Abstract
The tumor suppressor p53 consists of four 393-residue chains, each of which has two natively unfolded (N- and C-terminal) and two folded (core and tetramerization) domains. Their structural organization is poorly characterized as the protein tends to aggregate, has defied crystallization, and is at the limits of NMR studies. We first stabilized the protein by mutation to make it more suitable for extended study and then acquired NMR spectra on full-length protein and various combinations of shorter domain constructs. The NMR spectrum (15N,1H transverse relaxation optimized spectroscopy) of full-length p53 was close to that expected from the sum of the spectra of isolated individual domains. However, patterns of changes in chemical shifts revealed unexpected interactions between the core domains. We used the NMR data as constraints in docking algorithms and found a previously uncharacterized self-complementary surface for the association of core domains into dimers within the tetrameric complex. Binding to DNA requires about a 70° rotation to break those subunit interactions and form the known protein:protein interface in the p53–DNA complex. We verified the interactions by the effects of mutation on DNA binding. Spectroscopic, biophysical, and mutational data conspired to give a picture of the p53 tetramer as a dimer of loosely tethered core dimers of appropriate symmetry to be poised to bind target DNA.
Keywords: modular proteins, NMR, protein–protein interactions
The tumor suppressor p53 is a complex multifunctional protein that acts as a transcription factor in response to oncogenic and other stresses. It is at the centre of a multitude of networks in the cell, binds to DNA and a large number of protein signaling factors that modulate its activity, and is a subject to control by extensive posttranslational modification. Its activity is crucial in the prevention of cancer by inducing cell cycle arrest and apoptosis in response to oncogenic signals (1, 2). Each of its monomeric chains of 393 residues is comprised of several functional domains: the N-terminal domain (residues 1–93) comprises a transactivation domain (residues 1–60) (3) and a proline-rich regulatory domain (residues 64–92) (4), the DNA-binding core domain (CD) (residues 94–312) (5), the tetramerization domain (residues 324–355) (6), and the C-terminal negative regulatory domain (residues 360–393) (7). It undergoes a reversible equilibrium to form tetramers (8). The crystal structures of isolated human core domain bound to DNA (5), and of several oncogenic mutants, have been solved (9). The tetramerization domain is also well characterized by NMR (6, 10) and crystallography (11). The isolated N-terminal domain is natively unfolded (11, 12), apart from the formation of a nascent helix in the region 15–30 (11), which becomes fully helical when bound to Mdm2 (13) or in a membrane environment (14). The C-terminal negative regulatory domain of p53 is also unstructured (15, 16). The unstructured regions probably prevent the full-length protein being crystallized, leaving NMR as the only technique for obtaining high-resolution structural data on the full-length complex. There have been initial NMR studies on engineered dimeric p53 constructs with an impaired tetramerization domain comprising the core and tetramerization domains (CTetD), the core plus tetramerization plus C-terminal domains, and full-length protein (15, 17). Here, we present an NMR analysis of full-length tetrameric p53 (flp53) whose core domains have been engineered to be stable for extensive NMR studies (18) and which give excellent NMR spectra. The CD common to all our constructs had four mutations in the core domain that stabilize it by 3 kcal·mol−1 (18). Its structure is identical to that of wild type, apart from the mutated side chains (19), and the flp53 version is fully active in cells (unpublished data). We compared the spectra with those of isolated core and N-terminal domains and the CTetD construct to search for interdomain interactions and regions of structure that are affected by oligomerization. We located a previously undetected CD–CD dimer interface.
Results and Discussion
Oligomerization State of p53.
The isolated CD is monomeric under NMR conditions (20), as shown by analytical ultracentrifugation and with NMR relaxation measurements (spin-echo) being consistent with a Mr of 24,000. We found by analytical ultracentrifugation that flp53 was in equilibrium between dimers and tetramers with a dissociation constant of 250 ± 150 nM (per p53 tetramer) at 25°C, and 150 ± 100 nM at 10°C at the ionic strength used in NMR studies. The dissociation constant of CTetD was 220 ± 70 nM (per p53 tetramer) at 10°C. At the concentrations used in NMR spectroscopy (100–150 μM of monomeric units), >95% of the monomers of both constructs were in the form of tetramer.
