Abstract
Hypoxia-inducible factor (HIF) constitutes a target in therapeutic angiogenesis. HIF-1α functions as a sensor of hypoxia and induces expression of vascular endothelial growth factor (VEGF), which then induces angiogenesis. To explore the potential of HIF-1α gene therapy in stimulating wound healing, we delivered a gene encoding a stabilized form of HIF-1α, lacking the oxygen-sensitive degradation domain, namely HIF-1αΔODD, by using a previously characterized peptide-based gene delivery vector in fibrin as a surgical matrix. The peptide vector consisted of multiple domains: (i) A cysteine-flanked lysine hexamer provided DNA interactions that were stable extracellularly but destabilized intracellularly after reduction of the formed disulfide bonds. This DNA-binding domain was fused to either (ii) a fibrin-binding peptide for entrapment within the matrix or (iii) a nuclear localization sequence for efficient nuclear targeting. The HIF-1αΔODD gene was expressed and translocated to the nucleus under normoxic conditions, leading to up-regulation of vascular endothelial growth factor (VEGF)-A165 mRNA and protein levels in vitro. When the peptide-DNA nanoparticles entrapped in fibrin matrices were applied to full-thickness dermal wounds in the mouse (10 μg per wound in 30 μl of fibrin), angiogenesis was increased comparably strongly to that induced by VEGF-A165 protein (1.25 μg per wound in 30 μl of fibrin). However, the maturity of the vessels induced by HIF-1αΔODD was significantly higher than that induced by VEGF-A165 protein, as shown by stabilization of the neovessels with smooth muscle. Nonviral, local administration of this potent angiogenesis-inducing gene by using this peptide vector represents a powerful approach in tissue engineering and therapeutic angiogenesis.
Keywords: gene delivery, nonviral vector, polycation, fibrin
Angiogenesis is important in a number of approaches in tissue engineering and in the treatment of several pathologies directly (1, 2). In cell transplantation therapies, it may be necessary to induce angiogenesis before or concurrently with cell transplantation to support increased metabolic load at the transplant site. In pathologies such as chronic dermal wounds and myocardial ischemia, induction of angiogenesis per se may have a therapeutic effect. Toward these ends, a number of angiogeneic proteins, such as vascular endothelial growth factor (VEGF) and placental growth factor have been widely explored, both by administration as proteins and by gene therapy (1–3). Cell-based therapies for induction of angiogenesis have also been explored, e.g., by isolation and implantation of endothelial progenitor cells (4). A key challenge in angiogenesis remains the induction of mature, stable vessels. To do so, several strategies have been followed (5), including multiple growth factor delivery (6, 7) and delivery of growth factors at ultralow levels (8–10).
An additional attractive approach toward induction of stable, mature blood vessels is to administer a transcription factor that is primarily responsible for detection of hypoxia and induction of production of VEGF, along with a number of other proangiogenic proteins. The transcription factor hypoxia-inducible factor (HIF) plays this central role in the induction of angiogenesis. One major form is HIF-1 (11, 12), which consists of a heterodimer of HIF-1α and HIF-1β. Both subunits are constitutively expressed. HIF-1β is translocated into the nucleus, whereas HIF-1α possesses an oxygen-sensitive degradation domain (ODD), spanning from residues 401 to 603 (13). This domain is prolyl hydroxylated in an oxygen-dependent manner (14), leading to binding of the von Hippel–Lindau protein, which then targets HIF-1α for ubiquitination and degradation in the proteosome (13). As such, under normoxia, HIF-1α is rapidly degraded in the cytoplasm and its nuclear localization is competitively inhibited, whereas under hypoxia, the factor is free to enter the nucleus and dimerize with HIF-1β to induce gene expression.
Interference with the process of HIF-1α degradation under normoxia can induce effects related to hypoxia. When wild-type HIF-1α was overexpressed ≈100-fold, nuclear translocation was observed and VEGF expression was induced (15). When the ODD was deleted (HIF-1αΔODD), the protein was expressed and nuclearly localized under normoxic conditions leading to an up-regulation of VEGF and other genes (13). When transgenic animals were produced to express HIF-1αΔODD in their skin, marked hypervascularity was induced, importantly not at the expense of hyperpermeability (16). An alternative engineered HIF-1α variant has been constructed, consisting of the DNA-binding and dimerization domains of HIF-1α, but with the transactivation domain of herpes simplex virus (VP16) (17). This HIF-1α/VP16 construct has been shown to induce improved perfusion in ischemia models in the rabbit hindlimb (17). In porcine models of myocardial ischemia, HIV-1α/VP16 induced improved perfusion and left ventricular function when delivered with an adenoviral vector but not by naked plasmid DNA (18). In skeletal muscle, HIF-1α/VP16 was shown to induce a more mature angiogeneic response compared with VEGF when DNA encoding either one was administered with an adeno-associated virus vector (19). These results point to HIF-1α being a particularly interesting target protein in angiogenesis, particularly when using a variant form that is stable under normoxia and, thus, is constitutively translocated into the nucleus.
