swi3 SWIRM domain that are used for DNA recognition are required for an important subset of Swi3 functions in vivo.
Taken together, these studies suggest that one function of the Swi3 SWIRM domain is to target DNA within nucleosomes and that the α4 DNA recognition helix and the flanking loops (L4 and L5) within the helix–turn–helix directly participates in nucleic acid interaction.
Discussion
Here we present the structure of the Swi3 SWIRM domain, in vitro evidence that the SWIRM binds DNA and nucleosomes and contributes to Swi3 protein stability and SWI/SNF complex integrity, and in vivo evidence that the SWIRM domain plays an essential role in SWI/SNF function. Interestingly, our model of the Swi3 SWIRM domain revealed structural homology to helix–turn–helix DNA-binding domains, suggesting that the Swi3 SWIRM may bind DNA in an analogous manner. Because helix–turn–helix domains interact with only one face of the DNA, they are well suited for interaction with nucleosomal DNA, as one face is occluded by the histone octamer in nucleosomes (1). Consistent with this notion, we provide evidence that the SWIRM domains of Swi3 and Rsc8 bind both naked and nucleosomal DNA with comparable affinities. Therefore, we suggest that an important function of the SWIRM domain is to facilitate interaction of chromatin regulating complexes with nucleosomal DNA.
Evidence for SWIRM–DNA interaction in vivo emerged from our mutational studies. Mutagenesis of SWI3 yielded several SWIRM mutations (swi3S) that conferred moderate phenotypes yet encoded relatively stable Swi3S protein derivatives in vivo. Three such mutations were swi3D374A, swi3K383D/K387D, and swi3N392A. Interestingly, these mutations map to the α4-L5 region of the Swi3 SWIRM domain structure, a region corresponding to the C-terminal end of the DNA-recognition helix in the helix–turn–helix motif. Consistent with the hypothesis that the α4 helix of the Swi3 SWIRM domain is used for DNA binding, modeling of the Swi3 SWIRM domain bound to DNA, using the Myb/DNA complex for superposition shows that the D374, K383, K387, and D392 side chains are in position to interact with the DNA (Fig. 4C). This observation is consistent with SWIRM–DNA interactions occurring in a manner analogous to other helix–turn–helix DNA-binding proteins. However, we observed that the Rsc8 SWIRM domain bound free DNA and nucleosomes with a 10-fold reduced affinity relative to the SWIRM domain of Swi3, suggesting that additional RSC components may contribute to DNA binding or that the Rsc8 SWIRM domain may bind other ligands.
Our work reveals an additional role for the SWIRM domain in maintaining Swi3 and Rsc8 stability and SWI/SNF and RSC complex integrity. Swi3 derivatives lacking the SWIRM domain or bearing selected point mutations were present at highly reduced levels and reduced SWI/SNF complex integrity. Moreover, similar results were also obtained with many rsc8S mutations (data not shown), suggesting a conserved structural role for the SWIRM domain in ATPase remodeling enzymes. The use of the SWIRM domain both for DNA binding and for association with other core components of the remodeler complex is consistent with the known role of Swi3 and its homologs as central scaffolding subunits of SWI/SNF family remodelers (22–24).
The histone code hypothesis purports that enzymes that add or remove posttranslational histone modifications work in a coordinated fashion with the protein modules that recognize these modifications (5). A corollary is that these chromatin regulatory proteins must be recruited to nucleosomes to exert their activities. Although this recruitment could work through histone tail binding, it may additionally involve regulated protein association with DNA. Consistent with this notion, Grune et al. (25) have shown that the remodeling ATPase ISWI bears a helix-turn-helix motif termed the SLIDE domain that also interacts with nucleosomal DNA. Given the multitude of domains present on remodeling and modifying complexes, we suggest that DNA-binding domains such as the SWIRM domain may also be used in particular contexts, and in combination with histone tail binding domains, to enable chromatin remodelers and chromatin modification enzymes to associate with their proper nucleosome targets in vivo for the appropriate regulation of gene expression.
Materials and Methods
Protein Expression and Purification.