NMR Spectra.
The Mr of the flp53 tetramer of 170,000 is well above the limit of conventional NMR spectroscopy. Difficulties are increased by the spectral line width heterogeneity arising from the differences in internal dynamics among folded and unfolded p53 domains. Consequently, a conventional 15N,1H heteronuclear single quantum correlation (15N,1H-HSQC) spectrum of flp53 has the unstructured N- and C-terminal domains clearly visible (15, 16, 21) whereas the core and tetramerization domain signals are missing. By optimizing conditions, including >98% deuteration of all non-amide protons to reduce the proton density, we were able to obtain and assign good 15N,1H-TROSY spectra (TROSY, transverse relaxation-optimized spectroscopy) (22) of the whole protein (Fig. 1a).
Fig. 1.
NMR spectra of full-length p53 and fragments. (a) 15N,1H-TROSY spectrum of flp53 (red) with the 15N,1H-TROSY spectrum of core domain (blue) superimposed. (b) 15N,1H-TROSY spectrum of core plus tetramerization domain construct (red) with the 15N,1H-TROSY spectrum of core domain (blue) superimposed. (c) 15N,1H-TROSY of DNA (24-mer) bound to flp53 (blue), and of flp53 alone (blue).
The quality of the spectra gave clues about the rigidity and hence organization of the structures. Firstly, the core domains in flp53 clearly had less mobility than isolated CDs because they could not be resolved in HSQC spectra. However, the TROSY spectrum of the core domain in flp53 was too sharp for all four CDs to be rigidly immobilized in the tetramer of 160 kDa. Indeed, the addition of a specific 24-mer DNA that is known to bind all four CDs tightly led to the spectrum of the CD being no longer resolved by TROSY under the experimental conditions (Fig. 1c) but requiring cross-correlated relaxation-enhanced polarization transfer–TROSY, which is suitable for higher molecular masses. The core domains in flp53 must have freedom as either loosely coupled pairs of dimers or partly constrained monomers.
We assigned all of the resonances that were significantly shifted and most of the spectrum of the isolated thermostable domain (Fig. 1a) from standard triple-resonance experiments and our previous assignment of the wild-type p53 core (23). The 15N,1H-HSQC and 15N,1H-TROSY spectra from isolated, natively unfolded, N- and C-terminal domains were, in general, superposable with their equivalents in the 15N,1H-TROSY spectrum (data not shown). Although most signals from the CDs were superposable (Fig. 1a), there were some large changes in chemical shifts of some residues, consistent with subunit interactions (Fig. 2). We can identify some of the important interactions by comparing the different fragments and using existing structural information.
Fig. 2.
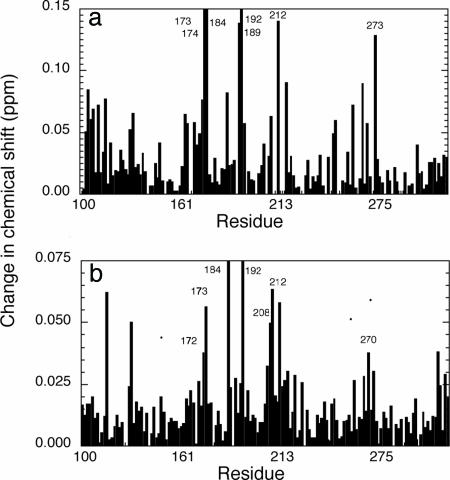
Changes in chemical shift of full-length p53 relative to its fragments. These are combined 15N/1H chemical shift perturbation maps as defined by (δΔIH2 + (δΔ15N2/5)2)0.5 (30). (a) flp53 minus CD. (b) Core plus tetramerization domain minus CD. The means and standard deviations are 0.025 ± 0.027, 0.011 ± 0.011, and 0.021 ± 0.018. The corresponding values for the bottom 50% changes in shift are 0.008 ± 0.004 and 0.007 ± 0.003.