A number of nonviral approaches to gene delivery have been described in tissue engineering, mostly based on polycations such as polylysine, polyhistidine, or polyethylene imine (20–24). These polycations, with the exception of those based on histidine, induce high levels of transfection, but are also associated with relatively high levels of cytotoxicity, which can limit their usefulness for in vivo transfection. To avoid this toxic polycationic effect, Rice and colleagues (25–27) have developed oligomeric cationic peptides, typically 15 lysine residues in length. Because these peptides form DNA complexes that are not highly stable, Rice and colleagues (26) further crosslinked them by disulfide bonding in the mildly oxidative extracellular environment, which also permits destabilization of the complex after endocytosis, when the endosomal environment becomes reductive during endosomal maturation.
We have described an extension of Rice's concept of cysteine-stabilized, lysine-based peptides for gene delivery (28). We have used even shorter lysine oligomers, namely lysine hexamers, in addition to two pH-sensitive charges derived from flanking histidine residues. These electrostatic DNA-binding peptides were flanked by cysteine residues for extracellular stabilization by disulfide bonding. Two bifunctional peptides were constructed based on this DNA-binding domain. The first, the majority peptide, also comprised a coagulation transglutaminase factor XIIIa substrate sequence, usually found at the N terminus of the protein α2-plasmin inhibitor. The second peptide, the minority peptide, comprised the DNA-binding domain in addition to a nuclear localization sequence derived from simian virus 40 large T antigen. The peptides were used at a ratio 1,000:1 to form stable cross-linked DNA-peptide condensates with a mean diameter of 164 nm and a size distribution from 43 to 204 nm. Such aggregates showed similar stability compared with condensates formed between DNA and high molecular weight polyL-lysine.
This study aimed to explore the efficacy of this previously described nonviral, peptide-based, gene delivery vector (28) to release a proangiogenic gene from a fibrin matrix directly into a dermal wound. Based on the important role of HIF-1α in induction of proangiogeneic proteins, including VEGF, plasmid DNA encoding the stabilized variant HIF-1αΔODD was delivered, and both the quantity and quality of neovessels was analyzed.
Results
We aimed to evaluate a previously described peptide- and matrix-based gene delivery vector (28) for induction of angiogenesis in a dermal wound model. Fibrin served as a surgically relevant cell invasion matrix. We have previously demonstrated that factor XIIIa substrate peptides on DNA-peptide nanoparticles allow covalent binding to the fibrin matrix during fibrin coagulation and prevents DNA loss from the matrix over time (28). Based on the encouraging results in gene delivery in vitro, we constructed two forms of HIF-1α, namely the wild type and a variant in which the ODD was deleted, HIF-1αΔODD. The deletion mutant was formed by using splicing by overlapping extension-PCR technique and cloned into the vector pEGFP-N1; sequencing confirmed the identity of the two clones.
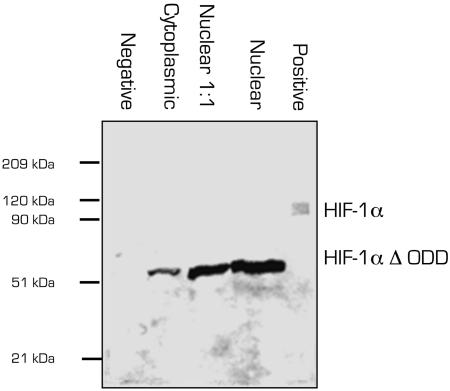
Western blot analysis, shown in Fig. 1, confirmed that the molecular mass of the HIF-1αΔODD mutant was ≈40 kDa lower than that of the wild-type HIF-1α. Consistent with the findings by Huang et al. (13), under normoxic conditions, the HIF-1αΔODD was found predominantly in the nuclear fraction, suggesting its active form. To assess whether this nuclear localization exerts an influence on VEGF-A165 expression, we analyzed VEGF-A165 mRNA levels by quantitative real-time PCR. Total VEGF-A165 mRNA was elevated ≈3-fold (to 3.08 ± 0.14), a factor comparable to that observed in nontransfected cells exposed to hypoxia for 16 h of culture (2.32 ± 0.30). We also measured VEGF-A165 protein expression after transfection with HIF-1αΔODD in culture under normoxic conditions. As shown in Fig. 2, we detected ≈7-fold higher protein levels in cells transfected with HIF-1αΔODD than when transfected with the empty plasmid or not at all, an effect that was stable for at least 5 days after transfection.
Fig. 1.
Expression of HIF-1α in transfected cells. Lanes: 1, negative control (cells transfected with empty vector); 2, cytoplasmic fraction of HIF-1αΔODD-transfected cells; 3 and 4, nuclear fractions of HIF-1αΔODD-transfected cells in different dilutions; 5, positive control (wild-type HIF-1α).