The SWIRM domains from Swi3 and Rsc8 were PCR-amplified from yeast genomic DNA, cloned into the pRSET-A expression vector, and expressed as a 6× His-tagged fusion protein in BL21(DE3) (Gold) (Novagen) cells. SWIRM-domain expressing cells were grown in LB media containing 100 μg/ml ampicillin at 37°C until the OD595 reached a value of 0.7, at which time 1 mM IPTG was added, the temperature was decreased to 15°C, and the cells were grown overnight. Cells were harvested by centrifugation and stored frozen at −20°C.
For protein purification, the cell pellet was resuspended in a buffer containing 20 mM Hepes (pH 7.5) and 500 mM NaCl and lysed by sonication. Both the Swi3 and Rsc8 SWIRM domains were purified from the soluble protein fraction using Ni2+-NTA agarose and Superdex 75 size exclusion chromatography (Amersham Pharmacia). Size exclusion chromatography was carried out in a buffer containing 20 mM citric acid (pH 4.5), 200 mM NaCl, and 5 mM 2-mercaptoethanol. Both proteins were concentrated to ≈1 mg/ml by using an Amicon concentrator (Millipore). The Rsc8 SWIRM domain was stored at −80°C until further use, whereas the Swi3 SWIRM domain was used immediately after purification.
The SeMet-derivatized Swi3 SWIRM domain was prepared as described above except that BL834(DE3) (Novagen) cells were used and grown in minimal media supplemented with SeMet. The GST-Swi3 SWIRM domain was prepared by cloning the corresponding coding sequence into the pGex4T-1 vector. All site-directed mutagenesis was performed by using the QuikChange mutagenesis kit according to the manufacturer’s protocol (Invitrogen), and all GST-tagged proteins were purified according to the manufacturer’s protocol (Novagen).
Crystallization, Data Collection, and Structural Analysis.
Crystals of the Swi3 SWIRM domain were prepared at room temperature by using the hanging drop vapor diffusion method by equilibration over a reservoir containing 0.1 M Tris·HCl (pH 8.5), 10% PEG 8000. Crystals grew over several days to typical dimensions of 200 × 80 × 80 μm, and cryoprotected by serial transfer to a reservoir solution supplemented with a final glycerol concentration of 35%. Both native and SeMet multiwavelength anomalous dispersion data sets were collected at BNL at beamline X25, and the data were processed and scaled with the program hkl2000 (HKL Research). The selenium sites were located and refined with the program solve (26), the experimental electron density improved by solvent flattering and the initial model automatically built with the program resolve (27). The model was adjusted manually with the program o (28) and refined against the native data by using the program cns (29). The final model was refined to 1.4-Å resolution with no outliers of the Rhamachandran plot (Table 1).
Fluorescence Polarization Protein–DNA Binding Assay.
Four-way-junction cruciform DNA and double-stranded DNA duplex samples were prepared according to Bianchi et al. (19). Two-fold serial dilutions of the recombinant Swi3 and Rsc8 SWIRM domains (from 8 μM to 0.03 nM and 9 μM to 0.56 nM, respectively), were incubated with 1 nM of fluorescein-labeled DNA substrate in a buffer containing 20 mM Hepes 7.5, 50 mM NaCl, and 100 μg/ml BSA in a total volume of 100 μl in borosilicate glass cuvettes. Each binding reaction was allowed to equilibrate for 30 min before fluorescence measurements using a BEACON 2000 Fluorescence Polarization System. Each experiment was preformed in duplicate, and the millipolarization value (mP) was plotted against a log scale of the protein concentration. The data were fitted to a nonlinear dose–response logistical transition [y = a0 + a1/(1 + x/a2)a3)] using the Levenberg–Marquardt algorithm within the slidewrite software package, where the a2 coefficient is the dissociation constant (KD).
Electrophoresis Mobility Shift Protein–DNA Binding Assay.
Two-fold serial dilutions of the Swi3 and Rsc8 SWIRM domains (from 1.6 μM to 7.8 nM and 1.8 μM to 9.0 nM, respectively) were incubated with 6.7 nM recombinant yeast mononucleosome or DNA only in a buffer containing 20 mM Hepes 7.5, 50 mM NaCl, and 10% glycerol in a total volume of 10 μl and equilibrated for 20 min at room temperature before analysis by mobility-shift gel electrophoresis. Gel electrophoresis of the samples were carried out by using 6% native polyacrylamide gel at 5°C in 0.5× TBE buffer and the gel stained with SYBR Green (Cambrex).