Organization of Core Domain Dimers in Free p53.
Tetrameric p53 binds to a consensus DNA site containing two copies of the decameric motif RRRC(A/T)|(T/A)GYYY (R, purine; Y, pyrimidine) separated by up to 13 bp (24, 25). The full-length consensus sequence contains two half sites in the form of four inverted 5-bp quarter sites (→ ← → ←). This imposes a symmetry relationship on the arrangement of subunits (6, 21, 26, 27). NMR and mutagenesis data on the binding of core domains are consistent with a head-to-head pair of core domains bound to a ten base pair sequence of DNA (20, 21, 28). The interactions are via the self-complementary association of the short helix 178–182 (20, 21, 28) and a region around 243 (20). This mode of binding has been directly observed by Shakked and coworkers (M. Kitayner, H. Rozenberg, H. Kessler, D. Rabinovich, and Z. Shakked, unpublished work), who have solved the crystal structure of p53 core domains bound to DNA oligomers, each of which contains two half sites, and which associate to form a tetrameric complex. The main residues that interact across the stronger of the two surfaces in the complex are 177, 178, 180, 181, 243, and 244 (Fig. 3a). We checked here the relevance of the crystal structure of the CD–DNA complex to that of tetrameric flp53 bound to natural DNA recognition sequences by measuring the effects of mutation on DNA binding affinity using fluorescence anisotropy (29): mutation of G244A weakened binding 8- and 6-fold; and R181E weakened binding 10- and 8-fold, with p21 and gadd45 DNA sequences, respectively, in qualitative agreement with studies on isolated core domains binding to DNA containing two half sites (28).
Fig. 3.
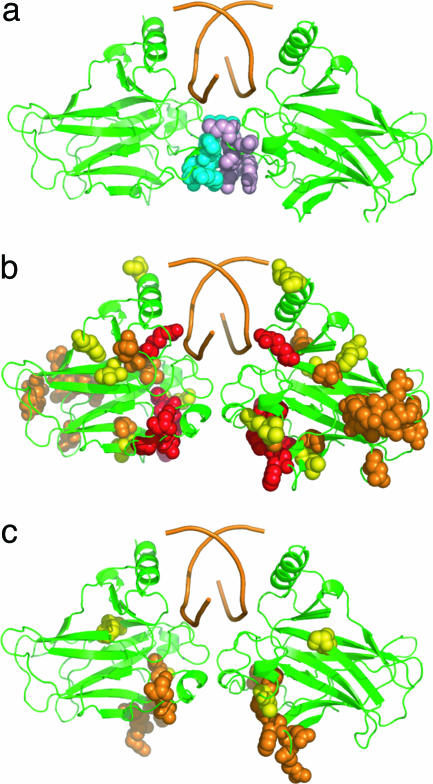
Location of changes in chemical shift on the structure of a core-domain DNA complex. (a) Parent complex (M. Kitayner, H. Rozenberg, H. Kessler, D. Rabinovich, and Z. Shakked, unpublished work) with interacting residues shown as cyan and pink spheres. (b) Chemical shifts in flp53 that were changed by >0.1 ppm (red), between 0.05 and 0.1 ppm (orange) and between 0.04 and 0.05 ppm (yellow). (c) Chemical shifts in CTetD that were changed by between 0.05 and 0.1 ppm (orange) and between 0.04 and 0.05 ppm (yellow).
Core-Domain–Core-Domain Interactions.