Fig. 2.
VEGF-A165 protein expression after transfection of 293T3 cells with HIF-1αΔODD. White bars, transfected with empty plasmid; black bars, transfected with HIF-1αΔODD; gray bars, nontransfected.
To investigate the angiogenic response to fibrin matrices modified with HIF-1αΔODD containing peptide-DNA nanoparticles (10 μg DNA per wound) or VEGF-A165 (1.25 μg per wound), we analyzed induction of angiogenesis in full thickness excision dermal wounds on the backs of mice. The mice received native fibrin matrices, or fibrin containing VEGF-A165 protein or HIF-1αΔODD peptide-DNA nanoparticles.
Endothelial cells were identified by an endothelial cell-specific surface marker CD31 (Fig. 3 A–C). Increased expression of CD31 was found in wounds treated with HIF-1αΔODD peptide-DNA nanoparticles and VEGF-A165 protein treated samples, respectively. In samples treated with fibrin alone (Fig. 3C), only a minor angiogenic response was observed, whereas in samples treated with HIF-1αΔODD gene (Fig. 3A) or VEGF-A165 protein (Fig. 3B) significantly more CD31-positive structures were observed (Fig. 3D, black bars). These results indicate that the quantity of induced angiogenic response after treatment with either the peptide-DNA nanoparticles encoding HIF-1αΔODD or with VEGF-A165 in fibrin matrices resulted in an increase of the number of vascular structures.
Fig. 3.
In vivo evaluation of angiogenesis 7 days after treatment with different matrices. Endothelial cell-specific CD31 was determined (A–C) and colocalized with smooth muscle cells (anti-CD31, red; anti-SMA, green) in (E–G). Fibrin matrices containing HIF-1αΔODD peptide-DNA nanoparticles (A and E) or VEGF-A165 protein (B and F), or plain fibrin matrices were used (C and G). (Scale bars: A–C, 200 μm; E–G, 25 μm.) (D) Quantification of CD31-positive and CD31 and SMA dual-positive vascular structures 7 days after treatment with different matrices. (H) Ratio of CD31-positive to SMA and CD31 dually positive vascular structures, indicating more mature vascular structures that form. Values are mean ± SEM from three to six independent experiments. ∗, P ≤ 0.05 for values different from fibrin (CD31); ∗∗, P ≤ 0.05 for values different than fibrin (CD31 and SMA).
As an approach by which to characterize vessel quality, tissue sections were probed for the presence of mature vessels, indicated by staining for both CD31, as an endothelial marker, and smooth muscle actin (SMA), as an indicator for the presence of smooth muscle cells around the vessels. Single labeling for CD31 indicated nascent vasculature, whereas dual staining for CD31 and SMA indicated more mature vascular structures, as illustrated in Fig. 3 E–G. A higher number of dual-stained structures was particularly apparent in tissues treated with HIF-1αΔODD peptide-DNA nanoparticles (Fig. 3E). Indeed, when the ratio of SMA and CD31-double positive to CD31-positive vascular structures was determined, an indication of the maturity of the angiogenic response could be obtained, as shown in Fig. 3 G and H. Although the total amount of angiogenesis within plain fibrin matrices was low, the vessels that did form were fairly mature, because 38 ± 4% of the structures showed dual-labeling for CD31 and SMA. Although a large increase in angiogenesis was observed in specimens treated with either VEGF-A165 protein or with peptide-DNA nanoparticles encoding for HIF-1αΔODD, the fraction of vessels that were more mature was ≈4-fold higher in wounds treated with HIF-1αΔODD (49 ± 5%) as compared with that obtained with VEGF-A165 protein administration from similar fibrin matrices (12 ± 3%) (Fig. 3H).
Discussion
Most tissue engineering approaches toward tissue replacement and repair require an underlying process of angiogenesis. Moreover, in many applications of in situ tissue engineering, such as induction of cellular ingrowth into a defect for tissue repair, the angiogenic response may be paramount.
Induction of angiogenesis by using biochemical stimulation has been plagued by the complexities of obtaining a mature vascular morphology and function, i.e., vessels that are well perfused, yet not hyperpermeable (1–3). Indeed, induction of angiogenesis with VEGF induces a vascular response that is unphysiologically permeable in the absence of application of some external maturation factor to accompany the VEGF signal (29). To redress this limitation, bioengineering approaches to deliver multiple growth factors to provide both angiogenic and maturation signals (6, 7), as well as approaches to deliver such factors at ultralow physiological concentrations (9, 10), have been explored.
As an alternative to local administration of one, or even multiple, growth factors, the approach of delivering HIF-1α, a transcription factor that is responsible for regulation of a number of proangiogenic signals has been presented (5, 11, 12). HIF-1α is known to play an important role in tissue response to hypoxia, and two variants have been designed that provide signaling even under conditions of normoxia. Animals in which wild-type HIF-1α was conditionally removed from the endothelium were observed to display a delayed wound healing phenotype, directly suggesting a role in angiogenesis in dermal tissue repair (30). Results from transgenic animals (16), hindlimb (17), and myocardial ischemia (18) models, and skeletal muscle gene delivery morphometric analyses (19) support that a mature angiogenic response may be obtainable through gene therapy with a form of HIF-1α that is stable under normoxic conditions.
