Abstract
Telomerase-mediated telomere addition counteracts telomere shortening due to incomplete DNA replication. Short telomeres are the preferred substrate for telomere addition by telomerase; however, the mechanism by which telomerase recognizes short telomeres is unclear. In yeast, the Ataxia telangiectasia mutated (Atm) homolog, Tel1, is necessary for normal telomere length regulation likely by altering telomere structure, allowing telomerase recruitment to short telomeres. To examine the role of Atm in establishing preference for elongation of short telomeres in mice, we examined telomerase-mediated elongation of short dysfunctional telomeres in the presence or absence of Atm. Here we show that Atm is dispensable for elongation of short telomeres by telomerase, suggesting that telomerase recruitment in mammalian cells and in yeast may be regulated differently.
Keywords: DNA damage, chromosome fusion, telomerase recruitment
Ataxia telangiectasia mutated (Atm) is the major sensor of DNA double-strand breaks in mammalian cells (1). Cells that are deficient in Atm display sensitivity to DNA-damaging agents and defective DNA repair. Atm also plays a role in mammalian telomere function (2–4). In contrast, the yeast homolog of Atm, Tel1, plays a minor role in the DNA damage response and a larger role in telomere function. Tel1 plays a role in damage response only in strains where the major DNA damage response pathways are disrupted. In yeast, Tel1 activity is critical for telomere length regulation. Deletion of Tel1 results in telomere hyperrecombination, telomere fusion, chromosome loss, and progressive telomere shortening (5, 6). However, after many generations of shortening, telomere length stabilizes at ≈50 bp, 15% of normal length (5). The remaining telomere length is maintained by the activity of the Atm-related (Atr) homolog Mec1. Simultaneous deletion of both TEL1 and MEC1 results in progressive and complete loss of telomeric sequences (7). This ever-shorter telomere phenotype is similar to that observed when telomerase components are deleted; however, in Δtel1Δmec1 strains telomerase activity is intact (8). Taken together, these data suggest that Tel1 is the major regulator of telomerase access to the telomere, with Mec1 playing a compensatory role in Δtel1 strains. Interestingly, the requirement for Tel1 and Mec1 to maintain telomere length can be bypassed. Targeting telomerase to the telomere by means of fusion to the telomere-binding protein Cdc13 restores telomere length maintenance in both Δtel1 and Δtel1Δmec1 cells (9). Furthermore, mutations that affect the telomere-binding proteins Rif1 and Rif2, which negatively regulate telomere length, also allow telomere elongation in Δtel1 and Δtel1Δmec1 mutants (8, 10). Taken together, these data suggest that, in yeast, Tel1 is important in the recruitment of telomerase to the telomere. It is unclear, however, whether mammalian Atm plays any role in telomerase recruitment.
Telomerase preferentially elongates the shortest telomeres in yeast, mouse, and human cells (11–13). Telomeres shorten progressively during cellular division in the absence of telomerase. Mating wild-type yeast to telomerase-null yeast with short telomeres results in preferential elongation of short telomeres (11), consistent with what was previously observed in mice (12). Using intergenerational crosses of mTR+/− mice (mouse telomerase RNA) having long telomeres to mTR−/− G6 (sixth generation in the absence of telomerase) short-telomere mice, we found that short dysfunctional telomeres were preferentially elongated in telomerase-positive, mTR+/− intergenerational F1 (iF1) mice but not in telomerase-negative, mTR−/− iF1 littermates (12).