We first compared the 15N,1H-TROSY spectrum of flp53 with the 15N,1H-TROSY spectrum of CD (Fig. 2a). There were very large shifts of >0.1 ppm [as defined by the combined 1H and 15N shifts ((δΔIH2 + (δΔ15N2/5)2)0.5 (30)] for six residues: 173 and 192 (which could not be resolved because they had moved into highly overlapped regions and/or shifted too much from the reference position), and 174, 212, 189, and 273 (Fig. 2a, red). In addition, there were several other residues that had significant chemical shifts of >0.04 ppm, a level that Fesik and coworkers deem significant (30). To distinguish the shifts caused by interactions between core domains from those of other interactions, we compared the 15N,1H-TROSY spectrum of the CTetD with isolated CDs (Fig. 2b). The changes in shift were smaller, with fewer residues with significant changes: residues 184 and 192 were so shifted that we were unable to reassign them; residues 173 (now identifiable), 208, 209, and 212 were changed by 0.05–0.06 ppm; and residues 172 and 270 were shifted by 0.038 ppm (Fig. 2b).
We superposed the chemical shift data of CTetD versus CD on the crystal structure of the CD–DNA complex (Fig. 3c). Six of the residues, 172, 173, 184, 192, 208, 209, and 212, form a contiguous patch. Four of the five most strongly shifted signals in flp53, residues 174, 192, 189, and 212, are either the same or are part of that patch. Residue 184 is not involved directly in interactions but did move during an energy refinement procedure (below) and does appear to be flexible in some crystal structures (A. Joerger and A.R.F., unpublished work). The chemical-shift data imply close contact across the interface, perhaps accompanied by some induced conformational changes. Importantly, none of the residues that are involved in the dimer interface in the DNA complex are implicated by NMR, because their signals remained in the reference position in the CTetD construct. It is easy to over-interpret changes in chemical shifts because of induced indirect effects. However, a well defined pattern of changes is significant.
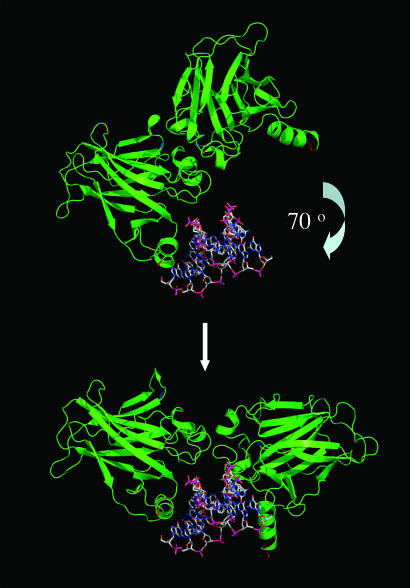
The interaction surface implied from the NMR data per se is consistent with the proposed overall symmetry of the DNA–CD dimer complex (21) and found in the crystal structure (M. Kitayner, H. Rozenberg, H. Kessler, D. Rabinovich, and Z. Shakked, unpublished work) but involves a large rotation of the subunits in the complex from the DNA, breaking the interactions between 243 and 244, and 180 and 181 (Fig. 4). A more extensive surface of interaction is formed contiguous to that in the protein–DNA complex. The unligated p53 tetramer has its DNA-binding site more open than in the DNA-bound complex and is poised to bind DNA by wrapping around it.
Fig. 4.
Geometric relationship between the surface implicated in CD–CD interactions in the unligated protein and in the complex with DNA.
Modeling the Domain Interactions.
We initially modeled the interaction between the two core domains to obtain a low-resolution picture of the overall geometry of the interaction (Fig. 4). However, we were able to obtain a plausible detailed model of the interaction (Fig. 5). Two subunits of core domain were docked by using 3d-dock suite v.2.0 (31) and three of the NMR constraints, residues 172, 192, and 212. The resulting complex was further optimized with the moe (Chemical Computing Group) energy minimization routine. The two interacting surfaces were self-complementary and approximately planar. The modeled interface buried 1,700 Å2 of solvent-accessible surface area, which is over three times larger than the area buried by the dimer in the DNA-bound structure. Val-172 and Phe-212 from each subunit made van der Waals’ contacts and formed a hydrophobic patch in the modeled interface. Glu-171 and Arg-174 formed an intermolecular salt bridge. Similar interactions are frequently observed in dimer interfaces and contribute highly to specificity. Both Glu-171 and Arg-174 are highly conserved in p53 orthologs from different species, suggesting that they might have an essential function, such as the one proposed. Residues 174, 192, 209, and 212 underwent large conformational changes to satisfy the shape and electrostatic complementarity of the new dimer interface. The backbone conformation of residues 208, 209, 210, 211, and 212 changed during the simulation, and there were also small changes in the region around residue 184. These structural changes would account for the changes in chemical shift (29).