Our goal in this work was to explore a nonviral gene delivery vector developed in our laboratory (28) in local induction of angiogenesis by using a wound healing model. Here, we examined the angiogenic response in full-thickness dermal excision wounds of normal mice and compared the angiogenic responses after treatment with native fibrin matrices, VEGF-A165 protein admixed into fibrin, and peptide-DNA nanoparticle containing the engineered variant of HIF-1α lacking the ODD, i.e., HIF-1αΔODD (13), administered in fibrin. We explored nonviral gene delivery by using a matrix-based approach in which peptide-DNA nanoparticles were formed with peptides that comprised a minimal-length lysine hexamer domain for initial DNA binding and a pair of cysteine residues for stabilization of the nanoparticle by disulfide crosslinking (26, 28). We have previously shown that these nanoparticles can achieve very high transfection efficiency at remarkably low levels of cytotoxicity, probably due to the very low degree of polymerization of the cationic amino acids. Our objective here was to understand whether high transfection levels could be achieved in vivo by examining the quantity of angiogenesis induced in the dermal wound. Moreover, we aimed to induce an angiogenic response resulting in highly mature blood vessels through the use of HIF-1αΔODD.
A potent biological response of the peptide-DNA nanoparticles encoding HIF-1αΔODD was indeed observed in vivo. The fibrin platform that we chose allowed good wound healing, inducing some angiogenic response on its own. By comparison, however, a 2-fold angiogenic response was induced when HIF-1αΔODD gene was delivered and a 5-fold angiogenic response when VEGF-A165 protein was delivered. Even more interesting than the quantity of angiogenesis is the quality of the angiogenic response that was induced. Angiogenesis induced with the peptide-DNA nanoparticles encoding HIF-1αΔODD induced a 4-fold increase of more mature blood vessels compared with VEGF-A165 protein application, as judged from the amount of dual-stained vascular structures for SMA (as a smooth muscle marker) and CD31 (as an endothelial marker). This difference demonstrated a clear benefit of HIF-1αΔODD-based therapy as compared with VEGF-A165 protein therapy from a similar fibrin matrix. Our findings correspond nicely with previous studies when VEGF was bolus injected or systemic delivered: applied VEGF showed relatively low efficiency due to rapid clearance from the target site, and edema can result from VEGF's tendency to increase vascular permeability. Furthermore, high local levels of VEGF can reduce up-regulated formation of supernumerary but malformed vessels in hemangioma-like assemblies (31–33). A possible explanation for our favorable result might be that HIF-1αΔODD not only induces a proangiogenic stimulus mediated by a single growth factor such as delivered by a protein-based therapy but also initiates the entire strictly regulated and equilibrated cascade of events leading to formation of more mature vascular structures. Further experiments will be needed to show whether this increase in vascular structures after 1 week will be maintained. In addition, it remains to be determined whether fibrin-binding VEGF-A165 variants that have been designed and explored by our research group are able to increase the amount of mature blood vessels formed (9, 10). In addition, it needs to be determined how VEGF/platelet-derived growth factor sequential dual protein therapy increases the maturity of the blood vessels (6). Such comparative studies remain to be performed, preferably in animal models for delayed wound healing induced by underlying microvascular pathology, because it is associated with diabetes.
Materials and Methods
Cell Lines and Reagents.
Human umbilical vein endothelial cells were purchased from PromoCell (Heidelberg, Germany) and cultured in endothelial cell medium containing 2% FBS (PromoCell). Cells were used between passages 3 and 5. Human embryonic kidney 293 cells (293T3) were purchased from American Type Culture Collection (Manassas, VA) and cultured in DMEM containing 10% FBS and 1% antibiotic antimycotic (all from GIBCO/BRL, Invitrogen).
Cloning Procedures of HIF-1α and HIF-1α Variants.
Human umbilical vein endothelial cells were used for construction of the cDNA library. The DNA of interest was isolated by reverse transcription by using random hexamer primers (RT-PCR Kit; Roche, Rotkreuz, Switzerland) followed by PCR with gene-specific primers. Three fragments were produced and sequentially assembled. Table 1, which is published as supporting information on the PNAS web site, shows the primers used for construction of the HIF-1α variants. To clone the three fragments of wild-type HIF-1α into the vector pEGFPN1 (Clontech, Palo Alto, CA), they were digested with NheI, SalI, MroI, and BamHI, respectively. First, the SalI/MroI and the MroI/BamHI digested fragments were ligated into the SalI/BamHI digested vector, then in a second step, the NheI/SalI fragment was added to complete the gene. The variant HIF-1αΔODD (13) was constructed with splicing by overlapping extension-PCR as described in ref. 34, which allowed removal of amino acids 401–603, comprising the ODD. All constructs were verified by DNA sequencing (Microsynth, Balgach, Switzerland).