Results and Discussion
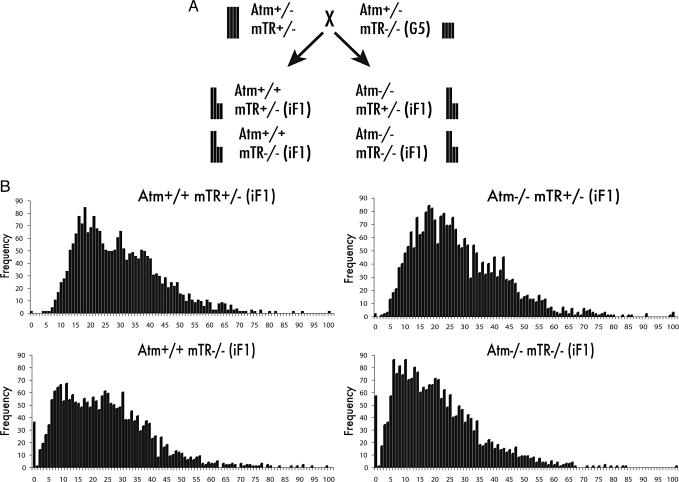
To directly test whether Atm is necessary for the elongation of critically short telomeres, we crossed Atm+/− mTR+/− animals with late-generation telomerase-null animals that were also heterozygous for Atm (Atm+/− mTR−/− G5) (Fig. 1A). Bone marrow was harvested from 8-wk-old progeny, and telomere length analysis was performed on metaphase spreads by quantitative FISH (14). In Atm+/+ mTR−/− iF1 offspring the frequency of critically short telomeres was very high, as expected. In contrast, in Atm+/+ mTR+/− iF1 progeny, critically short telomeres were not detected, consistent with our previous observations that the shortest telomeres are preferentially elongated (Fig. 1B) (12). Surprisingly, Atm was not required for elongation of critically short telomeres. Atm−/− mTR−/− iF1 animals displayed extensive critically short telomeres that were completely repaired in their telomerase-positive, Atm−/− mTR+/− iF1 littermates (Fig. 1B).
Fig. 1.
Telomerase lengthens critically short telomeres in the absence of Atm. (A) Diagram of the Atm+/− mTR+/− × Atm+/− mTR−/− G5 intergenerational cross. Vertical bars represent relative telomere lengths. Progeny from this cross inherit half long and half short telomeres, giving littermates equivalent average telomere length. (B) Telomere length analysis of primary bone marrow from 8-wk-old animals. Cy-3-labeled PNA probes were hybridized to metaphase spreads, and telomere signal intensities were quantified as described (14). Each histogram contains data from three animals. Telomere lengths are shown as frequency distributions of telomere fluorescence intensities. Critically short telomeres are defined as those telomeres of the “zero” class of the distribution.
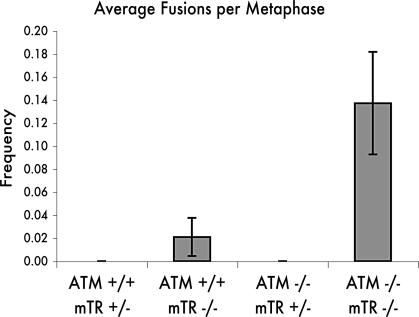
Atm is required for telomere capping in humans, mice, and yeast (2–4, 6, 15). To test whether Atm is required for normal telomere function in context of this intergeneration cross, we analyzed chromosome fusions in metaphase spreads from iF1 offspring. Telomerase-positive Atm+/+ mTR+/− and Atm−/− mTR+/− mice showed no chromosome fusions. Chromosome end-to-end fusions were detected in Atm+/+ mTR−/− cells as expected, and there was a significant elevation of the number of fusions in the Atm−/− mTR−/− cells (Fig. 2). This synergistic increase in chromosome fusions in the double-null mice further confirms the role of Atm in telomere capping and preferentially in capping the shortest telomeres in mice (3, 4).
Fig. 2.
Telomere end-to-end fusion. Metaphase spreads from primary bone marrow were analyzed for chromosome fusions. Atm+/+ mTR+/− (iF1) mice, n = 54; Atm+/+ mTR−/− (iF1) mice, n = 85; Atm−/− mTR+/− (iF1) mice, n = 54; Atm−/− mTR−/− (iF1) mice, n = 56. Significance of P = 0.016 was calculated by using a two-tailed t test between Atm+/+ mTR+/− and Atm−/− mTR−/−.
Our results demonstrate that Atm is not required for preferential action of telomerase on short telomeres. The lack of a requirement for Atm in the elongation of the shortest telomeres suggests that other signaling pathways must monitor telomere length in mammalian cells to allow the specific recruitment of telomerase to short telomeres. Atm and Tel1 play dramatically different roles in the DNA damage response in mammals and yeast. In mammalian cells, Atm mutations result in severe sensitivity to DNA damage (1). In contrast, tel1 mutants in yeast show sensitivity to DNA-damaging agents only in strains in which other components of the DNA damage pathways are mutant (16, 17). The difference in the roles of Tel1 and Atm in the DNA damage response may extend to telomerase recruitment. In mammalian cells where Atm does not have a specialized role in telomere length regulation, Atr may play the dominant role in telomerase recruitment.