Fig. 5.
Model of the dimer–dimer interface based on the NMR interactions, docking, and energy minimization. The key residues that interact in flp53, E171, V172, R174, and F212 are space-filled (C, gray; O, blue; N, red). The accessible surface area buried in the dimer interface in the modeled complex is 1,700 Å2, compared with 500 Å2 in the DNA-bound complex, with a gap volume index of 0.93. F212 interacts with V172 and F212 in the other subunit. E171 makes ionic interactions with R174 in the opposite subunit and R249 in its own subunit.
Probing the Surface by Mutagenesis.
The interactions are clearly weak because isolated core domains do not self-associate until concentrations approaching millimolar. Nevertheless, weak interactions can become greatly enhanced when added to those already present in an associated complex (the chelate effect). One can never be certain that the chemical shifts that are observed result from a mixture of conformations in rapid equilibrium. Model building is also not 100% reliable. Because of these caveats, we tested whether or not the interactions were real and contributed to self-association in flp53 by mutating them. The binding of p53 to DNA is cooperative with a Hill constant, h, of ≈1.8 (29). The Hill constant is always less than or equal to the number of molecules that associate to form a complex. The value of 1.8 was previously interpreted as reflecting a dimeric ground state for p53 associating to form a tetramer with DNA (29). We made radical mutations to the residues involved in the salt bridges in the model and measured DNA binding by adding aliquots of protein to a 1 nM solution of the p21 recognition element (29). Wild-type flp53 had a concentration (in terms of monomer) for 50% binding, P50 = 2.15 ± 0.12 nM and h = 1.7 ± 0.11; the charge reversal mutant R174E had P50 = 1.75 ± 0.25 nM and h = 2.29 ± 0.09; and the other charge reversal, E171R, had P50 = 2.1 ± 0.3 nM and h = 2.56 ± 0.05. Clearly, at the concentrations of protein during the titration region, 0.2–10 nM, the two mutants must be significantly in the form of monomers to have h > 2, and the wild type is less monomeric. The interactions involving E171 and R174 must clearly be stabilizing the equilibrium between monomers and dimers of p53.
In general, it is clearly feasible to analyze the structure of large multidomain proteins containing ordered and disordered regions by comparing high-resolution NMR spectral information on the well ordered domains with spectra on the isolated domains or smaller constructs, followed by verification by site-directed mutagenesis and complemented by computer modeling.
Experimental
Protein Purification.
We used the superstable mutant of p53 containing the following mutation in the CD: M133L/V203A/N239Y/N268D (18, 19) of p53 as pseudo-wild-type, which increased expression levels and sample stability. Isolated p53 CD, consisting of residues 94–312, and the p53 tetramerization plus C-terminal domain construct residues 313–393 were expressed and purified as described in refs. 16 and 23. p53 N-terminal domain, residues 1–93, was subcloned into the pRSET-HLT plasmid, expressed in Escherichia coli C41 (32), and purified by using standard His-tag purification protocols, followed by thrombin digestion, a second Ni-affinity chromatographic step to separate the HLT tag, and a final anion-exchange chromatography purification step. flp53 and CTetD (residues 94–360) were cloned into pET24a-HLTV plasmid. The protein was purified by using standard His-tag purification protocols, followed by tobacco etch virus protease digestion. The final purification step was heparin affinity chromatography. 15N,2H-labeled NMR samples were produced in M9 media made up with D2O and D6-glucose as a carbon source. The degree of deuteration, D, was estimated by mass spectrometry and quantified from the equation
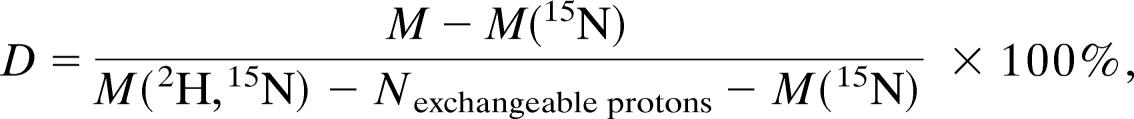 |
where M(15N) and M(2H,15N) are expected masses for 100% 15N and 2H,15N-labeled protein, and Nexchangeable protons is the number of protons that could be exchanged with water, i.e., protons of amides and polar side chain groups.