Transfection in Vitro.
293T3 cells and human umbilical vein endothelial cells were transfected by using the DEAE-dextran transfection method (35). As a control plasmid, pEGFPN1 (Clontech) reporter vector expressing green fluorescent protein was used. Before transfection, the DNA solution was prepared as follows: 1 μg of plasmid DNA and 16 μl of DEAE-dextran (Sigma D-9885, 10 mg/ml) per well were added to 10 mM PBS at pH 7.4 (total volume of 300 μl). Cells at ≈80% confluence were rinsed carefully with 10 mM PBS at pH 7.4, and the DNA-containing solution was added to the wells and incubated for 30 min at 37°C. Then, 3 ml of DMEM combined with 10% FBS and 6 μl of 50 mM chloroquine was added per well for 2.5 h at 37°C. The solution was aspirated and replaced by a solution consisting of 1.5 ml of DMEM/1% FBS/150 ml of DMSO (Sigma) per well. After 2.5 min of incubation, the solution was aspirated and cells were incubated for 2–4 days with 2 ml of DMEM with 2% FBS per well at 37°C and 5% CO2.
Nuclear and Cytoplasmic Extraction.
Nuclear extracts were prepared (36). In brief, 293T3 cells (confluent 25 cm2 flask, ≈106 cells) were scraped in 1 ml of ice-cold PBS, centrifuged for 2 min at 15,000 rpm, and resuspended in Buffer A (10 mM Hepes, pH 7.9/10 mM KCl/0.1 mM EDTA/1 mM DTT/0.5 mM PMSF). Again, cells were centrifuged at 15,000 rpm for 3 min in a Sigma 1–15 centrifuge with rotor 12124 (Osterode am, Harz, Germany), and the supernatants (cytosolic fraction) were saved for Western blotting and stored at −80°C. The pellets (containing the whole nuclei) were resuspended with Buffer B (20 mM Hepes, pH 7.9/0.4 M NaCl/1 mM EDTA/10% glycerol/1 mM DTT/0.5 mM PMSF) and placed on ice for 30 min. Nuclear debris was centrifuged at 15,000 rpm for 5 min, and supernatants (nuclear fraction) were collected. Protein concentration was determined by Bradford protein (Pierce) assay, and nuclear fractions were stored at −80°C until further use.
SDS/PAGE and Western Blotting.
SDS/PAGE was performed by using standard techniques (37). The proteins were transferred to Hybond ECL nitrocellulose membrane (Amersham Pharmacia Biosciences, Uppsala) by the wet blot tank procedure. Samples (2 μg of the nuclear or cytoplasmic extract) were run on 10% acrylamide SDS/PAGE gels under nonreducing conditions. Western blot analysis was done with a HIF-1α antibody (Novus Biologicals; Littleton, CO), at a dilution of 1:250, followed by a goat anti-mouse-horseradish peroxidase 1:5,000 diluted (DAKO, Glostrup, Denmark). Visualization was done by using ECL Western blotting detection reagent (Amersham Pharmacia Biosciences, Uppsala) and exposure to chemiluminescence film (Hyperfilm ECL; Amersham Pharmacia). As a positive control, a lane was loaded with wild-type HIF-1α that was provided with the antibody from the manufacturer.
RNA Isolation and RT-PCR.
For RT-PCR analyses, total RNA was isolated from 293T3 cells by using a RNeasy Mini Kit (Qiagen, Hombrechtikon, Switzerland). An additional treatment with RNase-free DNase (Qiagen) was performed according to the supplier's manual. The amount of RNA was determined in duplicate by using a spectrophotometer, and the ratio of 260 nm to 280 nm was determined to estimate the purity of the RNA. Only RNA of a purity ratio of 1.8–2.1 was used. RNA was stored at −80°C. For reverse transcription, 1 μg of RNA was used in a final volume of 20 μl by using a RT-PCR Kit and random hexamer primers (Roche, Basel, Switzerland). Samples were stored at −80°C until further use.
Real-Time PCR (TaqMan).
For quantification of total VEGF-A165 mRNA levels, cDNAs from previously described RT-PCRs were amplified by single-reporter real-time PCR by using the ABI Prism 7200 Sequence Detection System (Applied Biosystems, Foster City, CA). The specific primer probes for total VEGF-A165 (forward primer 5′-GCC AGC ACA TAG AGA GAA TGA GC, reverse primer 5′-ATC CGC ATG ATC TGC ATG G), and all reagents were supplied by Applied Biosystems. For PCRs, the SYBR-green Mastermix was used. As endogenous control, 18S cDNA was used for correction of the results with the comparative threshold cycle (TC) method for relative quantification as described by the manufacturer. The differences of the TC values of the sample and 18S cDNA were calculated (ΔTC). Relative expression levels were calculated by following the formula ΔΔTC = [ΔC (HIF-1α induced) − ΔTC (not HIF-1α induced)], and the value used to plot the relative expression was calculated by using the expression 2ΔΔ(TC).