Recently, the role of Atm and Atr in telomere length regulation in Arabidopsis was analyzed (18). In the absence of either Atm or Atr, Arabidopsis maintains normal telomere length over multiple generations. However, codeletion of Atm and Tert (the catalytic component of telomerase) leads to an earlier onset of telomere dysfunction despite an equivalent rate of telomere shortening across multiple generations compared with a Tert mutant alone. These results suggested that short telomeres and Atm deficiency cooperatively result in telomere dysfunction, consistent with experiments in mice (3). In addition, because Atr is not essential in Arabidopsis, the authors were able to show that Atr deficiency leads to an increased rate of telomere loss in atr/tert double knockouts. This increased shortening was proposed to be due to the combined effect of telomerase deficiency and failure to protect the telomere from nucleolytic attack in cells that lack Atr. Taken together, these results show that neither Atm nor Atr homologs are important for telomerase recruitment in Arabidopsis, consistent with the work we present here in Atm in mice.
Telomere uncapping due to Atm deficiency has been observed in every organism for which it has been tested (2–4, 6, 15, 18). This remarkable conservation of Atm homolog function at the telomere, taken together with our results, may suggest that Atm homologs in general are not involved in telomerase recruitment. The loss of Tel1 activity may result in short telomeres indirectly because of the uncapped state of the telomeres in ΔTel1 strains. In the uncapped state the telomere may be vulnerable to nucleolytic end processing, ultimately resulting in short telomeres. However, as discussed above, this effect is specific to yeast and is not observed in plants, mice, or humans.
The lack of a requirement for Atm in the elongation of the shortest telomeres suggests that other signaling pathways must monitor telomere length to allow the specific recruitment of telomerase to short telomeres. Understanding the pathways that recognize short telomeres and signal for telomerase recruitment will provide important molecular targets for cancer therapy.
Materials and Methods
Mice.
mTR+/− mice on C57BL/6 background were generated as described (19). Late-generation mTR−/− animals that are heterozygous for Atm (129S6/SvEvTac) were generated by intercrossing Atm+/− mTR+/− animals to generate Atm+/− mTR−/− G1 animals, then intercrossing Atm+/− mTR−/− G1 animals to get G2, and so forth to get Atm+/− mTR−/− G5 mice (20). Atm, mTR, iF1 mice analyzed here were on an equivalent genetic background. Atm and mTR animals were genotyped by PCR as described (19, 21).
Bone Marrow Cell Culture and Metaphase Spread Preparation.
Bone marrow cells were harvested from 8-wk-old animals and cultured in MarrowMAX media (GIBCO catalog no. 12260-014) for 24 h. Bone marrow cells were arrested in metaphase by using 0.1 μg/ml KaryoMAX Colcemid Solution (GIBCO catalog no. 15201-040) for 2 h, then harvested and swelled in prewarmed (37°C) 75 mM KCl hypotonic solution at room temperature for 10 min. Cells were then fixed with repeated exchanges in (3:1) methanol:acetic acid solution and dropped onto microscope slides over a boiling water bath.
Telomere Length and Chromosome Fusion Analysis.
Quantitative FISH analysis was performed by using Cy-3-labeled (CCCTAA)3 PNA probes (PE Biosystems) as described (14). Metaphase spreads were counterstained with DAPI. Images were acquired by using ip-lab software on a Zeiss Axioskop microscope. Chromosome fusions were scored when a continuous DAPI signal indicated an obvious chromosome fusion. Both analyses were performed double-blinded.
Acknowledgments
We thank members of the C.W.G. laboratory for critical reading of the manuscript and helpful comments. This work was supported by National Institutes of Health Grant P01 CA16519 (to C.W.G.).