Analytical Ultracentrifugation.
Equilibrium sedimentation experiments were performed on a Beckman XL-I ultracentrifuge by using Ti-60 rotor and 6-sector cells at speeds of 8,000, 11,000 and 15,000 rpm at 10°C and 25°C. The sample volume was 110 μl. Samples were considered to be at equilibrium as judged by a comparison of the several scans at each speed. For CTetD, the sample volume was reduced to 55 μl, and the speeds were 18,000 and 22,000 rpm at 10°C to reduce the time of the experiment and avoid aggregation. Buffer conditions were 25 mM sodium phosphate (pH 7.2), 150 mM KCl, and 1 mM DTT. Data were processed and analyzed by using ultraspin software, which is available from our web site (www.mrc-cpe.cam.ac.uk).
NMR.
Spectra were acquired with a Bruker Avance 800-MHz spectrometer equipped with a CryoProbe and single-axis gradients. HSQC and TROSY spectra (22) were recorded with protein in a buffer of 25 mM sodium phosphate (pH 7.2), 150 mM KCl, and 5 mM DTT at 20°C. In all experiments, concentrations of p53 or its domains were ≈100 μM. All spectra were externally referenced based on the position of the water peak.
DNA Binding.
The binding of mutants of flp53 to p21 and gadd45 recognition sequences were performed by using fluorescence anisotropy (29) at 10°C in buffer containing 25 mM imidazole chloride (pH 7.2), 213.4 mM NaCl, and 5 mM DTT. p53 protein at a concentration of 0.15–10 μM was titrated into fluorescein-labeled DNA (initial concentration of 1–3.5 nM or 12 nM for p21 and gadd45 DNA, respectively) in 30 increments of 2 μl followed by 70 increments of 6 μl.
Modeling.
A global scan of translational and rotational space of the possible orientations of the two molecules, constrained by surface complementarity and electrostatics, was performed by using ftdock (31, 33). Each possible complex was scored by using the rpscore program and an empirical pair potential matrix derived from nonhomologous interfaces observed in the Protein Data Bank. The high-scoring complexes were further filtered by using a three-point interatomic distance constraint defined by the residues 172, 192, and 212. The final putative open-state homodimer was scored highest according to the surface-complementarity score. Subsequent optimization of the dimer interface was performed to satisfy optimal van der Waals’ and electrostatic interactions between the interacting surfaces. Amino acid side chains at the interface were rebuilt de novo by using scwrl 3.0 (34). In the final step, the energy of the complex was minimized by using the charmm22 molecular force field distributed with moe software.
Acknowledgments
We thank Drs. Mark Bycroft, Andreas Joerger, and Alexey Murzin for advice and valuable comments; and Dr. Zipporah Shakked for communication of data before publication and for permission to use the coordinates of the DNA–CD complex. This work was supported by Cancer Research UK, the Medical Research Council, and European Community FP6 funding. A.A. is supported by Medical Research Council Grant G0100305. This publication reflects the authors’ views and not necessarily those of the European Community. The Community is not liable for any use made of the information.