VEGF-A165 ELISA.
Protein concentrations of soluble VEGF-A165 were determined by using the DuoSet ELISA development kit for human VEGF-A165 (R & D Systems). Aliquots (100 μl) of the 293T cell culture supernatants at confluence (≈1 × 106 cells) were obtained after 1, 3, and 5 days from triplicates of three independent experiments, and the amount of VEGF was determined and normalized to 1 ml of culture supernatant.
Peptide Vectors for in Vivo Transfection.
A system of peptides designed as multifunctional cassettes was used, as described in detail in ref. 28. Briefly, the system consisted of two bifunctional peptides: NQEQVSPLGGGCHKKKKKKHC [with fibrin-binding (single underline) and DNA-binding (double underline) domains] and CHKKKKKKHCGGGPKKKRKVEDPY [with a nuclear localization sequence (underline) and a DNA-binding domain (double underline)]. The two peptides were used in a ratio of 1,000:1.
Peptide-DNA Nanoparticles in Fibrin for in Vivo Transfection.
Peptide-DNA condensates were formed and incorporated into fibrin matrices as described in ref. 28. Nanoparticles were formed at a DNA concentration of 25 μg/ml and at a peptide-to-DNA ratio of 0.8 nmol peptide to 1 nmol DNA. When incorporated in fibrin, 10 μg of DNA was loaded into 30 μl of fibrin gels (for each wound), formed at a fibrinogen concentration of 10 mg/ml with 2 units/ml factor XIII (Baxter Bioscience, Vienna).
VEGF-A165 Protein in Fibrin for in Vivo Treatment.
VEGF-A165 was administered in a fibrin formulation. Protein (1.25 μg) (Peprotech, London) was mixed into 30 μl of fibrin gels (for each wound) as described above for the peptide–DNA complexes.
Animals.
Female 6- to 8-week-old BALB/c mice were obtained from Harlan Laboratories (Horst, The Netherlands). All procedures were approved by the Cantonal Veterinary Service Lausanne (see Fig. 4, which is published as supporting information on the PNAS web site).
Surgery.
Surgical areas were shaved, depilated, and wiped with 75% ethanol. Mice were anesthetized with 100 mg of ketamine and 5 mg of xylazine per kg body weight i.p., followed by a s.c. buprenorphine (0.05 mg/kg body weight). Two 5-mm-diameter full-thickness excision wounds were generated on the back of each animal by excising skin and panniculus carnosus as described in ref. 38. Into each wound, 30 μl of the fibrin formulation was administered. Two wounds were placed per animal. The wounds were covered with parafilm, Tegaderm (3M Co.), and Fixomull (Beiersdorf, Hamburg, Germany) after surgery to avoid drying. After 7 days, the animals were killed by ketamine overdose and cervical dislocation, and the wound area, including 2-mm margins, was fixed in 4% paraformaldehyde-PBS or 95% acidic ethanol overnight at 4°C. Seven-micrometer-thick paraffin sections from the midline of the wound were produced and histologically and histomorphometrically analyzed.
Hematoxylin and Eosin (HE) Staining.
The dried sections were immersed in 100% xylene for 20 min to remove the paraffin embedding, followed by a descending sequence of alcohol. The samples were washed in distilled water and stained with HE.
CD31 Staining.
The dried sections were rehydrated, washed in Tris-buffered saline (TBS; 10 mM Tris·HCl/150 mM NaCl, pH 7.4) for 10 min, immersed in 3% H2O2 for 15 min, and again washed in TBS for 10 min. After that, the sections were incubated overnight at 4°C with an anti-CD31 antibody (Pharmingen, San Jose, CA), diluted with TBS containing 5% newborn calf serum (GIBCO/BRL, Invitrogen)/0.01% NaN3 to working concentrations according to the manufacturers instructions. Incubation with biotinylated goat anti-rat polyclonal antibody (DAKO) was performed for 60 min followed by an incubation with 3-amino-9-ethylcarbazole (AEC)/horseradish peroxidase ready-to-use high sensitivity substrate solution (DAKO) for 45 min. AEC substrate solution for the peroxidase reaction resulted in a red color precipitation.
Immunofluorescence: CD31 and α-SMA Staining.