Glossary
Abbreviations:
- iF1
intergenerational F1
- Gn
nth generation in the absence of telomerase.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Shiloh Y. Nat. Rev. Cancer. 2003;3:155–168. doi: 10.1038/nrc1011. [DOI] [PubMed] [Google Scholar]
- 2.Metcalfe J. A., Parkhill J., Campbell L., Stacey M., Biggs P., Byrd P. J., Taylor A. M. Nat. Genet. 1996;13:350–353. doi: 10.1038/ng0796-350. [DOI] [PubMed] [Google Scholar]
- 3.Qi L., Strong M. A., Karim B. O., Armanios M., Huso D. L., Greider C. W. Cancer Res. 2003;63:8188–8196. [PubMed] [Google Scholar]
- 4.Wong K. K., Maser R. S., Bachoo R. M., Menon J., Carrasco D. R., Gu Y., Alt F. W., DePinho R. A. Nature. 2003;421:643–648. doi: 10.1038/nature01385. [DOI] [PubMed] [Google Scholar]
- 5.Greenwell P. W., Kronmal S. L., Porter S. E., Gassenhuber J., Obermaier B., Petes T. D. Cell. 1995;82:823–829. doi: 10.1016/0092-8674(95)90479-4. [DOI] [PubMed] [Google Scholar]
- 6.DuBois M. L., Haimberger Z. W., McIntosh M. W., Gottschling D. E. Genetics. 2002;161:995–1013. doi: 10.1093/genetics/161.3.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ritchie K. B., Mallory J. C., Petes T. D. Mol. Cell. Biol. 1999;19:6065–6075. doi: 10.1128/mcb.19.9.6065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan S. W., Chang J., Prescott J., Blackburn E. H. Curr. Biol. 2001;11:1240–1250. doi: 10.1016/s0960-9822(01)00391-8. [DOI] [PubMed] [Google Scholar]
- 9.Tsukamoto Y., Taggart A. K., Zakian V. A. Curr. Biol. 2001;11:1328–1335. doi: 10.1016/s0960-9822(01)00372-4. [DOI] [PubMed] [Google Scholar]
- 10.Ray A., Runge K. W. Proc. Natl. Acad. Sci. USA. 1999;96:15044–15049. doi: 10.1073/pnas.96.26.15044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Teixeira M. T., Arneric M., Sperisen P., Lingner J. Cell. 2004;117:323–335. doi: 10.1016/s0092-8674(04)00334-4. [DOI] [PubMed] [Google Scholar]
- 12.Hemann M. T., Strong M. A., Hao L. Y., Greider C. W. Cell. 2001;107:67–77. doi: 10.1016/s0092-8674(01)00504-9. [DOI] [PubMed] [Google Scholar]
- 13.Perrem K., Colgin L. M., Neumann A. A., Yeager T. R., Reddel R. R. Mol. Cell. Biol. 2001;21:3862–3875. doi: 10.1128/MCB.21.12.3862-3875.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lansdorp P. M., Verwoerd N. P., van de Rijke F. M., Dragowska V., Little M. T., Dirks R. W., Raap A. K., Tanke H. J. Hum. Mol. Genet. 1996;5:685–691. doi: 10.1093/hmg/5.5.685. [DOI] [PubMed] [Google Scholar]
- 15.Chan S. W., Blackburn E. H. Mol. Cell. 2003;11:1379–1387. doi: 10.1016/s1097-2765(03)00174-6. [DOI] [PubMed] [Google Scholar]
- 16.Morrow D. M., Tagle D. A., Shiloh Y., Collins F. S., Hieter P. Cell. 1995;82:831–840. doi: 10.1016/0092-8674(95)90480-8. [DOI] [PubMed] [Google Scholar]
- 17.Sanchez Y., Desany B. A., Jones W. J., Liu Q., Wang B., Elledge S. J. Science. 1996;271:357–360. doi: 10.1126/science.271.5247.357. [DOI] [PubMed] [Google Scholar]
- 18.Vespa L., Couvillion M., Spangler E., Shippen D. E. Genes Dev. 2005;19:2111–2115. doi: 10.1101/gad.1333805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blasco M. A., Lee H. W., Hande M. P., Samper E., Lansdorp P. M., DePinho R. A., Greider C. W. Cell. 1997;91:25–34. doi: 10.1016/s0092-8674(01)80006-4. [DOI] [PubMed] [Google Scholar]
- 20.Qi L., Strong M. A., Karim B. O., Huso D. L., Greider C. W. Nat. Cell. Biol. 2005;7:706–711. doi: 10.1038/ncb1276. [DOI] [PubMed] [Google Scholar]
- 21.Barlow C., Hirotsune S., Paylor R., Liyanage M., Eckhaus M., Collins F., Shiloh Y., Crawley J. N., Ried T., Tagle D., Wynshaw-Boris A. Cell. 1996;86:159–171. doi: 10.1016/s0092-8674(00)80086-0. [DOI] [PubMed] [Google Scholar]