Glossary
Abbreviations:
- CD
DNA-binding core domain
- CTetD
core and tetramerization domains
- HSQC
heteronuclear single quantum correlation
- TROSY
transverse relaxation-optimized spectroscopy.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Vogelstein B., Lane D., Levine A. J. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 2.Vousden K. H., Lu X. Nat. Rev. Cancer. 2002;2:594–604. doi: 10.1038/nrc864. [DOI] [PubMed] [Google Scholar]
- 3.Fields S., Jang S. K. Science. 1990;249:1046–1049. doi: 10.1126/science.2144363. [DOI] [PubMed] [Google Scholar]
- 4.Muller-Tiemann B. F., Halazonetis T. D., Elting J. J. Proc. Natl. Acad. Sci. USA. 1998;95:6079–6084. doi: 10.1073/pnas.95.11.6079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cho Y., Gorina S., Jeffrey P. D., Pavletich N. P. Science. 1994;265:346–355. doi: 10.1126/science.8023157. [DOI] [PubMed] [Google Scholar]
- 6.Clore G. M., Ernst J., Clubb R., Omichinski J. G., Kennedy W. M., Sakaguchi K., Appella E., Gronenborn A. M. Nat. Struct. Biol. 1995;2:321–333. doi: 10.1038/nsb0495-321. [DOI] [PubMed] [Google Scholar]
- 7.Ahn J., Prives C. Nat. Struct. Biol. 2001;8:730–732. doi: 10.1038/nsb0901-730. [DOI] [PubMed] [Google Scholar]
- 8.Sakaguchi K., Sakamoto H., Lewis M. S., Anderson C. W., Erickson J. W., Appella E., Xie D. Biochemistry. 1997;36:10117–10124. doi: 10.1021/bi970759w. [DOI] [PubMed] [Google Scholar]
- 9.Joerger A. C., Ang H. C., Veprintsev D. B., Blair C. M., Fersht A. R. J. Biol. Chem. 2005;280:16030–16037. doi: 10.1074/jbc.M500179200. [DOI] [PubMed] [Google Scholar]
- 10.Lee W., Harvey T. S., Yin Y., Yau P., Litchfield D., Arrowsmith C. H. Nat. Struct. Biol. 1994;1:877–890. doi: 10.1038/nsb1294-877. [DOI] [PubMed] [Google Scholar]
- 11.Lee H., Mok K. H., Muhandiram R., Park K. H., Suk J. E., Kim D. H., Chang J., Sung Y. C., Choi K. Y., Han K. H. J. Biol. Chem. 2000;275:29426–29432. doi: 10.1074/jbc.M003107200. [DOI] [PubMed] [Google Scholar]
- 12.Dawson R., Muller L., Dehner A., Klein C., Kessler H., Buchner J. J. Mol. Biol. 2003;332:1131–1141. doi: 10.1016/j.jmb.2003.08.008. [DOI] [PubMed] [Google Scholar]
- 13.Kussie P. H., Gorina S., Marechal V., Elenbaas B., Moreau J., Levine A. J., Pavletich N. P. Science. 1996;274:948–953. doi: 10.1126/science.274.5289.948. [DOI] [PubMed] [Google Scholar]
- 14.Rosal R., Pincus M. R., Brandt-Rauf P. W., Fine R. L., Michl J., Wang H. Biochemistry. 2004;43:1854–1861. doi: 10.1021/bi035718g. [DOI] [PubMed] [Google Scholar]
- 15.Ayed A., Mulder F. A. A., Yi G. S., Lu Y., Kay L. E., Arrowsmith C. H. Nature Struct. Biol. 2001;8:756–760. doi: 10.1038/nsb0901-756. [DOI] [PubMed] [Google Scholar]
- 16.Weinberg R. L., Freund S. M., Veprintsev D. B., Bycroft M., Fersht A. R. J. Mol. Biol. 2004;342:801–811. doi: 10.1016/j.jmb.2004.07.042. [DOI] [PubMed] [Google Scholar]
- 17.Mulder F. A., Ayed A., Yang D., Arrowsmith C. H., Kay L. E. J. Biomol. NMR. 2000;18:173–176. doi: 10.1023/a:1008317825976. [DOI] [PubMed] [Google Scholar]