The dried sections were rehydrated, washed three times in TBS for 10 min, and incubated overnight at 4°C with a FITC-labeled anti-α-SMA antibody (Sigma) diluted 1:500 with TBS containing 5% NCS/0.01% NaN3 according to the manufacturer's instructions. Anti-CD31 antibody (Pharmingen) was used as described above, but an Alexa546-conjugated goat anti-rat secondary antibody (Molecular Probes through Invitrogen) was used. After washing the slides three times in TBS for 10 min, DAPI counterstaining was performed (Molecular Probes) for 8 min at room temperature. The number of dually SMA-positive and CD31-positive vascular structures was counted per area (575 × 425 μm2) on wounds from four to six animals, corresponding to 6–11 individual slides per condition. The data represent mean values ± SEM and were normalized individually to the number of SMA-positive vascular structures per condition (equals 100% for fibrin, VEGF-A165protein, or HIF-1α-ΔODD).
Statistical Analysis.
The mean values were compared by one-way ANOVA by using origin 7.1 software. Statistical significance was accepted for P < 0.05.
Supplementary Material
Acknowledgments
We thank H. G. Schmökel for assistance with the small animal surgery and M. Pasquier for the expert histology. Partial funding was provided by the Swiss National Science Foundation.
Abbreviations
- HIF
hypoxia-inducible factor
- ODD
oxygen-dependent degradation domain
- SMA
smooth muscle actin
- TBS
Tris-buffered saline
- TC
threshold cycle
- VEGF
vascular endothelial growth factor.
Footnotes
Conflict of interest statement: The Swiss Federal Institute of Technology and the University of Zurich have filed for patents on certain aspects of the technology and have licensed these to a company in which they and J.A.H. hold equity.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Carmeliet P. J. Intern. Med. 2004;255:538–561. doi: 10.1111/j.1365-2796.2003.01297.x. [DOI] [PubMed] [Google Scholar]
- 2.Carmeliet P. Nat. Med. 2003;9:653–660. doi: 10.1038/nm0603-653. [DOI] [PubMed] [Google Scholar]
- 3.Ferrara N., Gerber H. P., LeCouter J. Nat. Med. 2003;9:669–676. doi: 10.1038/nm0603-669. [DOI] [PubMed] [Google Scholar]
- 4.Zisch A. H. Curr. Opin. Biotechnol. 2004;15:424–429. doi: 10.1016/j.copbio.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 5.Khan T. A., Sellke F. W., Laham R. J. Gene Ther. 2003;10:285–291. doi: 10.1038/sj.gt.3301969. [DOI] [PubMed] [Google Scholar]
- 6.Richardson T. P., Peters M. C., Ennett A. B., Mooney D. J. Nat. Biotechnol. 2001;19:1029–1034. doi: 10.1038/nbt1101-1029. [DOI] [PubMed] [Google Scholar]
- 7.Weber C. C., Cai H., Ehrbar M., Kubota H., Martiny-Baron G., Weber W., Djonov V., Weber E., Mallik A. S., Fussenegger M., et al. J. Biol. Chem. 2005;280:22445–22453. doi: 10.1074/jbc.M410367200. [DOI] [PubMed] [Google Scholar]
- 8.Zisch A. H., Lutolf M. P., Hubbell J. A. Cardiovasc. Pathol. 2003;12:295–310. doi: 10.1016/s1054-8807(03)00089-9. [DOI] [PubMed] [Google Scholar]
- 9.Ehrbar M., Djonov V. G., Schnell C., Tschanz S. A., Martiny-Baron G., Schenk U., Wood J., Burri P. H., Hubbell J. A., Zisch A. H. Circ. Res. 2004;94:1124–1132. doi: 10.1161/01.RES.0000126411.29641.08. [DOI] [PubMed] [Google Scholar]
- 10.Ehrbar M., Metters A., Zammaretti P., Hubbell J. A., Zisch A. H. J. Controlled Release. 2005;101:93–109. doi: 10.1016/j.jconrel.2004.07.018. [DOI] [PubMed] [Google Scholar]
- 11.Huang L. E., Bunn H. F. J. Biol. Chem. 2003;278:19575–19578. doi: 10.1074/jbc.R200030200. [DOI] [PubMed] [Google Scholar]
- 12.Hewitson K. S., Schofield C. J. Drug Discov. Today. 2004;9:704–711. doi: 10.1016/S1359-6446(04)03202-7. [DOI] [PubMed] [Google Scholar]
- 13.Huang L. E., Gu J., Schau M., Bunn H. F. Proc. Natl. Acad. Sci. USA. 1998;95:7987–7992. doi: 10.1073/pnas.95.14.7987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bruick R. K., McKnight S. L. Science. 2001;294:1337–1340. doi: 10.1126/science.1066373. [DOI] [PubMed] [Google Scholar]