Genes, Plasmids, and Site-Directed Mutagenesis for in Vivo Studies.
SWI3 was isolated from genomic DNA (YBC1895) by PCR amplification, and the clone included 1 kb of upstream DNA sequence. The amplicon was cloned into pRS315 with the Zero Blunt Topo Cloning kit (Invitrogen). The RSC8 genes on pRS426 and pRS315 were a gift from Marian Carlson (Columbia University, New York). Site-directed mutagenesis was performed as described above.
Extract Preparation and Immunoprecipitations.
To monitor SWI/SNF assembly in swi3S mutants, we prepared whole-cell extracts as described (30). Briefly, swi3Δ strains transformed with pRS315 bearing either SWI3 or swi3S mutant derivatives were grown in SC-Leu media at 30°C until they reached an OD600 of 1.0, shifted to 35°C for 4 h and harvested. Pellets were resuspended in 10% glycerol, 400 mM NaCl, 50 mM Tris·Cl (pH 7.5), 0.05% Nonidet P-40, 1 mM EDTA, and 0.1 mM 2-mercaptoethanol, and subjected to bead beating. The aqueous portion was isolated by centrifugation and used for IP. For IP analysis, αSWP73 antibody was immobilized on Protein Sepharose A beads and soluble protein extract was incubated with the Protein A antibody mixture for 4 h at 4°C. This mixture was washed three times with 1 ml of buffer containing 250 mM NaCl, 0.05% Tween-20, 10% glycerol, 1 mM EDTA, 50 mM Tris pH 7.5, and 1× protease inhibitors, and the proteins were liberated from the Sepharose beads by addition of 5 M urea and alnlyzed on 7.5% acrylamide SDS gel probed with αSWI3 antibody.
Supplementary Material
Acknowledgments
We thank the staff at National Synchrotron Light Source for assistance with data collection. This work was supported by National Institutes of Health Grants GM052880 (to R.M.), GM60415 (to B.R.C.), and CA24014 (to B.R.C.), the Howard Hughes Medical Institute (B.R.C.); and by a grant from the Commonwealth Universal Research Enhancement Program, Pennsylvania Department of Health (The Wistar Institute).
Glossary
Abbreviation:
- SeMet
selenomethionine.
Footnotes
Conflict of interest statement: No conflicts declared.
Data deposition: The structural coordinates reported in this paper have been deposited to the Protein Data Bank, www.pdb.org (PDB ID code 2FQ3).
References
- 1.Luger K. Curr. Opin. Genet Dev. 2003;13:127–135. doi: 10.1016/s0959-437x(03)00026-1. [DOI] [PubMed] [Google Scholar]
- 2.Wu C. J. Biol. Chem. 1997;272:28171–28174. doi: 10.1074/jbc.272.45.28171. [DOI] [PubMed] [Google Scholar]
- 3.Felsenfeld G., Boyes J., Chung J., Clark D., Studitsky V. Proc. Natl. Acad. Sci. USA. 1996;93:9384–9388. doi: 10.1073/pnas.93.18.9384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Narlikar G. J., Fan H. Y., Kingston R. E. Cell. 2002;108:475–487. doi: 10.1016/s0092-8674(02)00654-2. [DOI] [PubMed] [Google Scholar]
- 5.Jenuwein T., Allis C. D. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 6.Muchardt C., Yaniv M. J. Mol. Biol. 1999;293:187–198. doi: 10.1006/jmbi.1999.2999. [DOI] [PubMed] [Google Scholar]
- 7.Dhalluin C., Carlson J. E., Zeng L., He C., Aggarwal A. K., Zhou M. M. Nature. 1999;399:491–496. doi: 10.1038/20974. [DOI] [PubMed] [Google Scholar]