- 18.Nikolova P. V., Henckel J., Lane D. P., Fersht A. R. Proc. Natl. Acad. Sci. USA. 1998;95:14675–14680. doi: 10.1073/pnas.95.25.14675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Joerger A. C., Allen M. D., Fersht A. R. J. Biol. Chem. 2004;279:1291–1296. doi: 10.1074/jbc.M309732200. [DOI] [PubMed] [Google Scholar]
- 20.Rippin T. M., Freund S. M., Veprintsev D. B., Fersht A. R. J. Mol. Biol. 2002;319:351–358. doi: 10.1016/S0022-2836(02)00326-1. [DOI] [PubMed] [Google Scholar]
- 21.Klein C., Planker E., Diercks T., Kessler H., Kunkele K. P., Lang K., Hansen S., Schwaiger M. J. Biol. Chem. 2001:49020–49027. doi: 10.1074/jbc.M107516200. [DOI] [PubMed] [Google Scholar]
- 22.Pervushin K., Riek R., Wider G., Wuthrich K. Proc. Natl. Acad. Sci. USA. 1997;94:12366–12371. doi: 10.1073/pnas.94.23.12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wong K. B., DeDecker B. S., Freund S. M., Proctor M. R., Bycroft M., Fersht A. R. Proc. Natl. Acad. Sci. USA. 1999;96:8438–8442. doi: 10.1073/pnas.96.15.8438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.El-Deiry W. S., Kern S. E., Pietenpol J. A., Kinzler K. W., Vogelstein B. Nat. Genet. 1992;1:45–49. doi: 10.1038/ng0492-45. [DOI] [PubMed] [Google Scholar]
- 25.Funk W. D., Pak D. T., Karas R. H., Wright W. E., Shay J. W. Mol. Cell. Biol. 1992;12:2866–2871. doi: 10.1128/mcb.12.6.2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nagaich A. K., Zhurkin V. B., Sakamoto H., Gorin A. A., Clore G. M., Gronenborn A. M., Appella E., Harrington R. E. J. Biol. Chem. 1997;272:14830–14841. doi: 10.1074/jbc.272.23.14830. [DOI] [PubMed] [Google Scholar]
- 27.Lebrun A., Lavery R., Weinstein H. Protein Eng. 2001;14:233–243. doi: 10.1093/protein/14.4.233. [DOI] [PubMed] [Google Scholar]
- 28.Dehner A., Klein C., Hansen S., Muller L., Buchner J., Schwaiger M., Kessler H. Angew. Chem. Int. Ed. 2005;44:2–6. doi: 10.1002/anie.200501887. [DOI] [PubMed] [Google Scholar]
- 29.Weinberg R. L., Veprintsev D. B., Fersht A. R. J. Mol. Biol. 2004;341:1145–1159. doi: 10.1016/j.jmb.2004.06.071. [DOI] [PubMed] [Google Scholar]
- 30.Hajduk P. J., Dinges J., Miknis G. F., Merlock M., Middleton T., Kempf D. J., Egan D. A., Walter K. A., Robins T. S., Shuker S. B., et al. J. Med. Chem. 1997;40:3144–3150. doi: 10.1021/jm9703404. [DOI] [PubMed] [Google Scholar]
- 31.Gabb H. A., Jackson R. M., Sternberg M. J. E. J. Mol. Biol. 1997;272:106–120. doi: 10.1006/jmbi.1997.1203. [DOI] [PubMed] [Google Scholar]
- 32.Miroux B., Walker J. E. J. Mol. Biol. 1996;260:289–298. doi: 10.1006/jmbi.1996.0399. [DOI] [PubMed] [Google Scholar]
- 33.Moont G., Gabb H. A., Sternberg M. J. E. Proteins Struct. Funct. Genet. 1999;35:364–373. [PubMed] [Google Scholar]
- 34.Canutescu A. A., Shelenkov A. A., Dunbrack R. L. Protein Sci. 2003;12:2001–2014. doi: 10.1110/ps.03154503. [DOI] [PMC free article] [PubMed] [Google Scholar]