- 15.Hofer T., Desbaillets I., Hopfl G., Wenger R. H., Gassmann M. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2002;133:475–481. doi: 10.1016/s1532-0456(02)00117-5. [DOI] [PubMed] [Google Scholar]
- 16.Elson D. A., Thurston G., Huang L. E., Ginzinger D. G., McDonald D. M., Johnson R. S., Arbeit J. M. Genes Dev. 2001;15:2520–2532. doi: 10.1101/gad.914801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vincent K. A., Shyu K. G., Luo Y. X., Magner M., Tio R. A., Jiang C. W., Goldberg M. A., Akita G. Y., Gregory R. J., Isner J. M. Circulation. 2000;102:2255–2261. doi: 10.1161/01.cir.102.18.2255. [DOI] [PubMed] [Google Scholar]
- 18.Heinl-Green A., Radke P. W., Munkonge F. M., Frass O., Zhu J., Vincent K., Geddes D. M., Alton E. W. Eur. Heart J. 2005;26:1327–1332. doi: 10.1093/eurheartj/ehi223. [DOI] [PubMed] [Google Scholar]
- 19.Pajusola K., Künnapuu J., Vuorikoski S., Soronen J., André H., Pereira T., Korpisalo P., Ylä-Herttuala S., Poelliner L., Alitalo K. FASEB J. 2005;19:1365–1367. doi: 10.1096/fj.05-3720fje. [DOI] [PubMed] [Google Scholar]
- 20.Putnam D., Zelikin A. N., Izumrudov V. A., Langer R. Biomaterials. 2003;24:4425–4433. doi: 10.1016/s0142-9612(03)00341-7. [DOI] [PubMed] [Google Scholar]
- 21.Midoux P., LeCam E., Couland D., Delain E., Pichon C. Somatic Cell Mol. Genet. 2002;27:27–47. doi: 10.1023/a:1022931923153. [DOI] [PubMed] [Google Scholar]
- 22.Putnam D., Gentry C. A., Pack D. W., Langer R. Proc. Natl. Acad. Sci. USA. 2001;98:1200–1205. doi: 10.1073/pnas.031577698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pack D. W., Putnam D., Langer R. Biotechnol. Bioeng. 2000;67:217–223. [PubMed] [Google Scholar]
- 24.Demeneix B., Hassani Z., Behr J. P. Curr. Gene Ther. 2004;4:445–455. doi: 10.2174/1566523043345940. [DOI] [PubMed] [Google Scholar]
- 25.Kwok K. Y., Park Y., Yang Y. S., McKenzie D. L., Liu Y. H., Rice K. G. J. Pharm. Sci. 2003;92:1174–1185. doi: 10.1002/jps.10384. [DOI] [PubMed] [Google Scholar]
- 26.McKenzie D. L., Kwok K. Y., Rice K. G. J. Biol. Chem. 2000;275:9970–9977. doi: 10.1074/jbc.275.14.9970. [DOI] [PubMed] [Google Scholar]
- 27.McKenzie D. L., Smiley E., Kwok K. Y., Rice K. G. Bioconjugate Chem. 2000;11:901–909. doi: 10.1021/bc000056i. [DOI] [PubMed] [Google Scholar]
- 28.Trentin D., Hubbell J., Hall H. J. Controlled Release. 2005;102:263–275. doi: 10.1016/j.jconrel.2004.09.029. [DOI] [PubMed] [Google Scholar]
- 29.Lee R. J., Springer M. L., Blanco-Bose W. E., Shaw R., Ursell P. C., Blau H. M. Circulation. 2000;102:898–901. doi: 10.1161/01.cir.102.8.898. [DOI] [PubMed] [Google Scholar]
- 30.Tang N., Wang L., Esko J., Giordano F. J., Huang Y., Gerber H. P., Ferrara N., Johnson R. S. Cancer Cell. 2004;6:485–495. doi: 10.1016/j.ccr.2004.09.026. [DOI] [PubMed] [Google Scholar]
- 31.Carmeliet P. Nat. Med. 2000;6:1102–1103. doi: 10.1038/80430. [DOI] [PubMed] [Google Scholar]
- 32.Simons M., Ware J. A. Nat. Rev. Drug Discov. 2003;2:1–9. doi: 10.1038/nrd1226. [DOI] [PubMed] [Google Scholar]
- 33.Idris N., Haider H. K., Goh M. W. K., Sim E. K. W. Growth Factors. 2005;22:269–279. doi: 10.1080/08977190412331284344. [DOI] [PubMed] [Google Scholar]
- 34.Ho S. N., Hunt H. D., Horton R. M., Pullen J. K., Pease L. R. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 35.McCutchan J. H., Pagano J. S. J. Natl. Cancer Inst. 1968;41:351–357. [PubMed] [Google Scholar]
- 36.Andrews P. A., Jones J. A. Cancer Commun. 1991;3:93–102. doi: 10.3727/095535491820873524. [DOI] [PubMed] [Google Scholar]
- 37.Ausubel F. M., Katagiri F., Mindrinos M., Glazebrook J. Proc. Natl. Acad. Sci. USA. 1995;92:4189–4196. doi: 10.1073/pnas.92.10.4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Werner S., Smola H., Liao X., Longaker M. T., Krieg T., Hofschneider P. H., Williams L. T. Science. 1994;266:819–822. doi: 10.1126/science.7973639. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.