- 8.Owen D. J., Ornaghi P., Yang J. C., Lowe N., Evans P. R., Ballario P., Neuhaus D., Filetici P., Travers A. A. EMBO J. 2000;19:6141–6149. doi: 10.1093/emboj/19.22.6141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kasten M., Szerlong H., Erdjument-Bromage H., Tempst P., Werner M., Cairns B. R. EMBO J. 2004;23:1348–1359. doi: 10.1038/sj.emboj.7600143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nielsen P. R., Nietlispach D., Mott H. R., Callaghan J., Bannister A., Kouzarides T., Murzin A. G., Murzina N. V., Laue E. D. Nature. 2002;416:103–107. doi: 10.1038/nature722. [DOI] [PubMed] [Google Scholar]
- 11.Boyer L. A., Langer M. R., Crowley K. A., Tan S., Denu J. M., Peterson C. L. Mol. Cell. 2002;10:935–942. doi: 10.1016/s1097-2765(02)00634-2. [DOI] [PubMed] [Google Scholar]
- 12.Akhtar A., Zink D., Becker P. B. Nature. 2000;407:405–409. doi: 10.1038/35030169. [DOI] [PubMed] [Google Scholar]
- 13.Aravind L., Iyer L. M. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-8-research0039. RESEARCH0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi Y., Lan F., Matson C., Mulligan P., Whetstine J. R., Cole P. A., Casero R. A. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 15.Hakimi M. A., Dong Y., Lane W. S., Speicher D. W., Shiekhattar R. J. Biol. Chem. 2003;278:7234–7239. doi: 10.1074/jbc.M208992200. [DOI] [PubMed] [Google Scholar]
- 16.Holm L., Sander C. Trends Biochem. Sci. 1995;20:478–480. doi: 10.1016/s0968-0004(00)89105-7. [DOI] [PubMed] [Google Scholar]
- 17.Schwartz T., Rould M. A., Lowenhaupt K., Herbert A., Rich A. Science. 1999;284:1841–1845. doi: 10.1126/science.284.5421.1841. [DOI] [PubMed] [Google Scholar]
- 18.Wah D. A., Hirsch J. A., Dorner L. F., Schildkraut I., Aggarwal A. K. Nature. 1997;388:97–100. doi: 10.1038/40446. [DOI] [PubMed] [Google Scholar]
- 19.Bianchi M. E., Beltrame M., Paonessa G. Science. 1989;243:1056–1059. doi: 10.1126/science.2922595. [DOI] [PubMed] [Google Scholar]
- 20.Cairns B. R., Lorch Y., Li Y., Zhang M., Lacomis L., Erdjument-Bromage H., Tempst P., Du J., Laurent B., Kornberg R. D. Cell. 1996;87:1249–1260. doi: 10.1016/s0092-8674(00)81820-6. [DOI] [PubMed] [Google Scholar]
- 21.Carlson M., Osmond B. C., Botstein D. Genetics. 1981;98:25–40. doi: 10.1093/genetics/98.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Treich I., Carlson M. Mol. Cell Biol. 1997;17:1768–1775. doi: 10.1128/mcb.17.4.1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Treich I., Ho L., Carlson M. Nucleic Acids Res. 1998;26:3739–3745. doi: 10.1093/nar/26.16.3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peterson C. L., Herskowitz I. Cell. 1992;68:573–583. doi: 10.1016/0092-8674(92)90192-f. [DOI] [PubMed] [Google Scholar]
- 25.Grune T., Brzeski J., Eberharter A., Clapier C. R., Corona D. F., Becker P. B., Muller C. W. Mol. Cell. 2003;12:449–460. doi: 10.1016/s1097-2765(03)00273-9. [DOI] [PubMed] [Google Scholar]
- 26.Terwilliger T. C., Berendzen J. Acta Crystallogr. D. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Terwilliger T. C. Acta Crystallogr. D. 1999;55:1863–1871. doi: 10.1107/S0907444999010033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard Acta Crystallogr. A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 29.Brunger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., et al. Acta Crystallogr. D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 30.Cairns B. R., Schlichter A., Erdjument-Bromage H., Tempst P., Kornberg R. D., Winston F. Mol. Cell. 1999;4:715–723. doi: 10.1016/s1097-2765(00)80382-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.